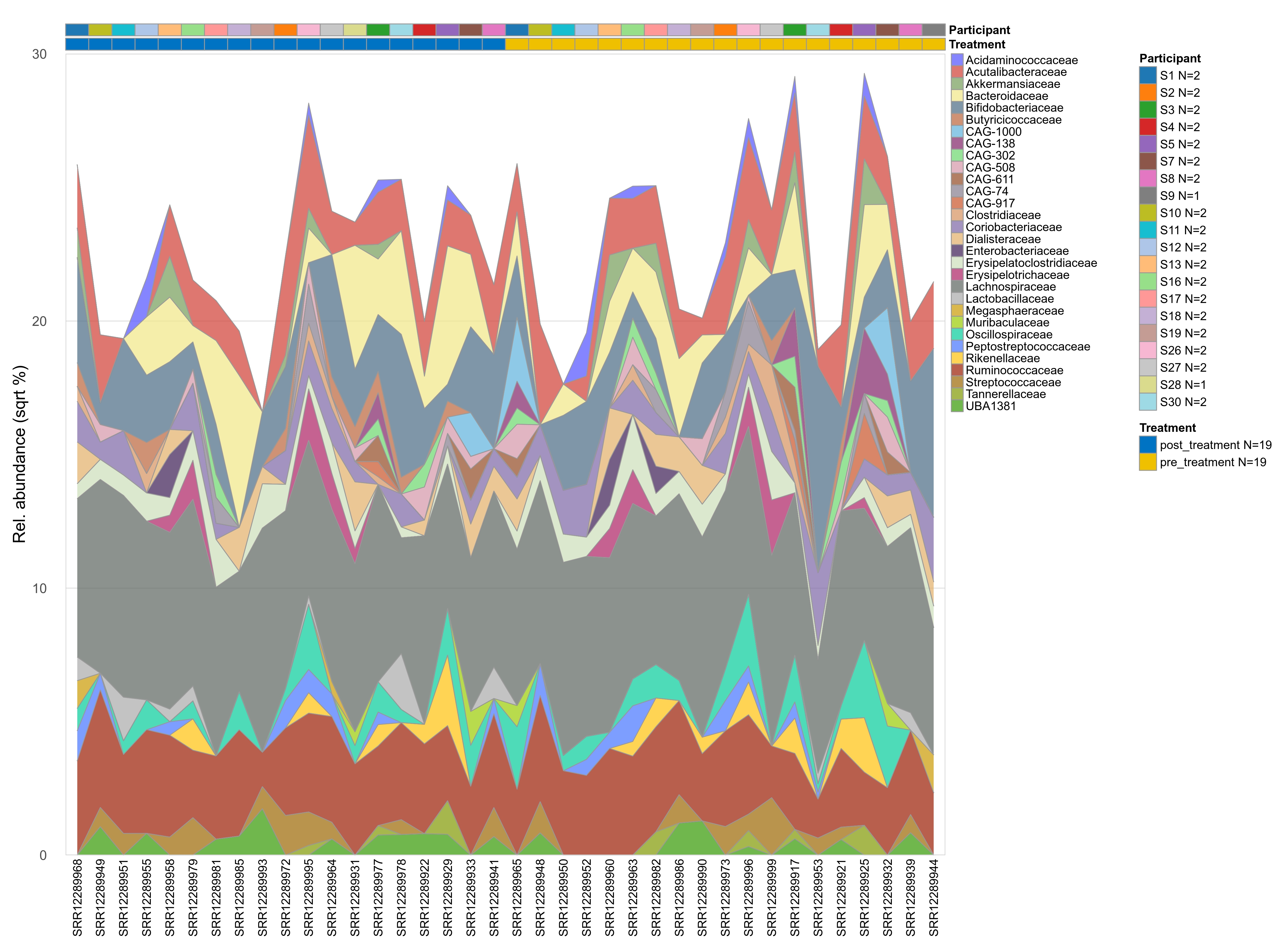
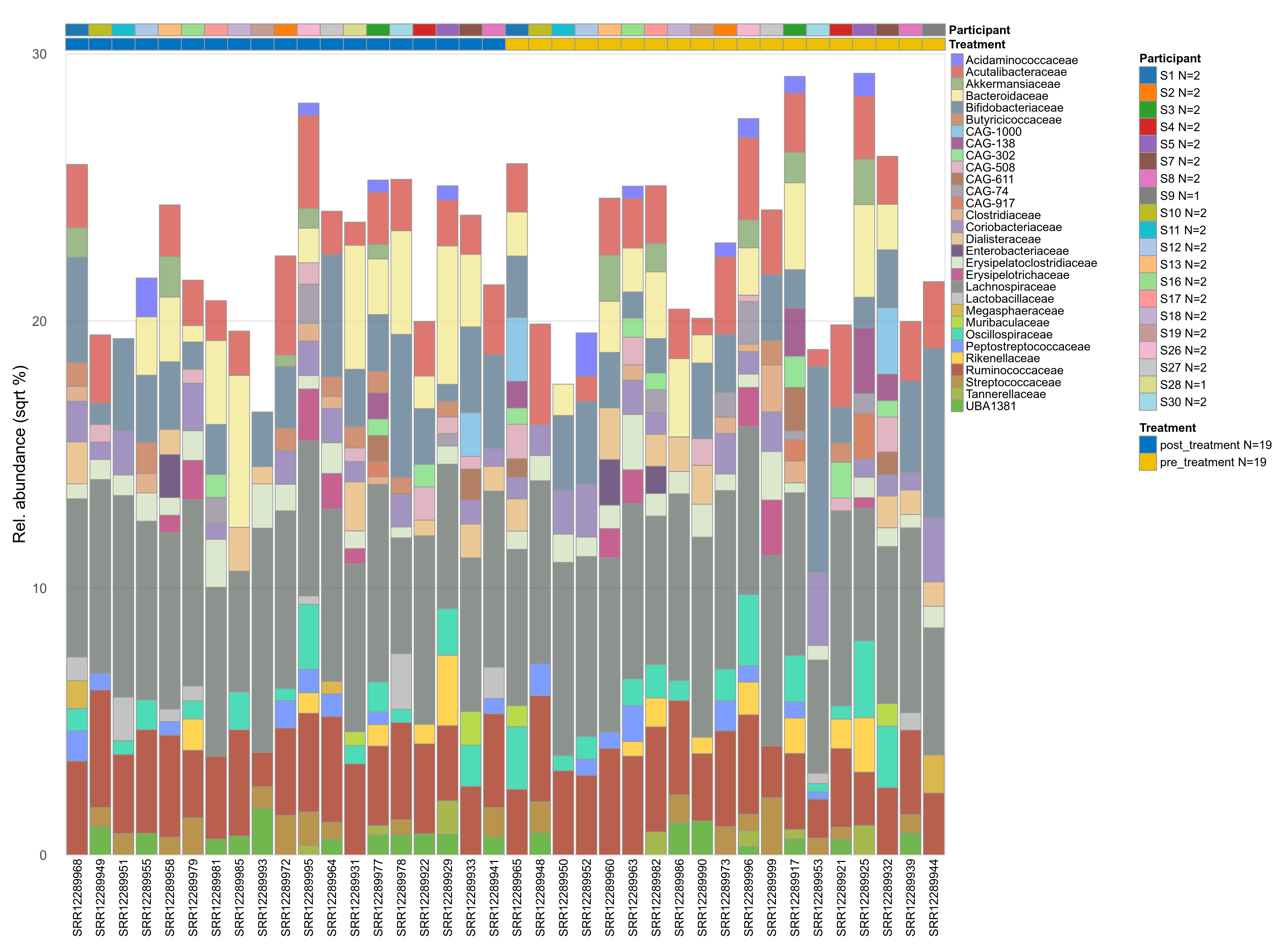
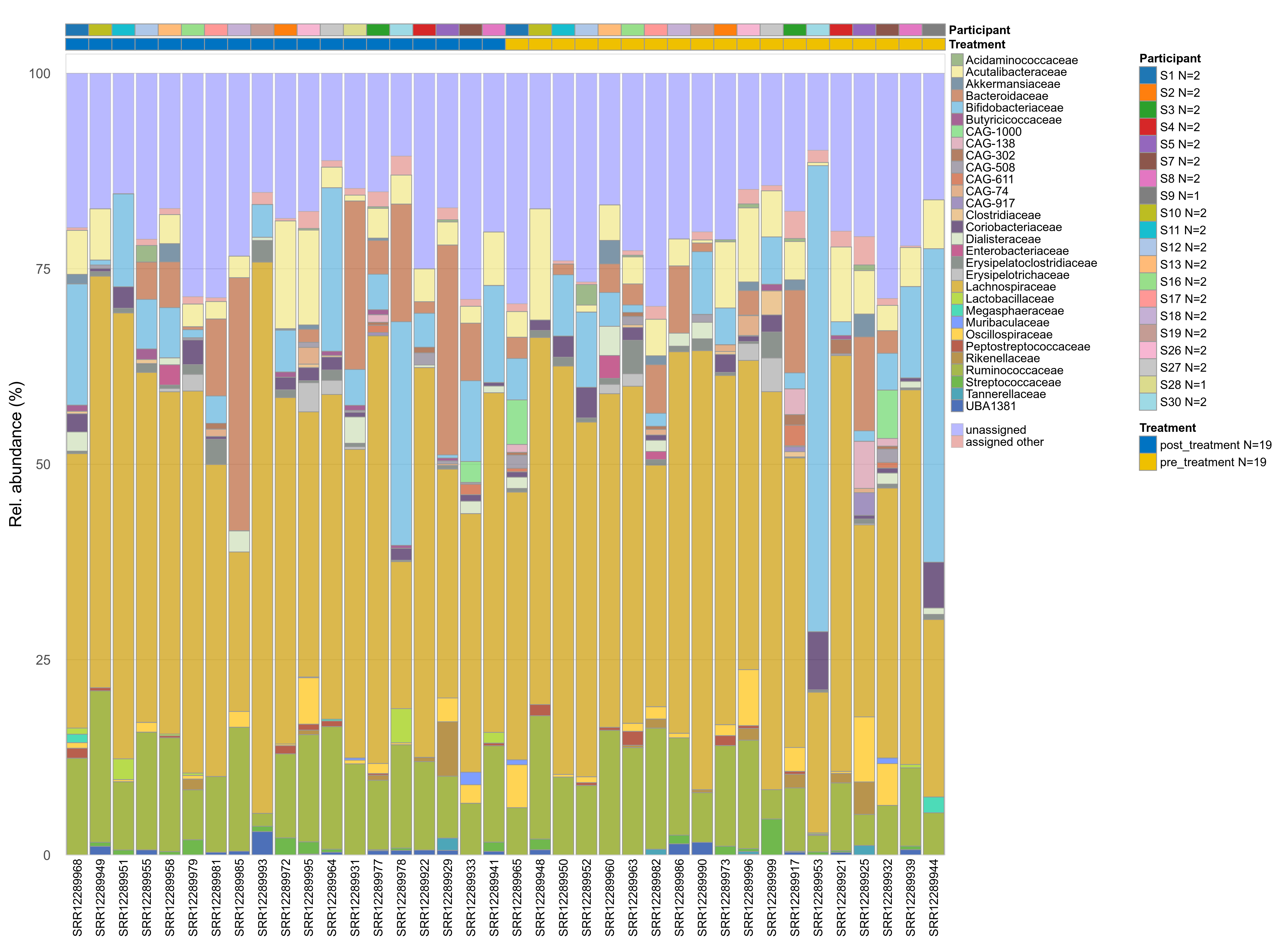
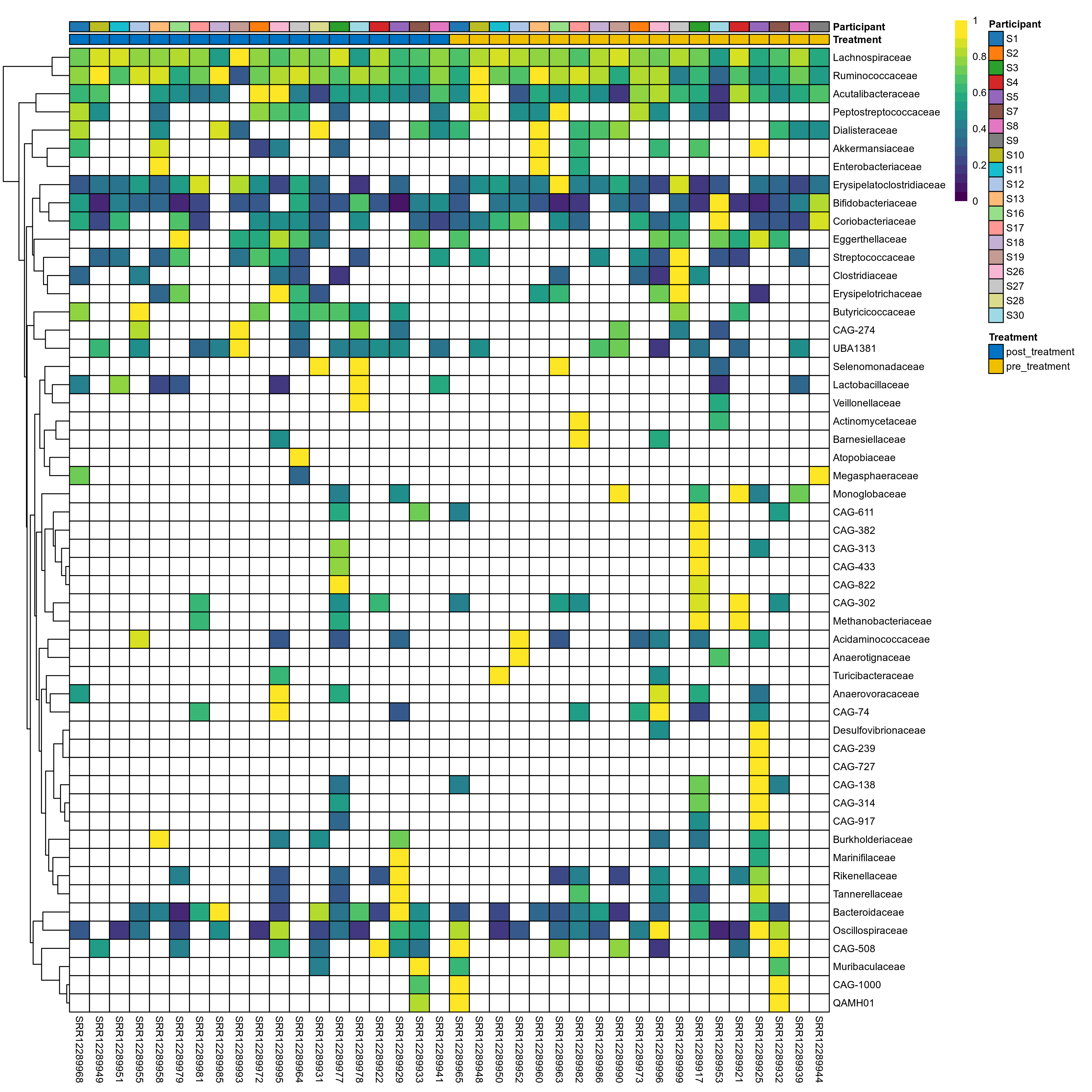
The charts below show the taxonomic composition of the analysed samples using different quantitative visualization techniques. Only the top most abundant families are shown.
Click here to open full-sized image in new window.
Click here to open full-sized image in new window.
Click here to open full-sized image in new window.
Click here to open full-sized image in new window.

Click here to open full-sized image in new window.
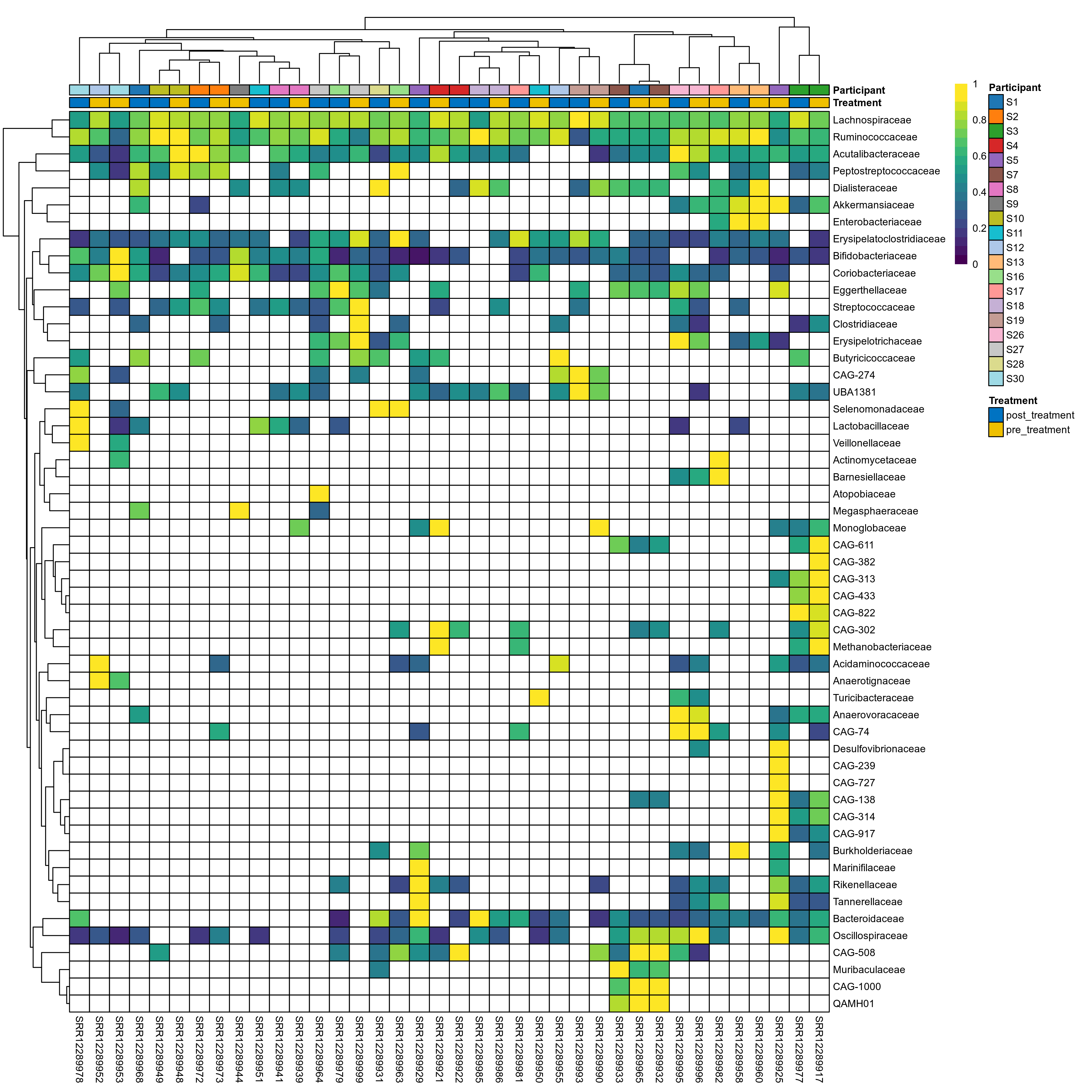
Click here to open full-sized image in new window.
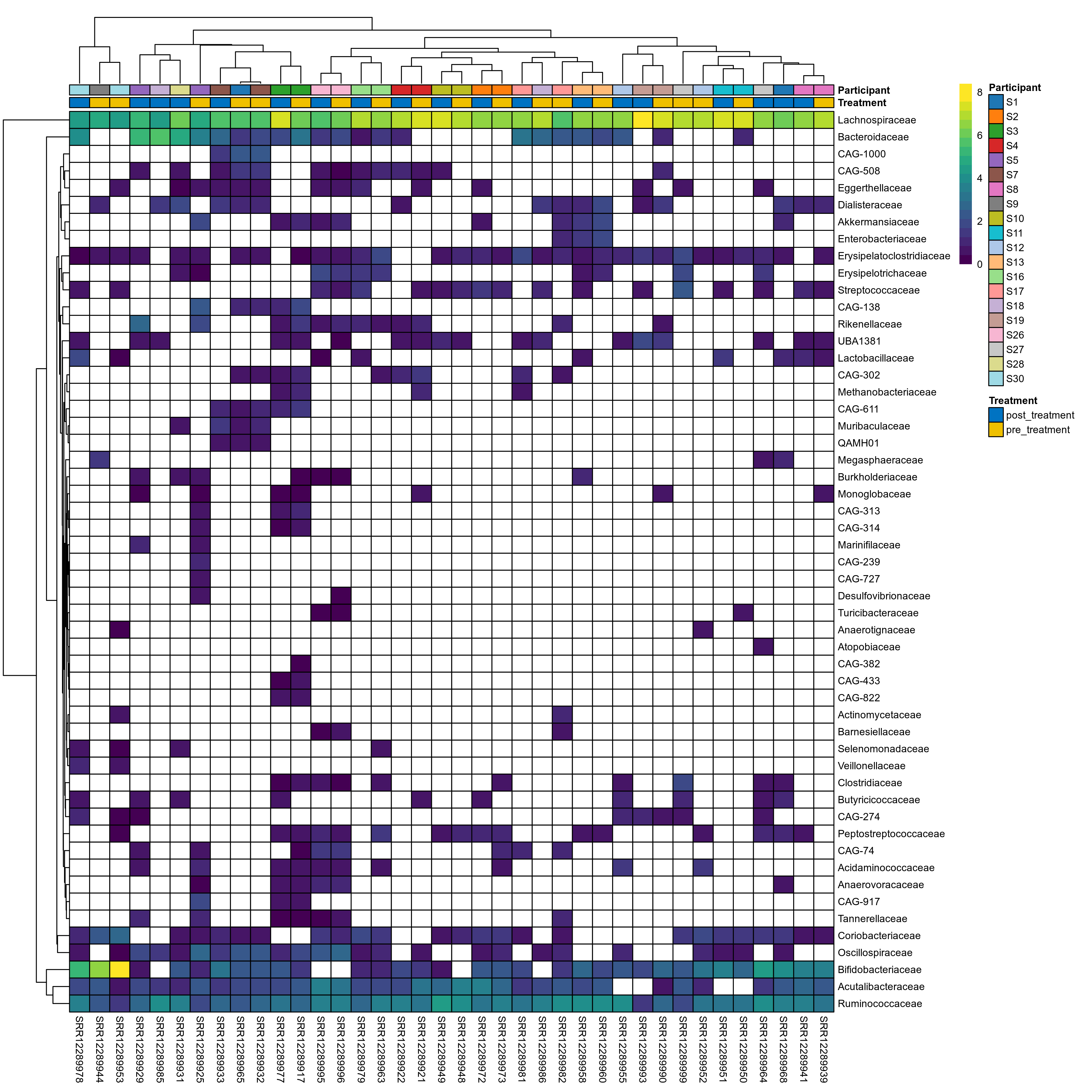
Click here to open full-sized image in new window.
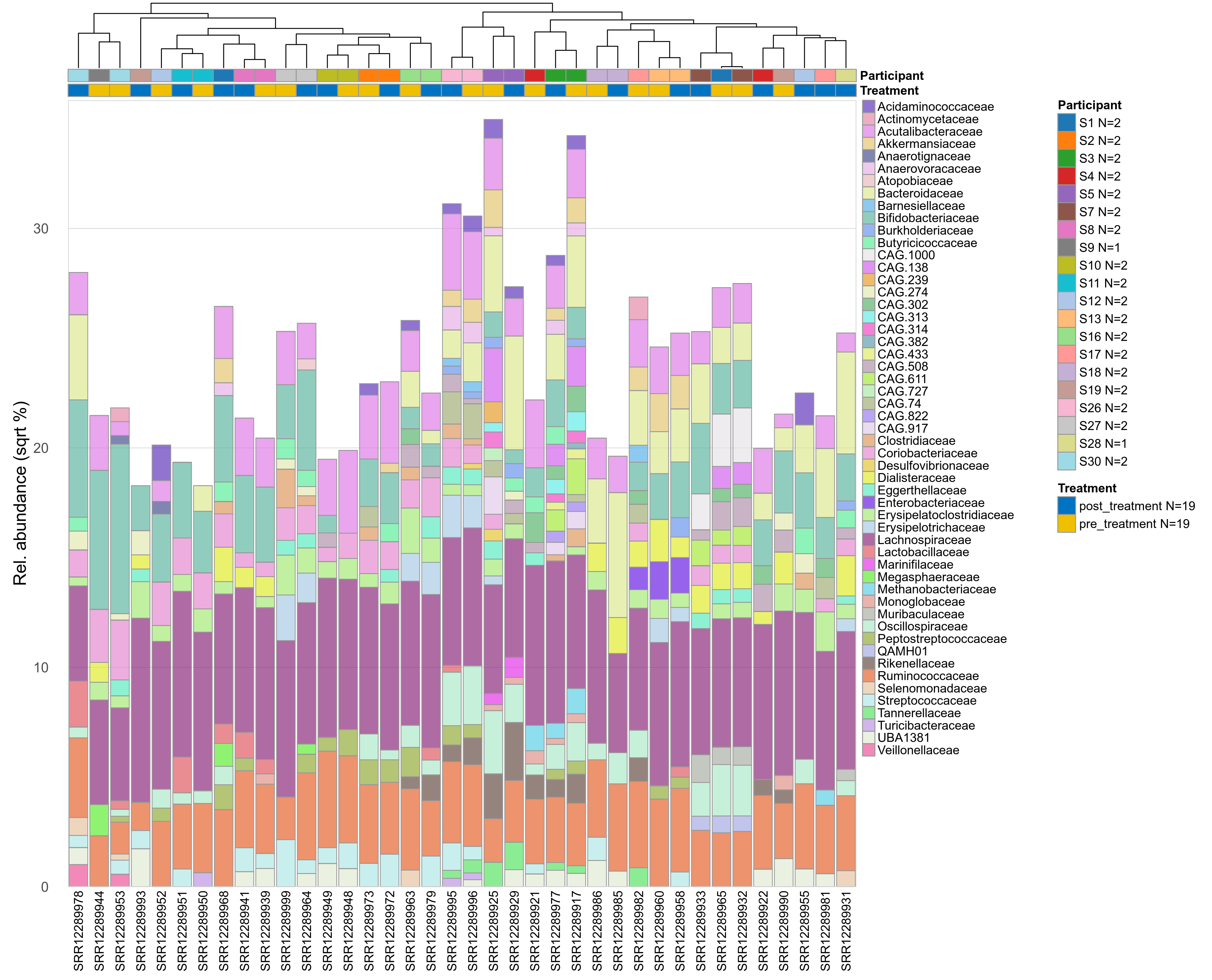
Click here to open full-sized image in new window.
Click here to open interactive barchart in new window.
This page provides an overview of the microbial alpha diversity of the analysed samples. Alpha diversity is measured by the Shannon index and species richnes. Richness simply quantifies the total number of families present in each sample. Shannon index additionally accounts for relative abundance and evenness of the families present and quantifies the entropy of microbial communties. Barcharts and boxplots present the mean diversity in each study group.
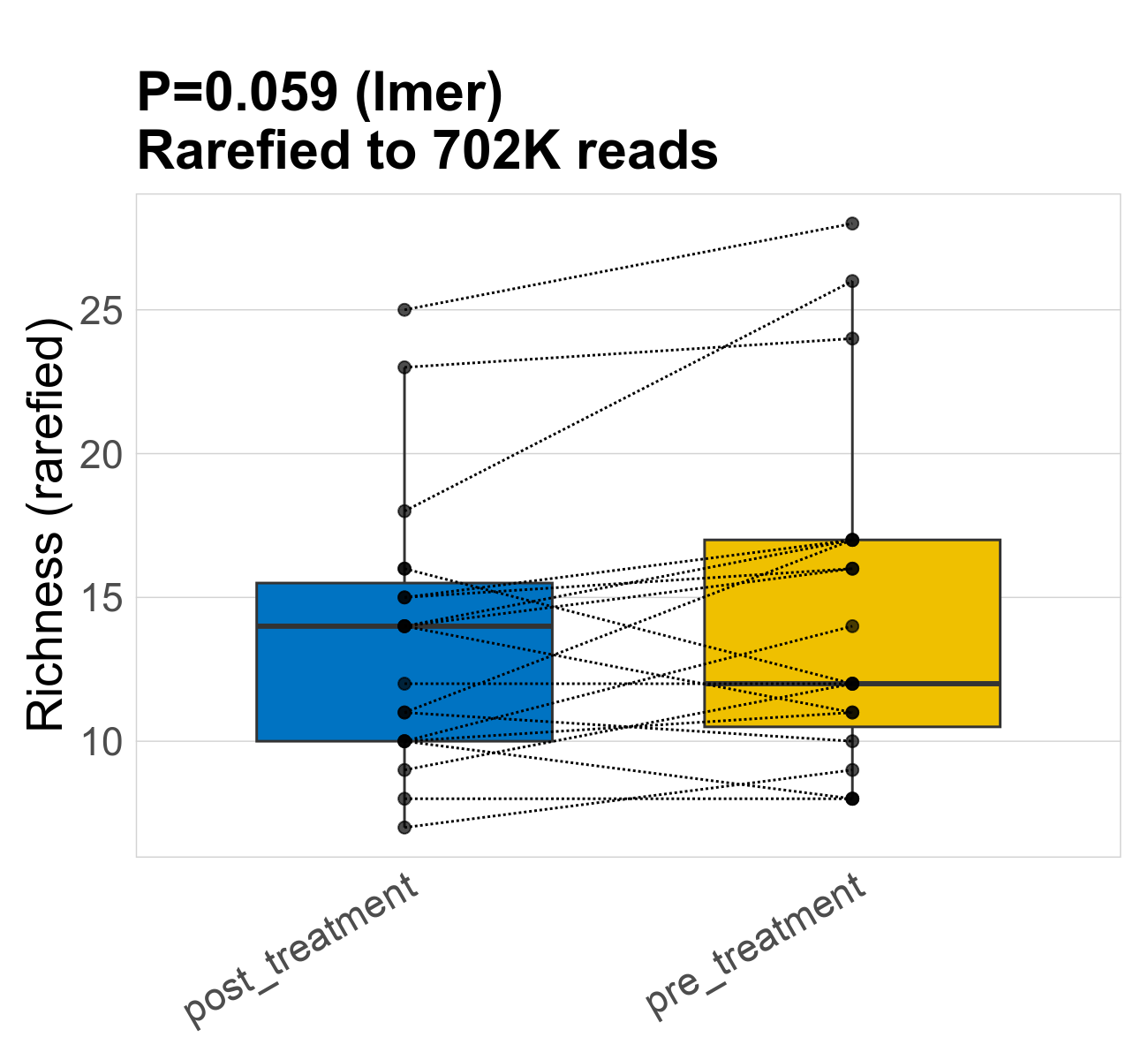
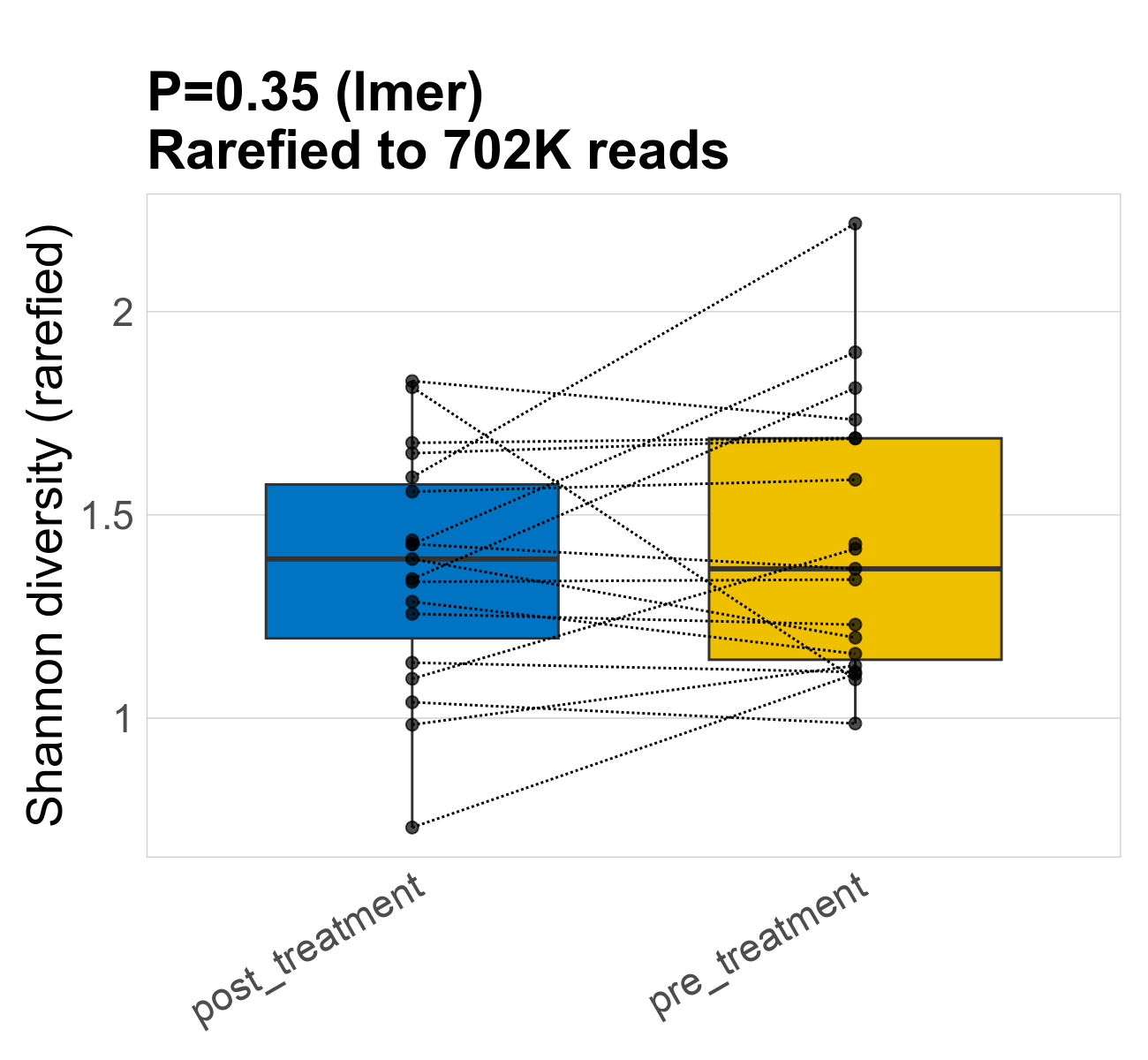
| Index | rarefiedTo | P lmer condition effect | Mean Pos | Mean Abundance | Median Abundance | Mean Treatmentpost_treatment | Median Treatmentpost_treatment | SD Treatmentpost_treatment | Mean Treatmentpre_treatment | Median Treatmentpre_treatment | SD Treatmentpre_treatment | Fold Change Log2(Treatmentpre_treatment/Treatmentpost_treatment) | Positive samples | Positive Treatmentpost_treatment | Positive Treatmentpre_treatment | Positive_Treatmentpost_treatment_percent | Positive_Treatmentpre_treatment_percent |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Shannon | 702268 | 0.35 | 1.4 | 1.4 | 1.4 | 1.4 | 1.4 | 0.29 | 1.4 | 1.4 | 0.33 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| Richness | 702268 | 0.059 | 14 | 14 | 13 | 14 | 14 | 4.7 | 15 | 12 | 6 | 0.1 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
Taxonomic profiles were analyzed using supervised and unsupervised multivariate methods. Profiles were ordinated using the unsupervised methods Principal Coordinates Analysis (PCoA), Non-Metric Multidimensional Scaling (NMDS) and Principal Component Analysis (PCA). PCoA and NMDS are related to PCA, but take dissimlarity matrices as input. PCoA and NMDS both attempt to represent the pairwise dissimlarities between samples in low dimensional space as close as possible. NMDS is a rank-based approach and therefore less effected by outliers.
The supervised methods Adonis and Redundancy analysis (RDA) were used to assess if variance in microbial community composition can be attributed to the study condition. NMDS, PCoA and Adonis were run on Bray-Curtis dissimilarities. A short introduction of the used methods can be found at the GUide to STatistical Analysis in Microbial Ecology (GUSTA ME). Sparse Partial Least Square Discriminant Analysis (sPLS-DA) from the MixMc package was additionaly used to extract features associated with the study condition.
Differentially abundant families were identified by ANOVA or LMER (linear mixed effect regression) of clr transformed relative abudances, Fisher's exact test and/or ALDEx2 (on families read counts). Fisher's exact test is used to test for differences in the detection rate, i.e. number of samples in which each family has been detected.
LMER is used for repeated measures data, using random effects to control for correlation between samples from the same subject. Fixed effects are included for treatment groups, time, and treatment over time, where appropriate. The LMER P values correspond to a nested model test of the significance of including the corresponding fixed effect.
ALDEx2 uses subsampling (Bayesian sampling) to estimate the underlying technical variation. For each subsample instance, center log-ratio transformed data is statistically compared across study groups and computed P values are corrected for multiple testing using the Benjamini–Hochberg procedure. The expected P value (mean P value) is reported, which are those that would likely have been observed if the same samples had been run multiple times. The expected values are reported for both the distribution of P values and for the distribution of Benjamini–Hochberg corrected values.
| Taxon | GTDB taxonomy | P lmer condition effect (sqrt) | FDR lmer condition effect (sqrt) | Pbonf lmer condition effect (sqrt) | P lmer condition effect (clr) | FDR lmer condition effect (clr) | Pbonf lmer condition effect (clr) | Mean Pos | Mean Abundance | Median Abundance | Mean Treatmentpost_treatment | Median Treatmentpost_treatment | SD Treatmentpost_treatment | Mean Treatmentpre_treatment | Median Treatmentpre_treatment | SD Treatmentpre_treatment | Fold Change Log2(Treatmentpre_treatment/Treatmentpost_treatment) | Positive samples | Positive Treatmentpost_treatment | Positive Treatmentpre_treatment | Positive_Treatmentpost_treatment_percent | Positive_Treatmentpre_treatment_percent |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
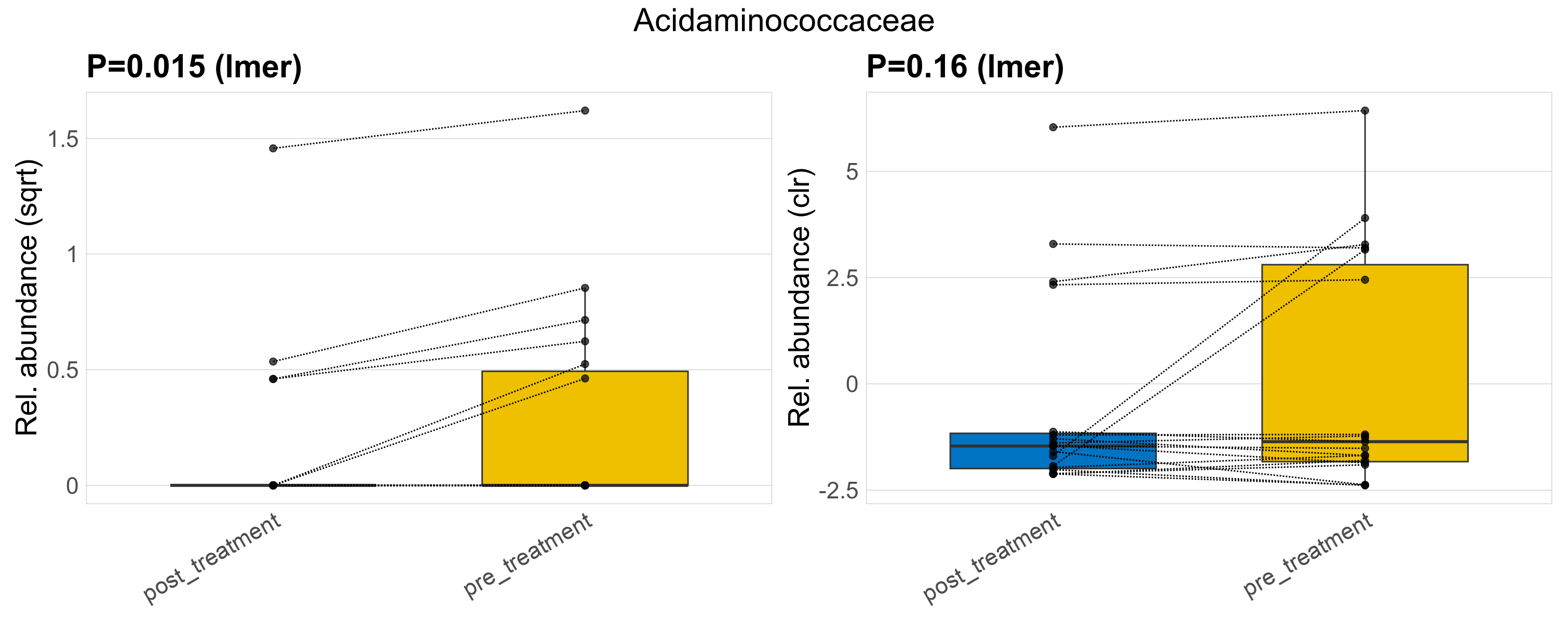
| Acidaminococcaceae | Acidaminococcaceae | 0.015 | 0.23 | 0.63 | 0.16 | 0.51 | 1 | 0.76 | 0.2 | 0 | 0.15 | 0 | 0.49 | 0.25 | 0 | 0.61 | 0.74 | 10 / 38 (26%) | 4 / 19 (21%) | 6 / 19 (32%) | 0.211 | 0.316 |
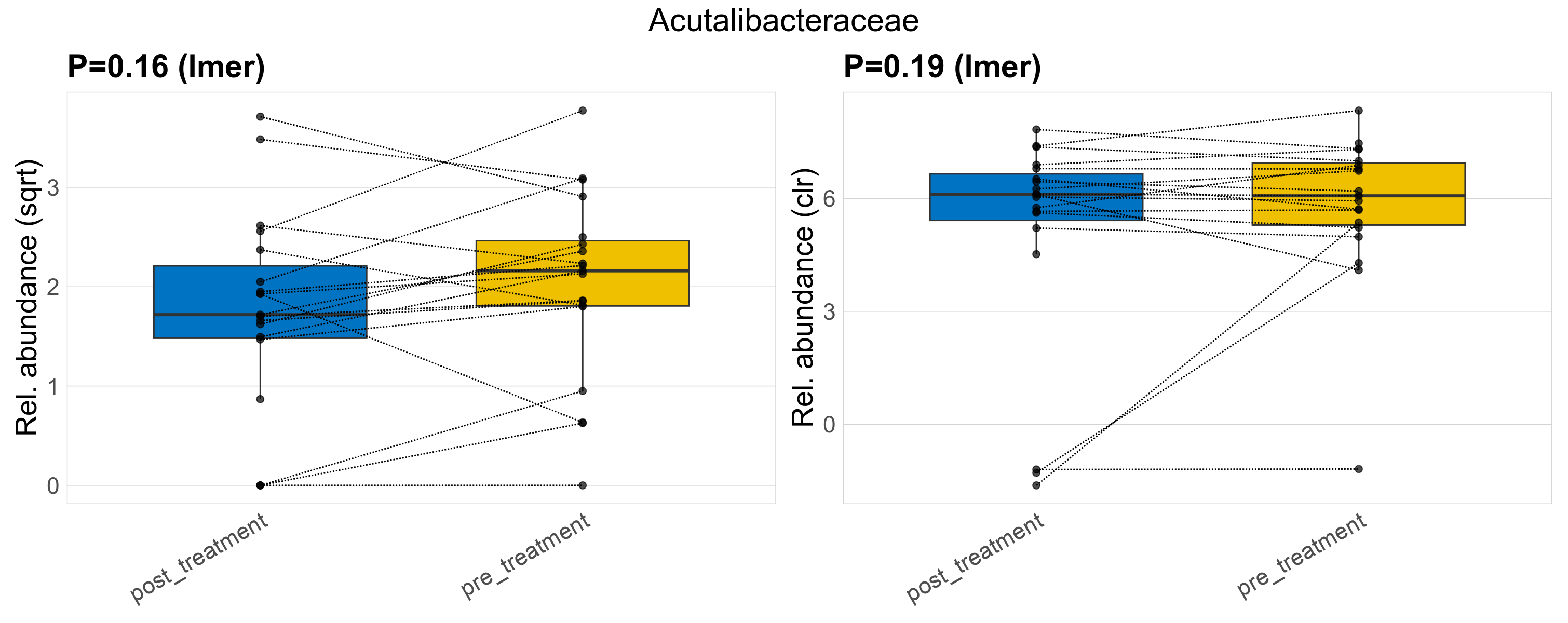
| Acutalibacteraceae | Acutalibacteraceae | 0.16 | 0.44 | 1 | 0.19 | 0.56 | 1 | 5 | 4.5 | 3.7 | 4 | 3 | 3.7 | 4.9 | 4.7 | 3.6 | 0.29 | 34 / 38 (89%) | 16 / 19 (84%) | 18 / 19 (95%) | 0.842 | 0.947 |
| Akkermansiaceae | Akkermansiaceae | 0.33 | 0.5 | 1 | 0.85 | 0.98 | 1 | 1.4 | 0.37 | 0 | 0.24 | 0 | 0.59 | 0.5 | 0 | 0.97 | 1.1 | 10 / 38 (26%) | 5 / 19 (26%) | 5 / 19 (26%) | 0.263 | 0.263 |
| Anaerovoracaceae | Anaerovoracaceae | 0.6 | 0.7 | 1 | 0.59 | 0.77 | 1 | 0.54 | 0.086 | 0 | 0.099 | 0 | 0.28 | 0.072 | 0 | 0.21 | -0.46 | 6 / 38 (16%) | 3 / 19 (16%) | 3 / 19 (16%) | 0.158 | 0.158 |
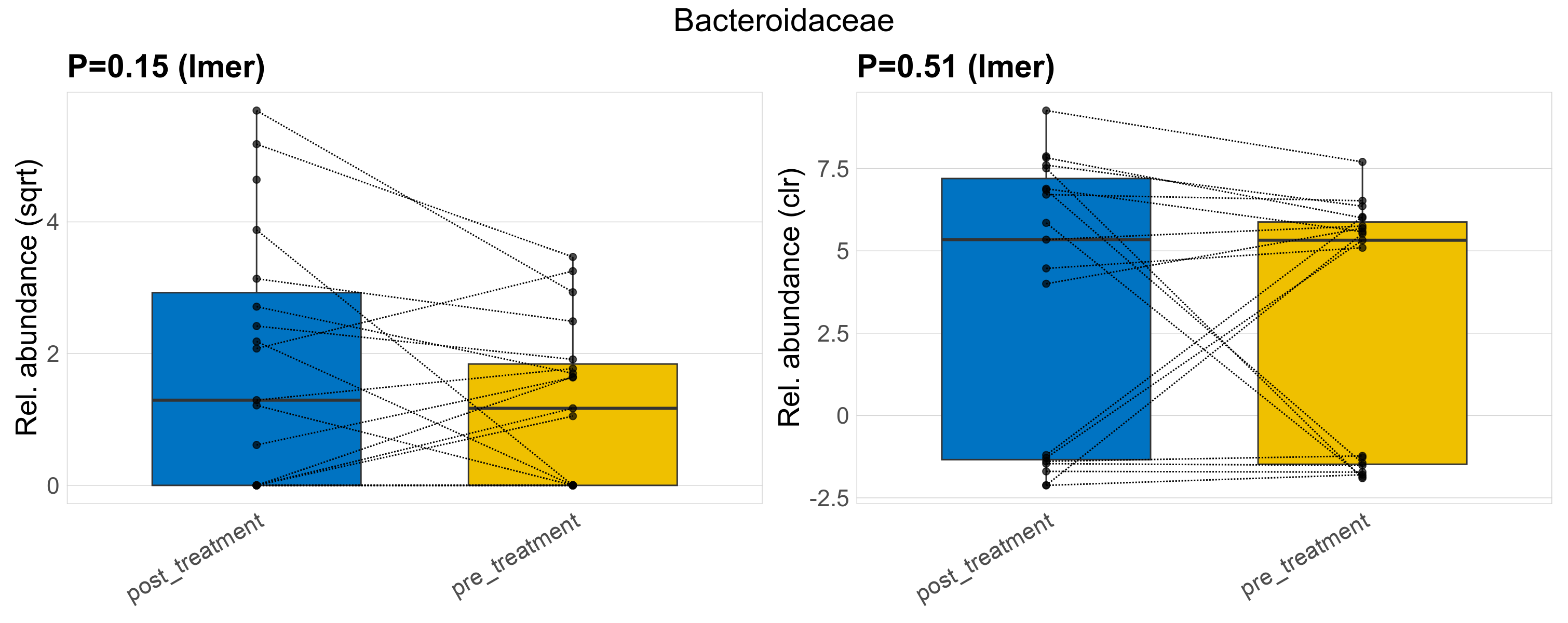
| Bacteroidaceae | Bacteroidaceae | 0.15 | 0.44 | 1 | 0.51 | 0.77 | 1 | 8.1 | 4.9 | 1.6 | 6.9 | 1.7 | 9.9 | 2.9 | 1.4 | 3.8 | -1.3 | 23 / 38 (61%) | 12 / 19 (63%) | 11 / 19 (58%) | 0.632 | 0.579 |
| Barnesiellaceae | Barnesiellaceae | 0.25 | 0.49 | 1 | 0.47 | 0.77 | 1 | 0.31 | 0.025 | 0 | 0.0066 | 0 | 0.029 | 0.043 | 0 | 0.14 | 2.7 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
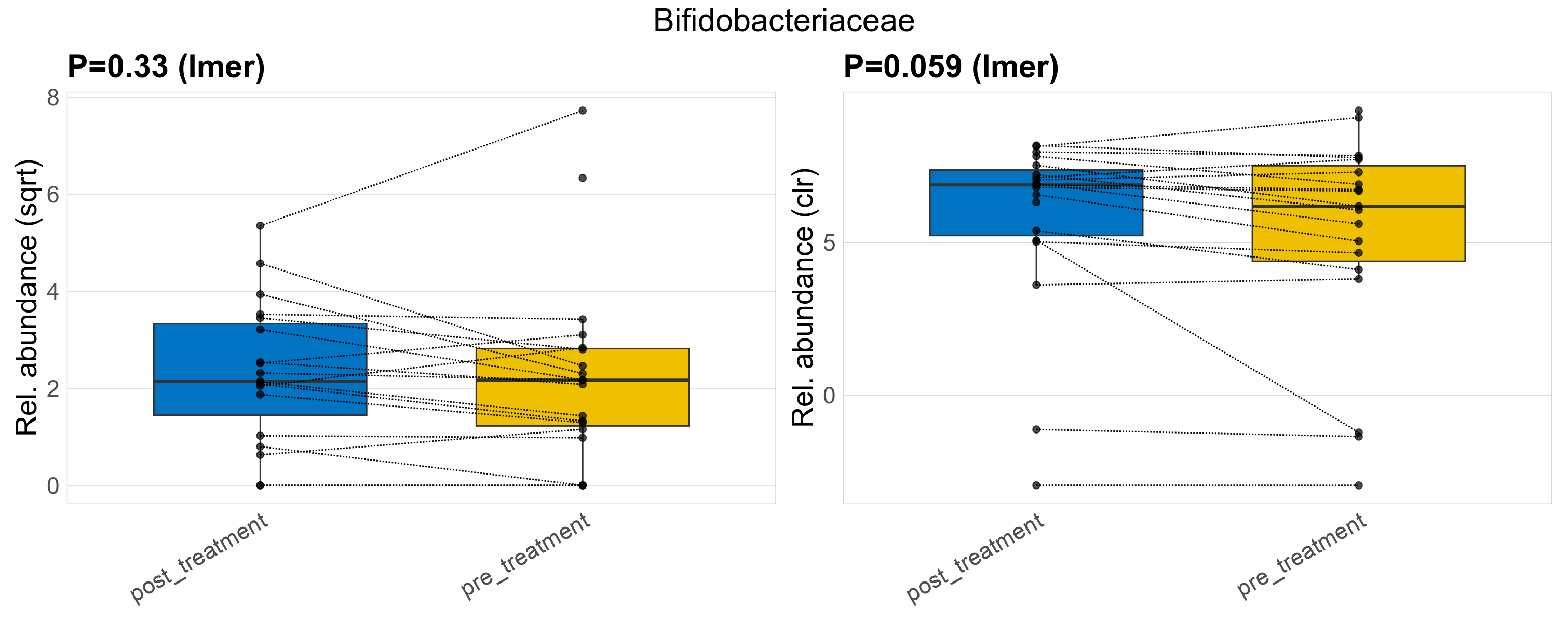
| Bifidobacteriaceae | Bifidobacteriaceae | 0.33 | 0.5 | 1 | 0.059 | 0.35 | 1 | 9.4 | 8.2 | 4.6 | 7.4 | 4.6 | 7.6 | 8.9 | 4.7 | 15 | 0.27 | 33 / 38 (87%) | 17 / 19 (89%) | 16 / 19 (84%) | 0.895 | 0.842 |
| Burkholderiaceae | Burkholderiaceae | 0.3 | 0.49 | 1 | 0.31 | 0.71 | 1 | 0.28 | 0.052 | 0 | 0.08 | 0 | 0.2 | 0.024 | 0 | 0.063 | -1.7 | 7 / 38 (18%) | 4 / 19 (21%) | 3 / 19 (16%) | 0.211 | 0.158 |
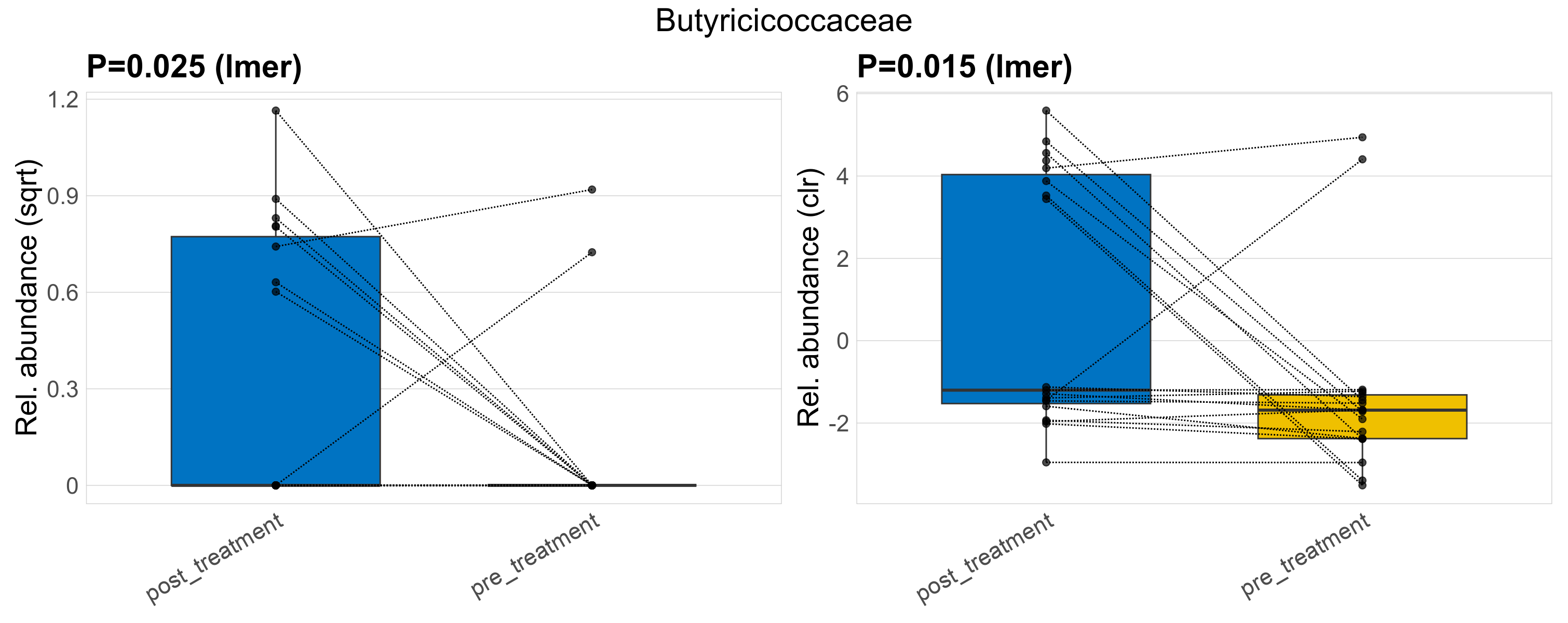
| Butyricicoccaceae | Butyricicoccaceae | 0.025 | 0.23 | 0.99 | 0.015 | 0.31 | 0.6 | 0.68 | 0.18 | 0 | 0.29 | 0 | 0.4 | 0.072 | 0 | 0.22 | -2 | 10 / 38 (26%) | 8 / 19 (42%) | 2 / 19 (11%) | 0.421 | 0.105 |
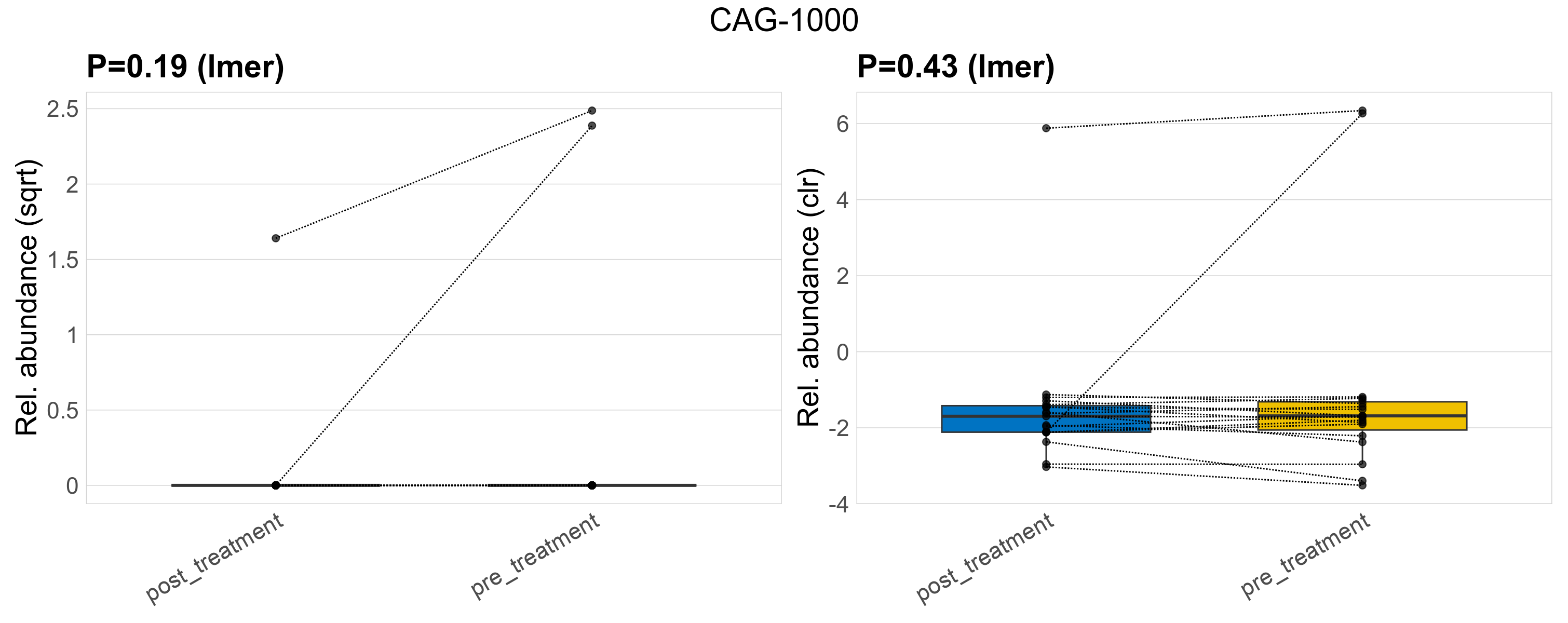
| CAG-1000 | CAG-1000 | 0.19 | 0.47 | 1 | 0.43 | 0.77 | 1 | 4.9 | 0.38 | 0 | 0.14 | 0 | 0.62 | 0.63 | 0 | 1.9 | 2.2 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
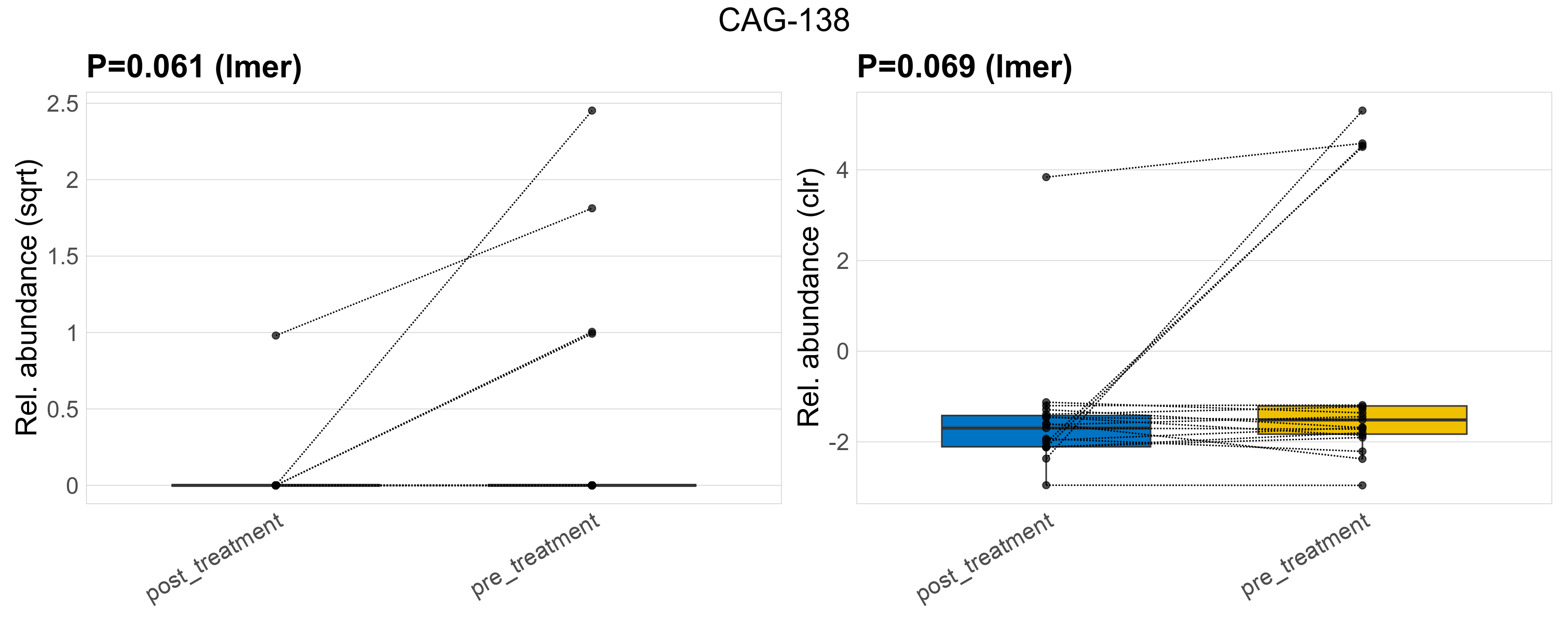
| CAG-138 | CAG-138 | 0.061 | 0.29 | 1 | 0.069 | 0.36 | 1 | 2.5 | 0.32 | 0 | 0.051 | 0 | 0.22 | 0.59 | 0 | 1.5 | 3.5 | 5 / 38 (13%) | 1 / 19 (5.3%) | 4 / 19 (21%) | 0.0526 | 0.211 |
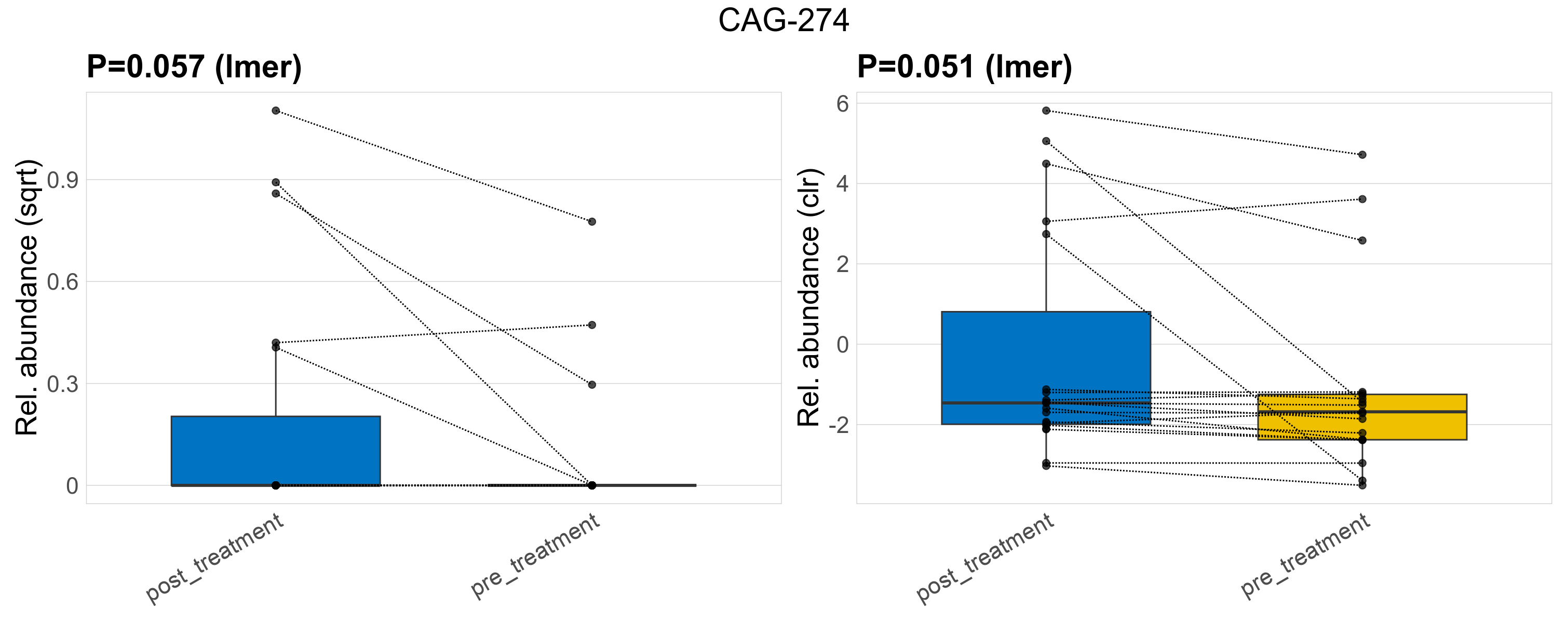
| CAG-274 | CAG-274 | 0.057 | 0.29 | 1 | 0.051 | 0.35 | 1 | 0.5 | 0.11 | 0 | 0.16 | 0 | 0.35 | 0.048 | 0 | 0.14 | -1.7 | 8 / 38 (21%) | 5 / 19 (26%) | 3 / 19 (16%) | 0.263 | 0.158 |
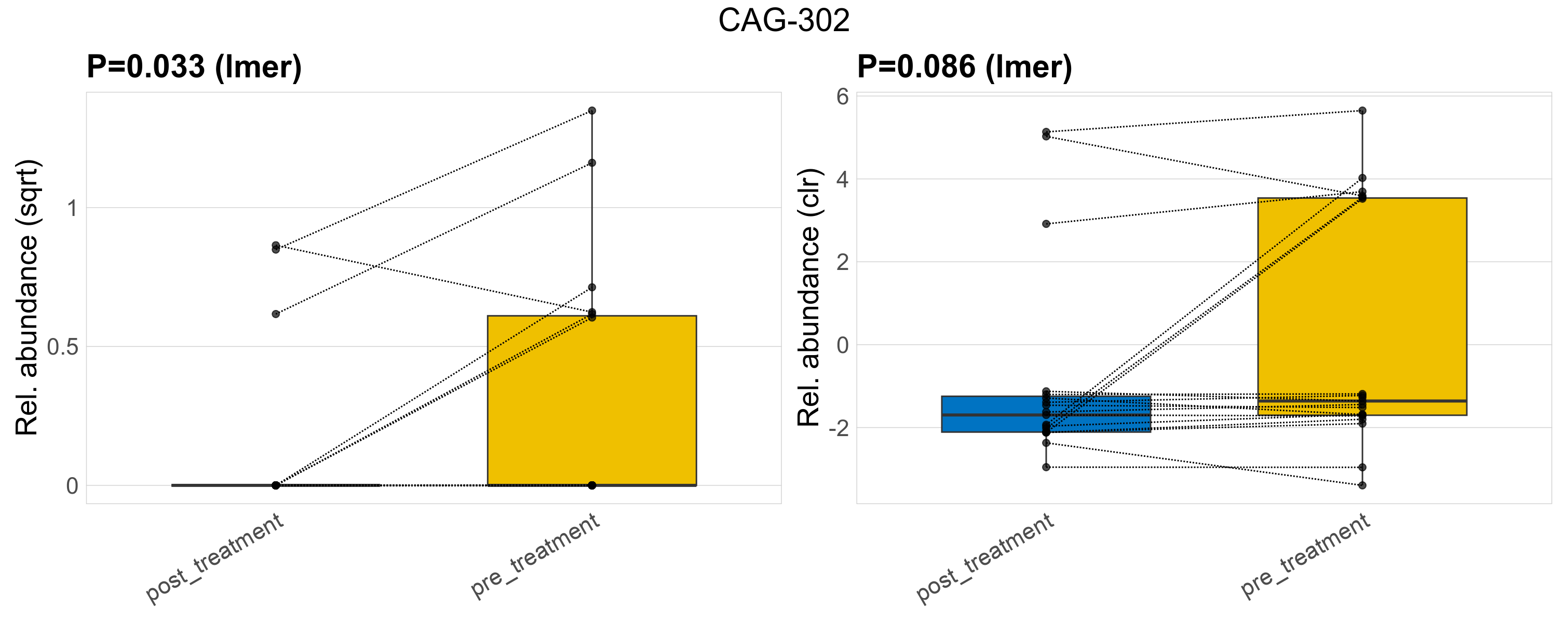
| CAG-302 | CAG-302 | 0.033 | 0.23 | 1 | 0.086 | 0.4 | 1 | 0.74 | 0.18 | 0 | 0.097 | 0 | 0.24 | 0.25 | 0 | 0.51 | 1.4 | 9 / 38 (24%) | 3 / 19 (16%) | 6 / 19 (32%) | 0.158 | 0.316 |
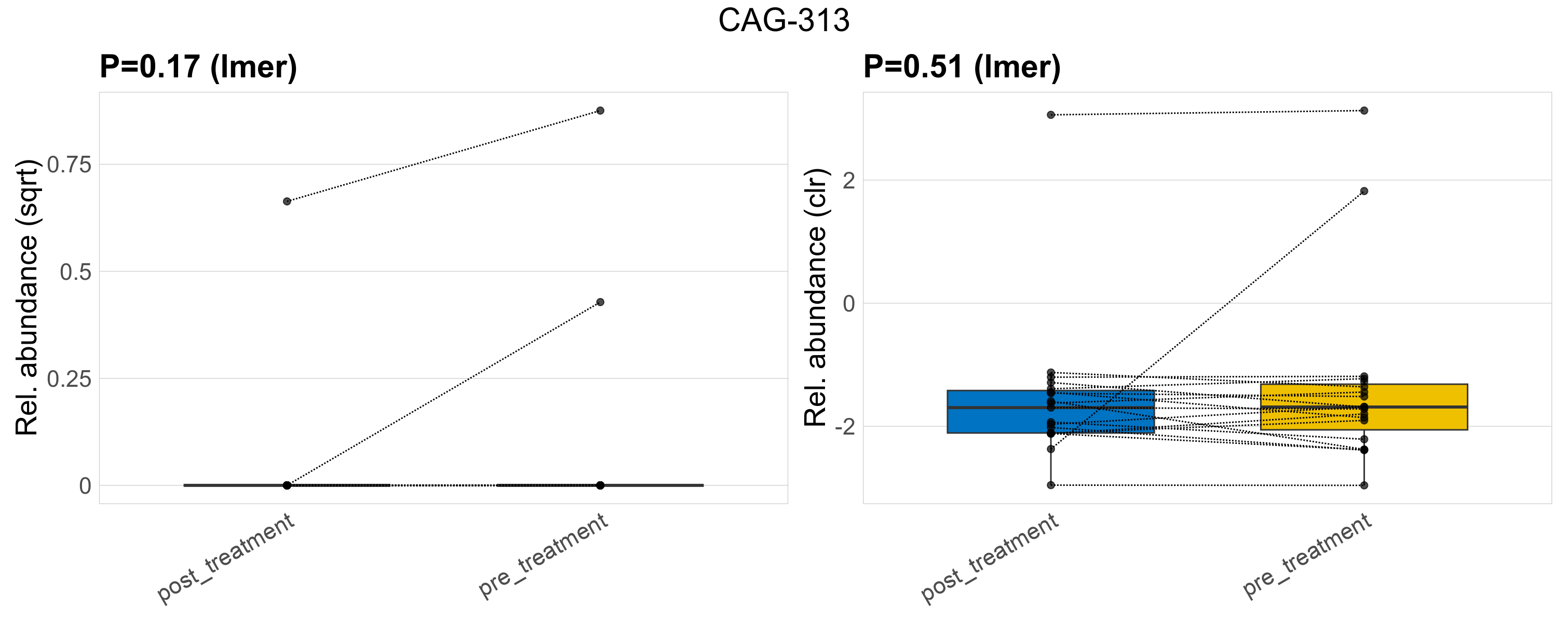
| CAG-313 | CAG-313 | 0.17 | 0.44 | 1 | 0.51 | 0.77 | 1 | 0.46 | 0.037 | 0 | 0.023 | 0 | 0.1 | 0.05 | 0 | 0.18 | 1.1 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| CAG-314 | CAG-314 | 0.22 | 0.49 | 1 | 0.43 | 0.77 | 1 | 0.32 | 0.025 | 0 | 0.0075 | 0 | 0.033 | 0.043 | 0 | 0.13 | 2.5 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| CAG-508 | CAG-508 | 0.79 | 0.87 | 1 | 0.74 | 0.91 | 1 | 0.73 | 0.25 | 0 | 0.2 | 0 | 0.38 | 0.3 | 0 | 0.58 | 0.58 | 13 / 38 (34%) | 7 / 19 (37%) | 6 / 19 (32%) | 0.368 | 0.316 |
| CAG-611 | CAG-611 | 0.29 | 0.49 | 1 | 0.56 | 0.77 | 1 | 1.2 | 0.16 | 0 | 0.12 | 0 | 0.37 | 0.2 | 0 | 0.62 | 0.74 | 5 / 38 (13%) | 2 / 19 (11%) | 3 / 19 (16%) | 0.105 | 0.158 |
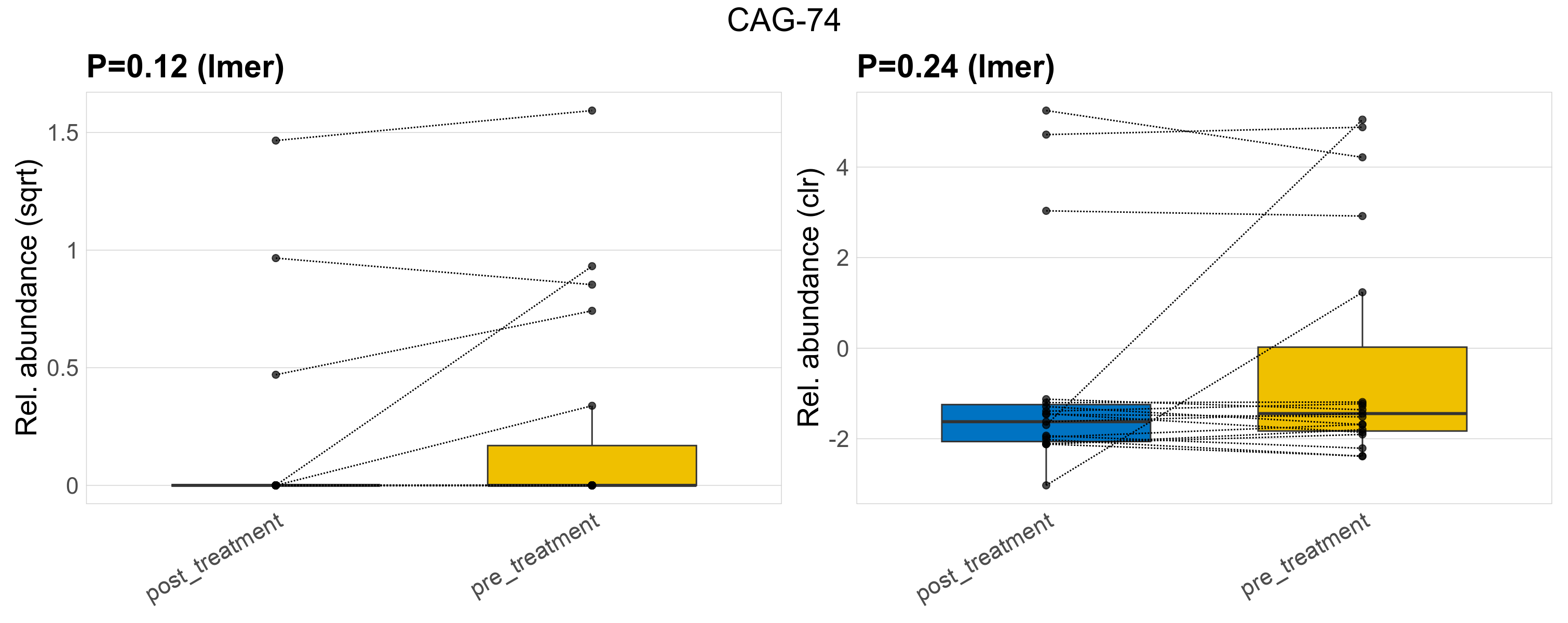
| CAG-74 | CAG-74 | 0.12 | 0.44 | 1 | 0.24 | 0.58 | 1 | 1 | 0.21 | 0 | 0.17 | 0 | 0.52 | 0.25 | 0 | 0.62 | 0.56 | 8 / 38 (21%) | 3 / 19 (16%) | 5 / 19 (26%) | 0.158 | 0.263 |
| CAG-917 | CAG-917 | 0.25 | 0.49 | 1 | 0.39 | 0.77 | 1 | 1.3 | 0.1 | 0 | 0.017 | 0 | 0.074 | 0.19 | 0 | 0.67 | 3.5 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| Clostridiaceae | Clostridiaceae | 0.48 | 0.63 | 1 | 0.98 | 1 | 1 | 0.61 | 0.16 | 0 | 0.082 | 0 | 0.16 | 0.24 | 0 | 0.71 | 1.5 | 10 / 38 (26%) | 5 / 19 (26%) | 5 / 19 (26%) | 0.263 | 0.263 |
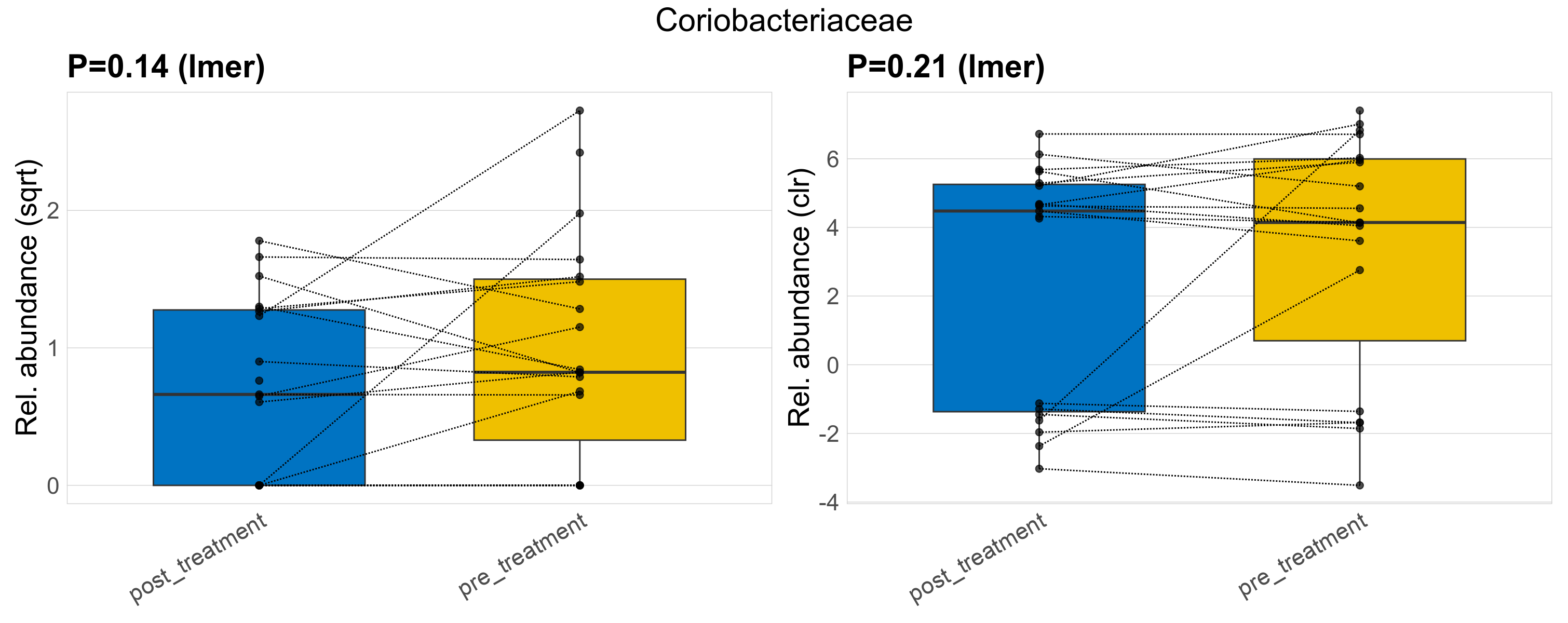
| Coriobacteriaceae | Coriobacteriaceae | 0.14 | 0.44 | 1 | 0.21 | 0.56 | 1 | 1.9 | 1.3 | 0.65 | 0.91 | 0.44 | 1 | 1.6 | 0.68 | 2.1 | 0.81 | 26 / 38 (68%) | 12 / 19 (63%) | 14 / 19 (74%) | 0.632 | 0.737 |
| Dialisteraceae | Dialisteraceae | 0.47 | 0.63 | 1 | 1 | 1 | 1 | 1.6 | 0.69 | 0 | 0.66 | 0 | 1.1 | 0.71 | 0 | 1 | 0.11 | 16 / 38 (42%) | 8 / 19 (42%) | 8 / 19 (42%) | 0.421 | 0.421 |
| Eggerthellaceae | Eggerthellaceae | 0.92 | 0.92 | 1 | 0.95 | 1 | 1 | 0.47 | 0.17 | 0 | 0.17 | 0 | 0.27 | 0.17 | 0 | 0.24 | 0 | 14 / 38 (37%) | 7 / 19 (37%) | 7 / 19 (37%) | 0.368 | 0.368 |
| Enterobacteriaceae | Enterobacteriaceae | 0.27 | 0.49 | 1 | 0.51 | 0.77 | 1 | 2.2 | 0.17 | 0 | 0.14 | 0 | 0.6 | 0.21 | 0 | 0.7 | 0.58 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
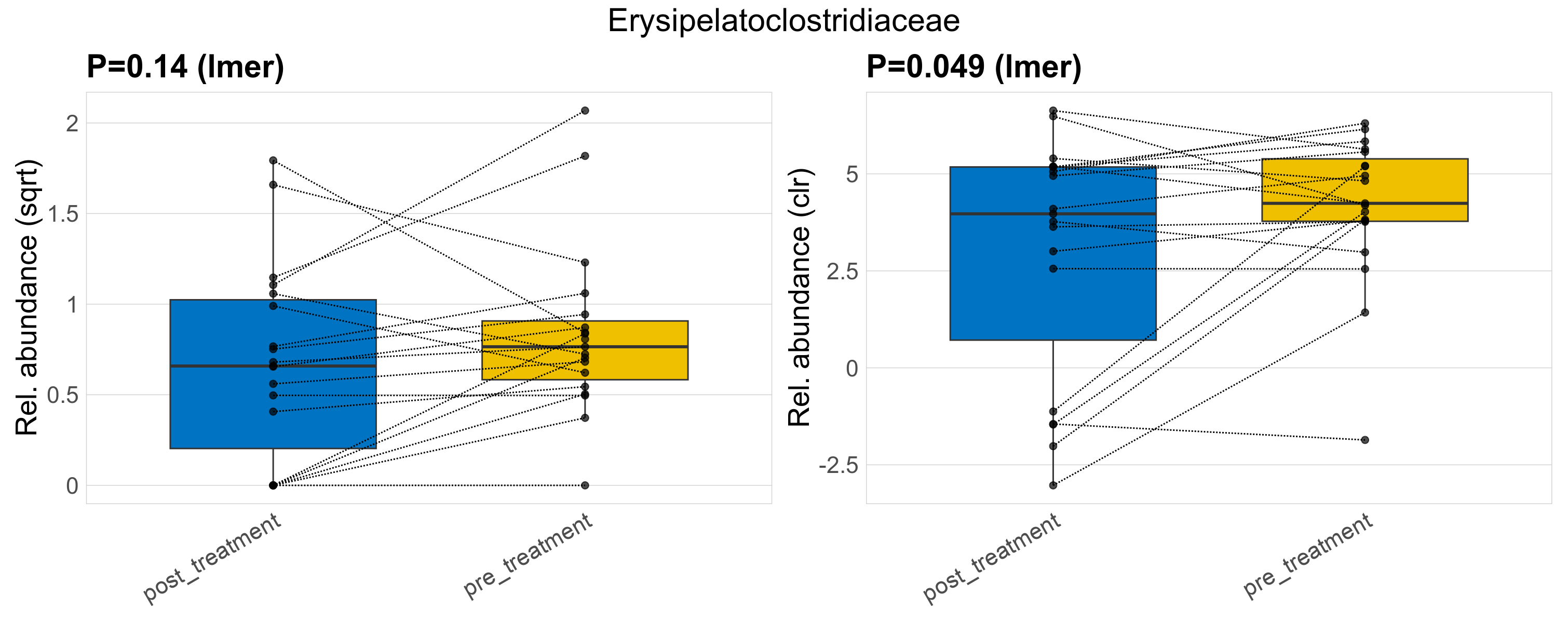
| Erysipelatoclostridiaceae | Erysipelatoclostridiaceae | 0.14 | 0.44 | 1 | 0.049 | 0.35 | 1 | 0.97 | 0.82 | 0.51 | 0.73 | 0.43 | 0.91 | 0.91 | 0.59 | 1.1 | 0.32 | 32 / 38 (84%) | 14 / 19 (74%) | 18 / 19 (95%) | 0.737 | 0.947 |
| Erysipelotrichaceae | Erysipelotrichaceae | 0.41 | 0.58 | 1 | 0.56 | 0.77 | 1 | 1.8 | 0.47 | 0 | 0.44 | 0 | 1 | 0.5 | 0 | 1.1 | 0.18 | 10 / 38 (26%) | 5 / 19 (26%) | 5 / 19 (26%) | 0.263 | 0.263 |
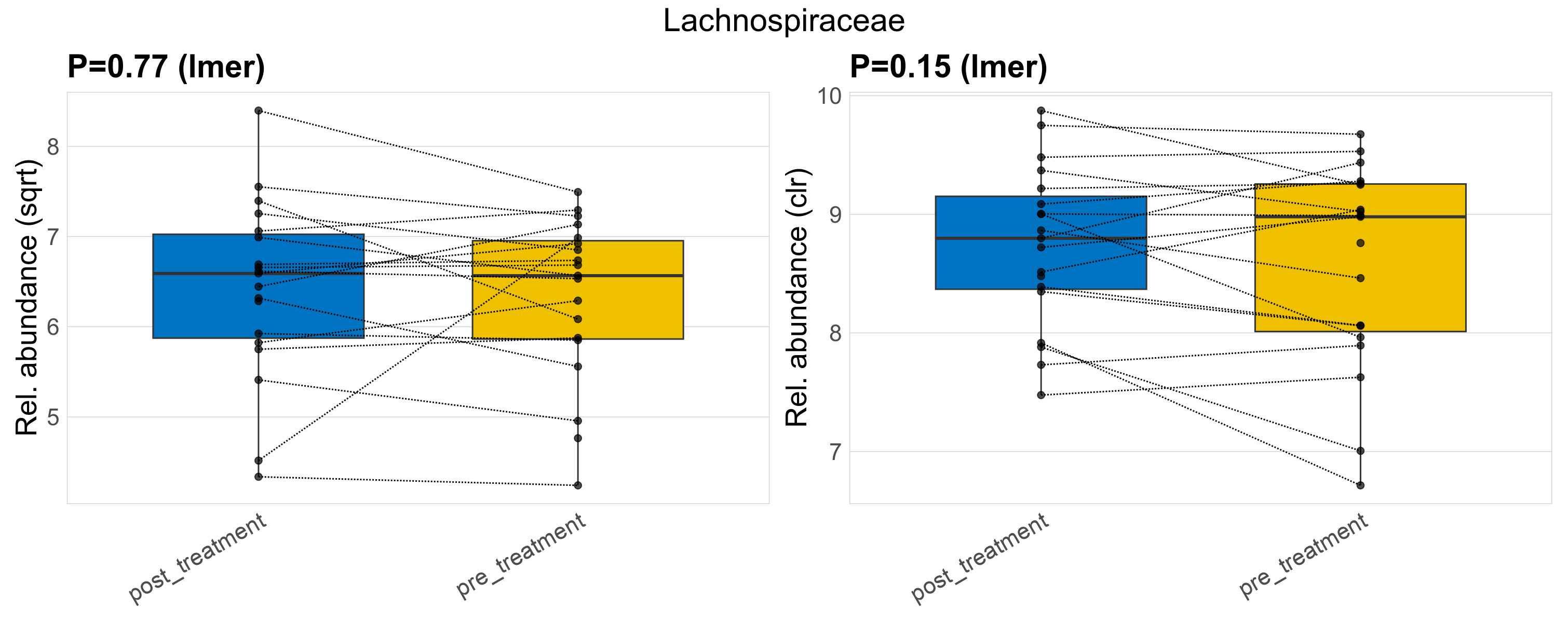
| Lachnospiraceae | Lachnospiraceae | 0.77 | 0.87 | 1 | 0.15 | 0.51 | 1 | 41 | 41 | 43 | 42 | 43 | 12 | 41 | 43 | 11 | -0.035 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
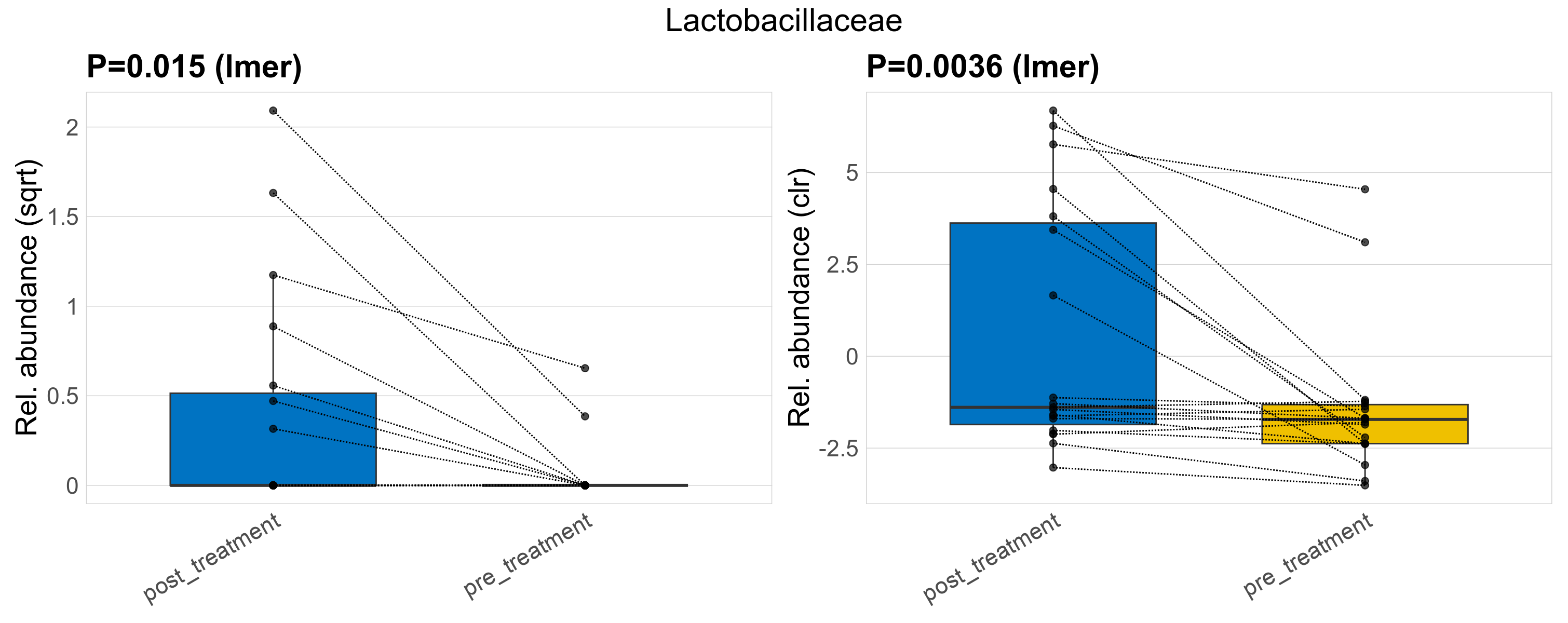
| Lactobacillaceae | Lactobacillaceae | 0.015 | 0.23 | 0.62 | 0.0036 | 0.15 | 0.15 | 1.2 | 0.27 | 0 | 0.52 | 0 | 1.2 | 0.03 | 0 | 0.1 | -4.1 | 9 / 38 (24%) | 7 / 19 (37%) | 2 / 19 (11%) | 0.368 | 0.105 |
| Megasphaeraceae | Megasphaeraceae | 0.53 | 0.66 | 1 | 0.4 | 0.77 | 1 | 1.1 | 0.087 | 0 | 0.069 | 0 | 0.25 | 0.11 | 0 | 0.46 | 0.67 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
| Methanobacteriaceae | Methanobacteriaceae | 0.52 | 0.66 | 1 | 0.96 | 1 | 1 | 0.91 | 0.095 | 0 | 0.051 | 0 | 0.15 | 0.14 | 0 | 0.42 | 1.5 | 4 / 38 (11%) | 2 / 19 (11%) | 2 / 19 (11%) | 0.105 | 0.105 |
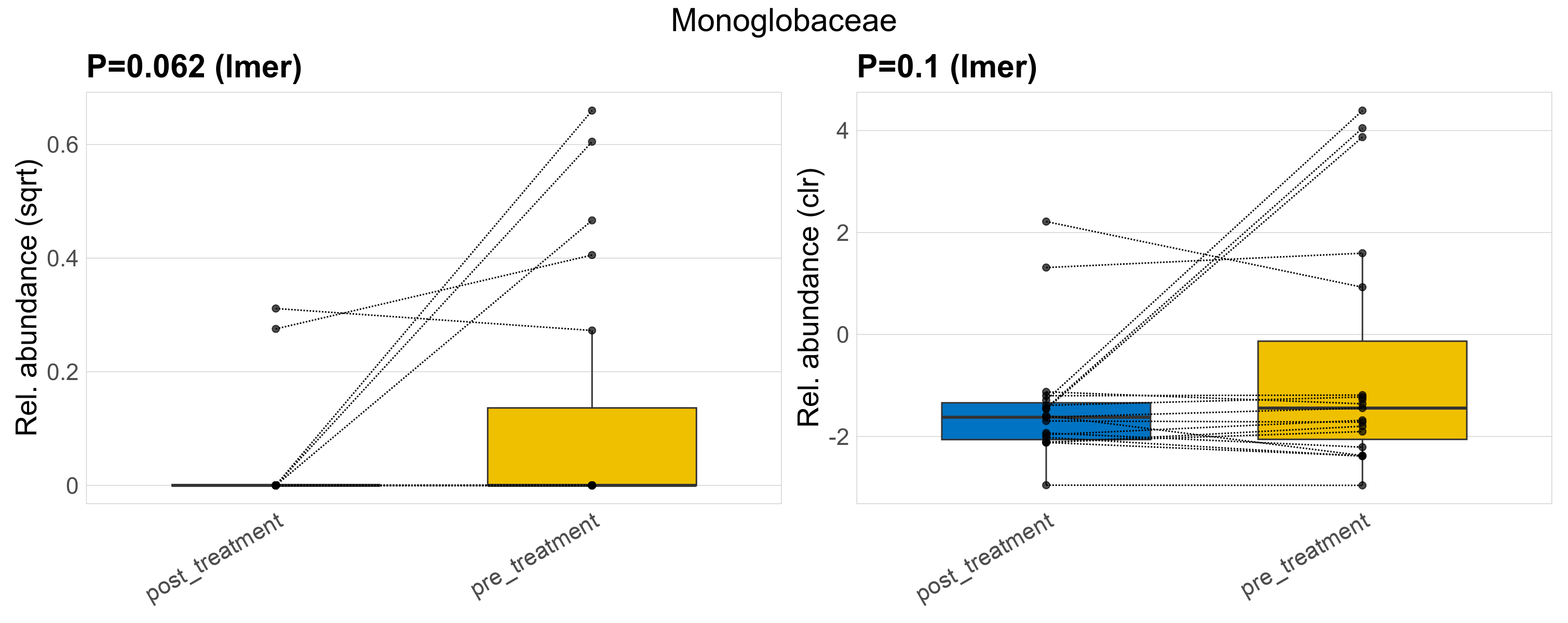
| Monoglobaceae | Monoglobaceae | 0.062 | 0.29 | 1 | 0.1 | 0.41 | 1 | 0.2 | 0.038 | 0 | 0.0091 | 0 | 0.027 | 0.066 | 0 | 0.13 | 2.9 | 7 / 38 (18%) | 2 / 19 (11%) | 5 / 19 (26%) | 0.105 | 0.263 |
| Muribaculaceae | Muribaculaceae | 0.81 | 0.87 | 1 | 0.87 | 0.98 | 1 | 0.8 | 0.085 | 0 | 0.098 | 0 | 0.37 | 0.071 | 0 | 0.21 | -0.46 | 4 / 38 (11%) | 2 / 19 (11%) | 2 / 19 (11%) | 0.105 | 0.105 |
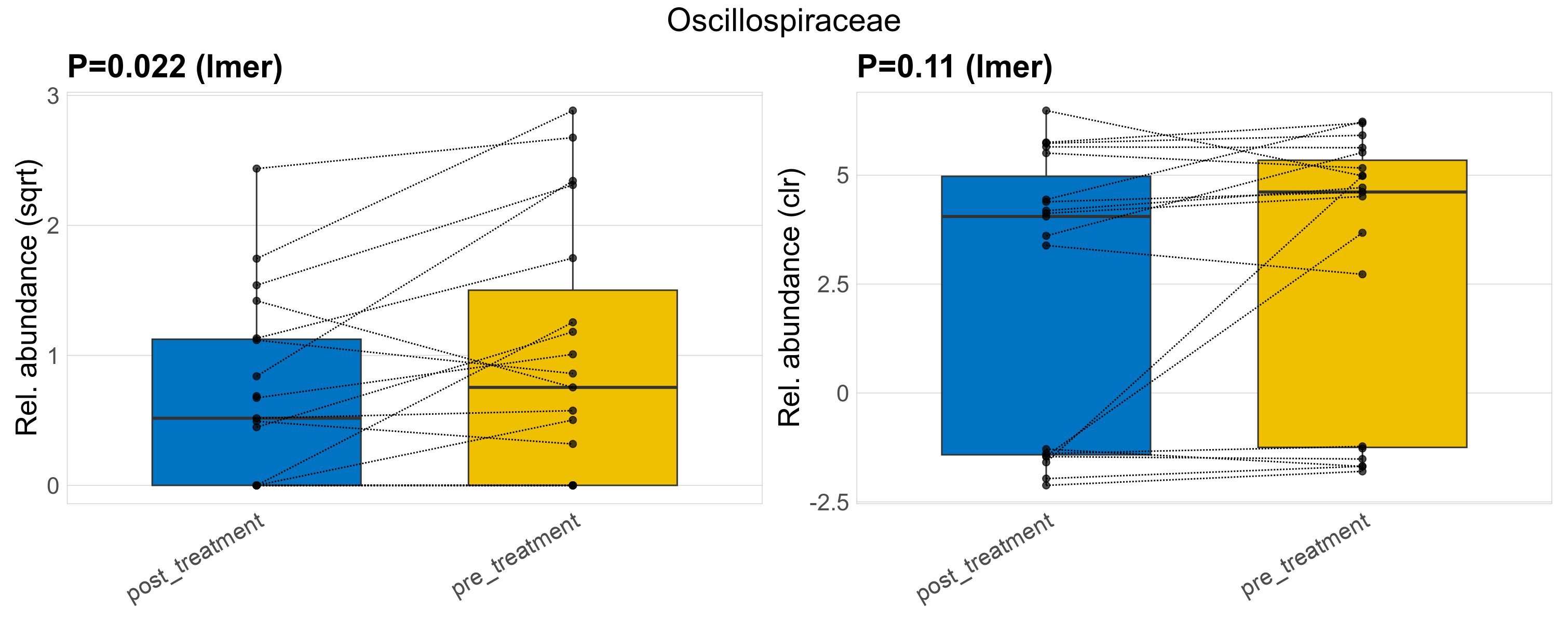
| Oscillospiraceae | Oscillospiraceae | 0.022 | 0.23 | 0.88 | 0.11 | 0.41 | 1 | 2.1 | 1.4 | 0.39 | 0.96 | 0.27 | 1.5 | 1.9 | 0.57 | 2.7 | 0.98 | 25 / 38 (66%) | 12 / 19 (63%) | 13 / 19 (68%) | 0.632 | 0.684 |
| Peptostreptococcaceae | Peptostreptococcaceae | 0.9 | 0.92 | 1 | 0.86 | 0.98 | 1 | 0.69 | 0.29 | 0 | 0.26 | 0 | 0.4 | 0.32 | 0 | 0.55 | 0.3 | 16 / 38 (42%) | 8 / 19 (42%) | 8 / 19 (42%) | 0.421 | 0.421 |
| QAMH01 | QAMH01 | 0.27 | 0.49 | 1 | 0.55 | 0.77 | 1 | 0.5 | 0.039 | 0 | 0.022 | 0 | 0.094 | 0.057 | 0 | 0.17 | 1.4 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
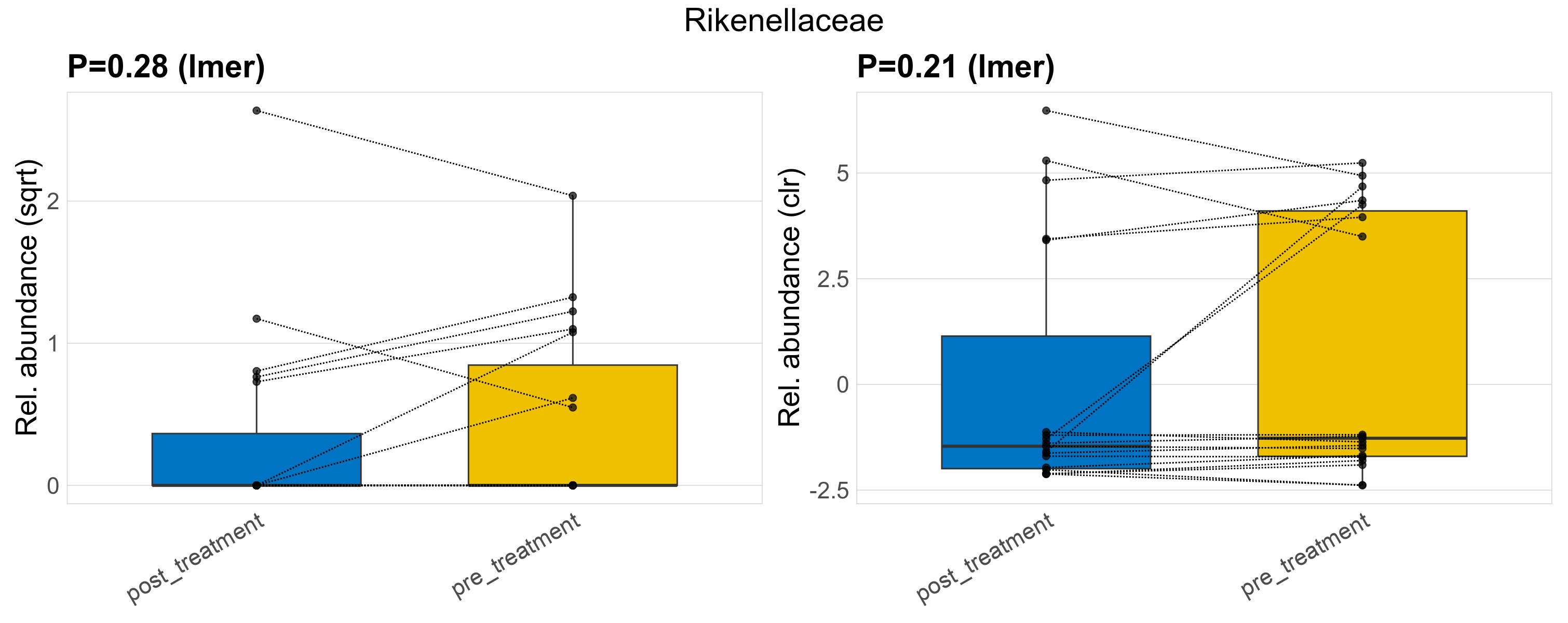
| Rikenellaceae | Rikenellaceae | 0.28 | 0.49 | 1 | 0.21 | 0.56 | 1 | 1.7 | 0.54 | 0 | 0.53 | 0 | 1.6 | 0.55 | 0 | 1.1 | 0.053 | 12 / 38 (32%) | 5 / 19 (26%) | 7 / 19 (37%) | 0.263 | 0.368 |
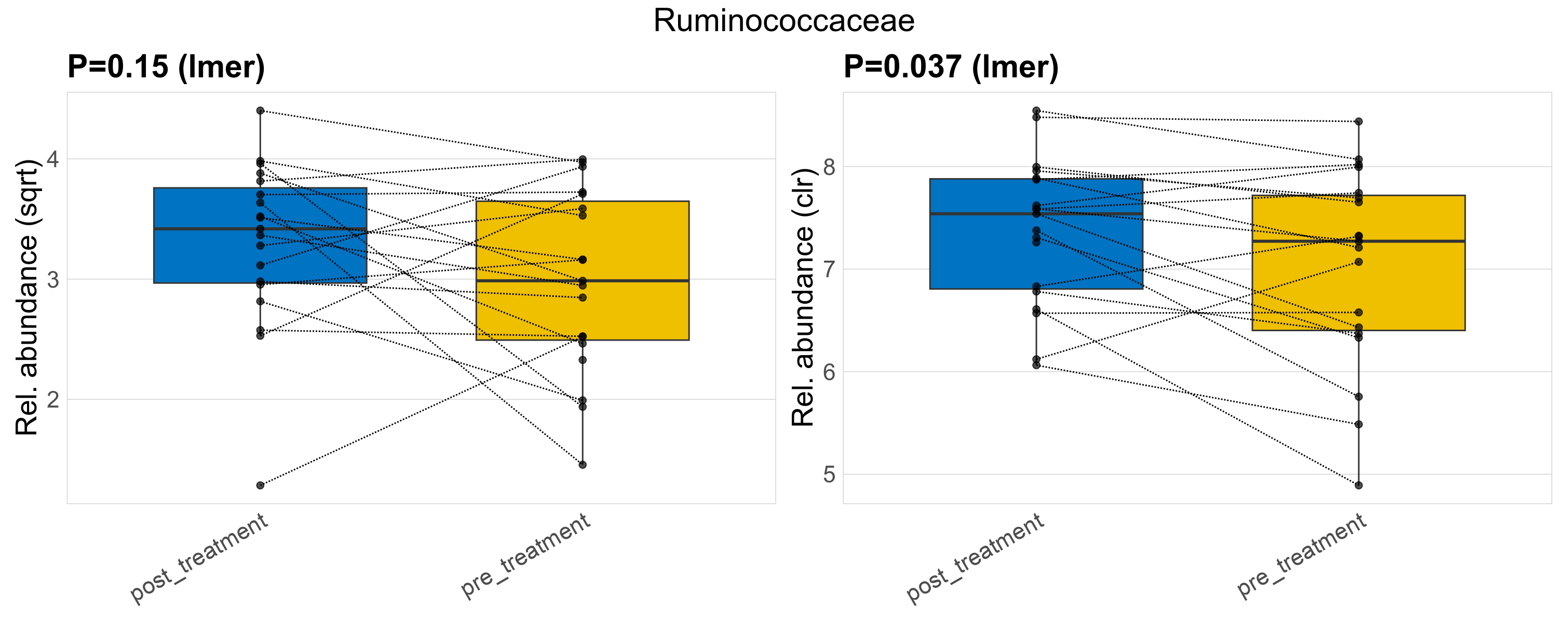
| Ruminococcaceae | Ruminococcaceae | 0.15 | 0.44 | 1 | 0.037 | 0.35 | 1 | 10 | 10 | 10 | 11 | 12 | 4.1 | 9.5 | 8.9 | 4.4 | -0.21 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| Selenomonadaceae | Selenomonadaceae | 0.83 | 0.87 | 1 | 0.91 | 1 | 1 | 0.46 | 0.049 | 0 | 0.063 | 0 | 0.19 | 0.035 | 0 | 0.13 | -0.85 | 4 / 38 (11%) | 2 / 19 (11%) | 2 / 19 (11%) | 0.105 | 0.105 |
| Streptococcaceae | Streptococcaceae | 0.58 | 0.7 | 1 | 0.32 | 0.71 | 1 | 1.1 | 0.52 | 0 | 0.52 | 0.31 | 0.71 | 0.51 | 0 | 1.1 | -0.028 | 18 / 38 (47%) | 10 / 19 (53%) | 8 / 19 (42%) | 0.526 | 0.421 |
| Tannerellaceae | Tannerellaceae | 0.28 | 0.49 | 1 | 0.47 | 0.77 | 1 | 0.62 | 0.11 | 0 | 0.096 | 0 | 0.36 | 0.13 | 0 | 0.33 | 0.44 | 7 / 38 (18%) | 3 / 19 (16%) | 4 / 19 (21%) | 0.158 | 0.211 |
| Turicibacteraceae | Turicibacteraceae | 0.38 | 0.55 | 1 | 0.65 | 0.83 | 1 | 0.22 | 0.017 | 0 | 0.0079 | 0 | 0.034 | 0.027 | 0 | 0.096 | 1.8 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
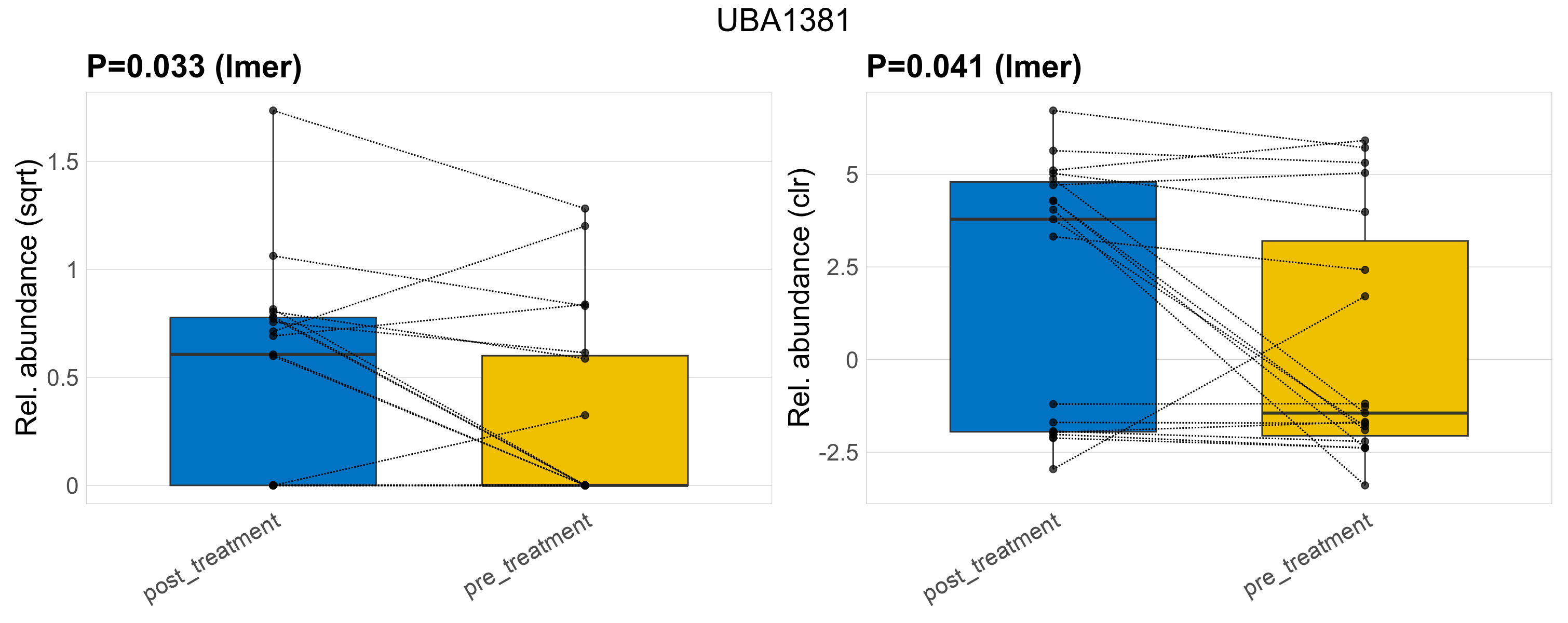
| UBA1381 | UBA1381 | 0.033 | 0.23 | 1 | 0.041 | 0.35 | 1 | 0.79 | 0.37 | 0 | 0.47 | 0.37 | 0.7 | 0.28 | 0 | 0.5 | -0.75 | 18 / 38 (47%) | 11 / 19 (58%) | 7 / 19 (37%) | 0.579 | 0.368 |
Click here to open full-sized image in new window.
Click here to open full-sized image in new window.
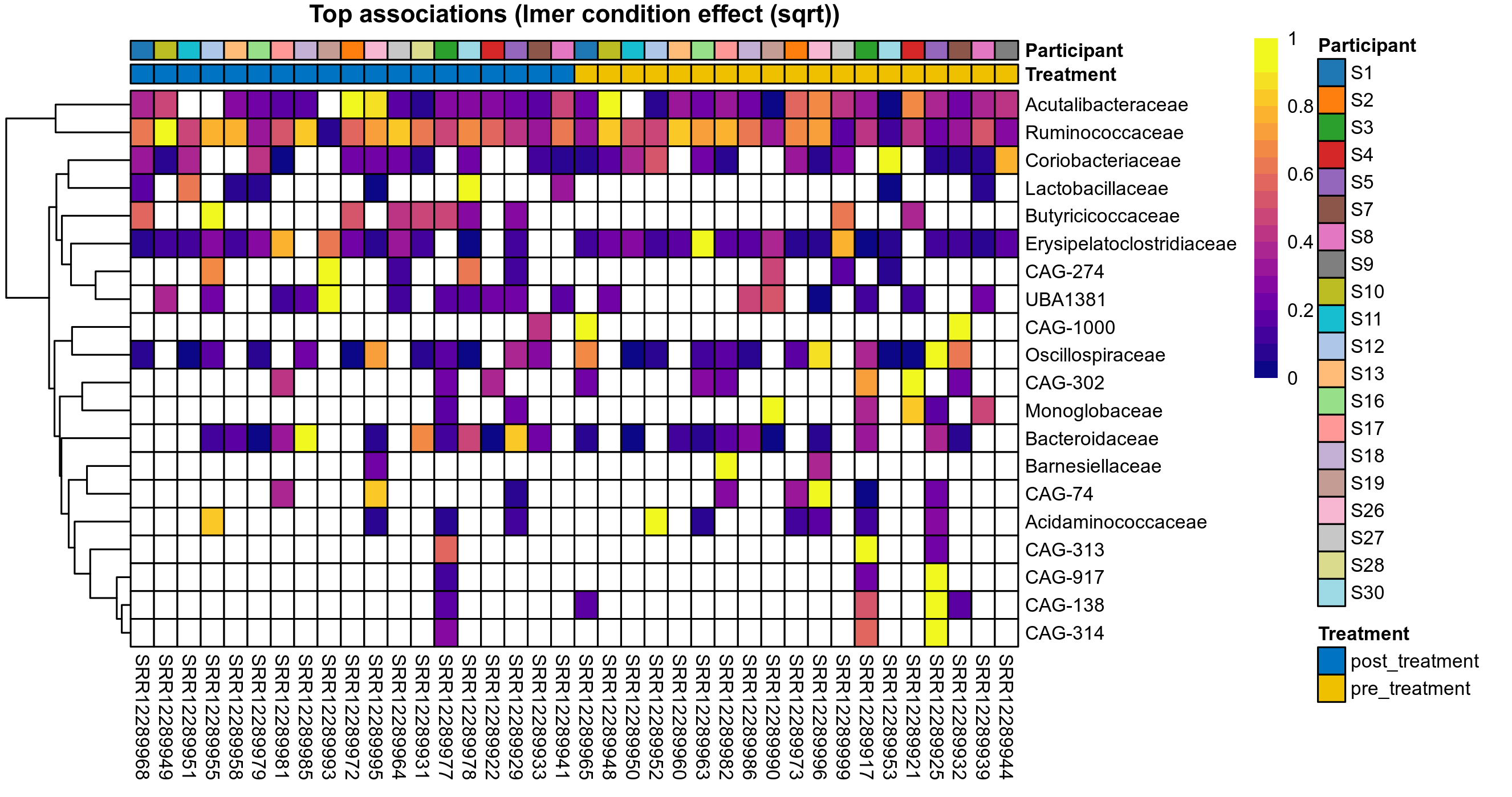
P lmer condition effect (sqrt): Differentially abundant species were identified by linear mixed effect regression without confounders. Random effects: Participant (intercept). Fixed effects: Treatment. Effect tested: Treatment.
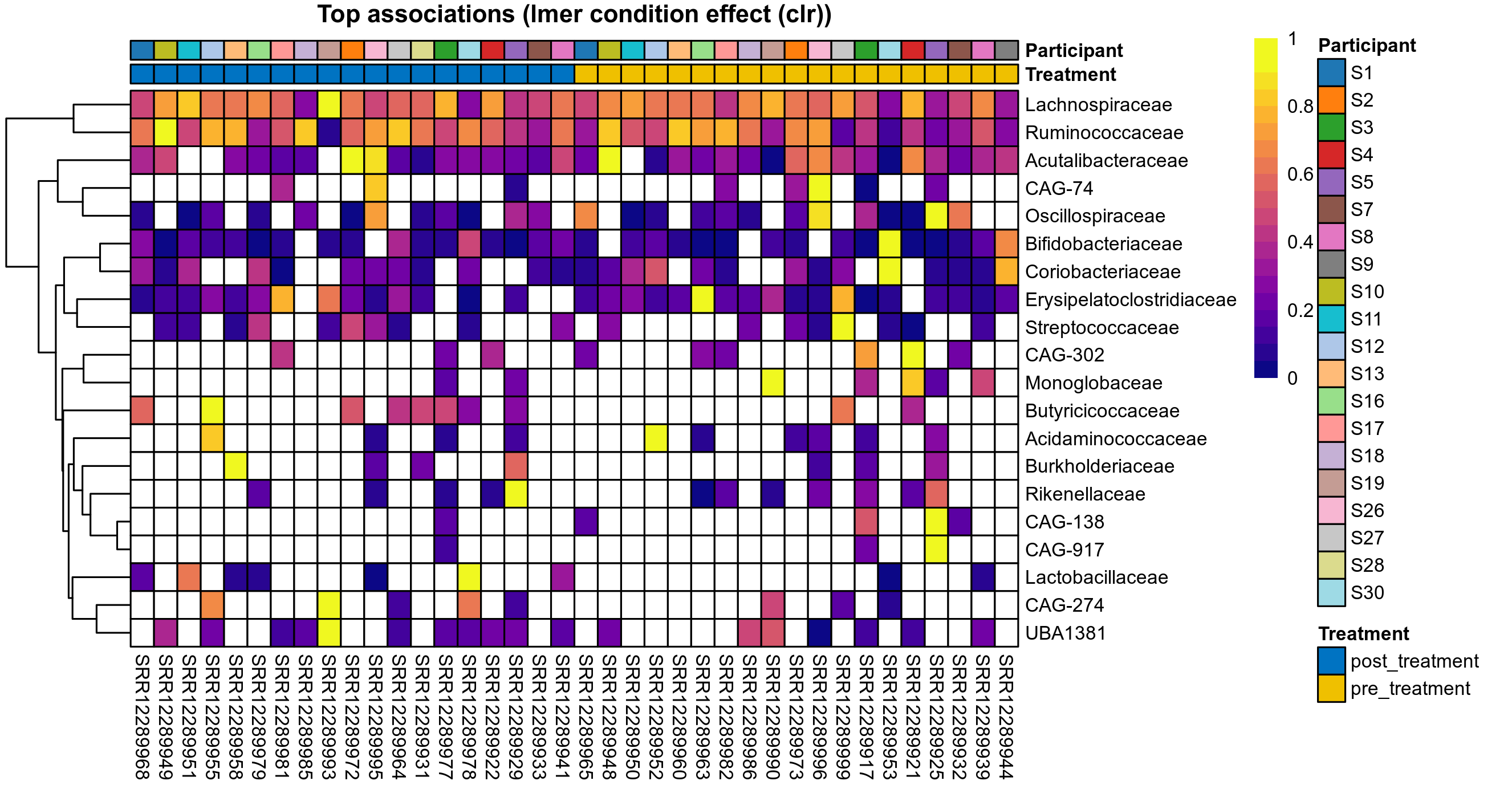
P lmer condition effect (clr): Differentially abundant species were identified by linear mixed effect regression without confounders. Random effects: Participant (intercept). Fixed effects: Treatment. Effect tested: Treatment.
The following plots present the distribution of the top most differentially abundant families across all applied statistical analysis. Plots are ordered alphabetically.