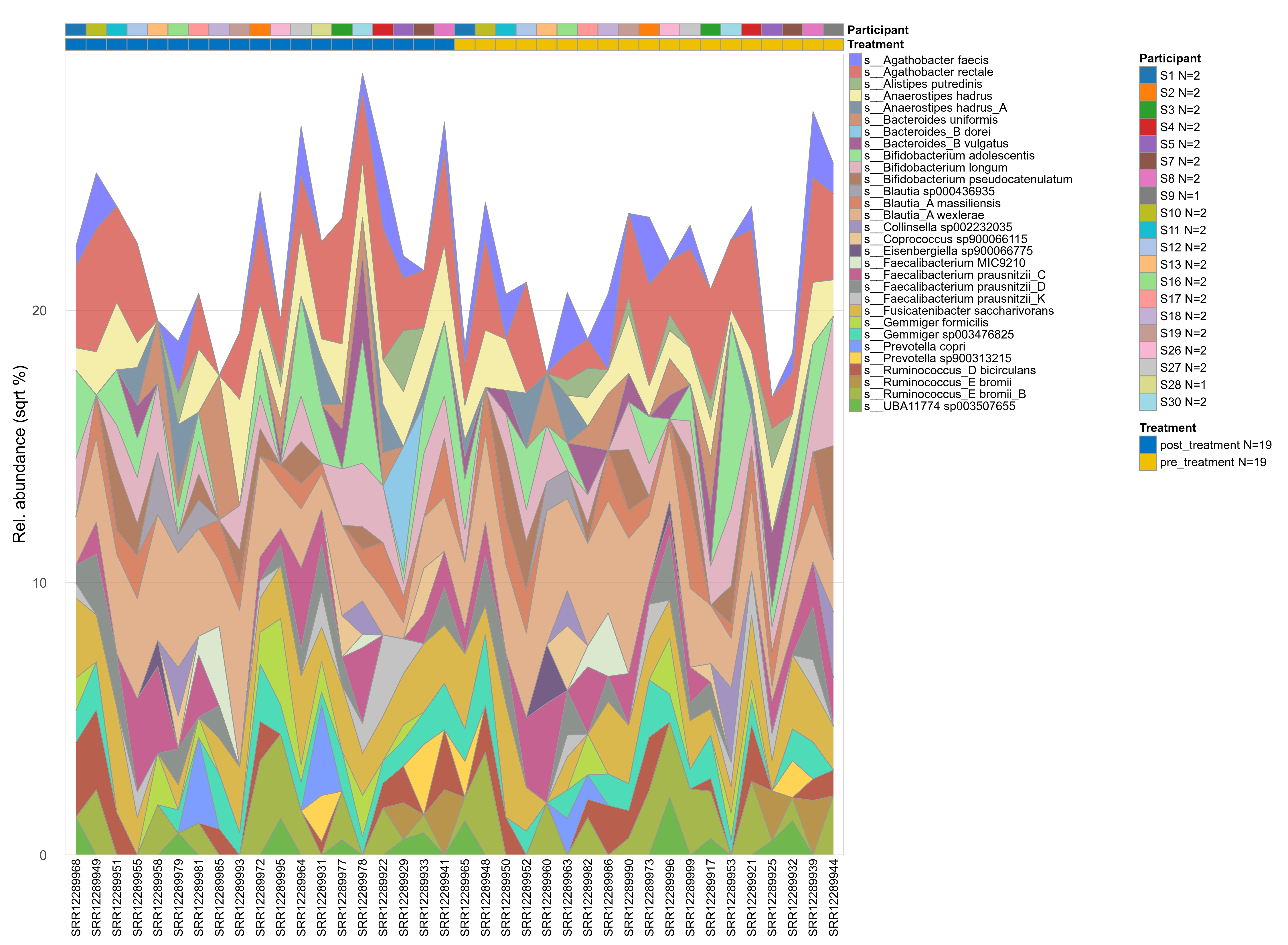
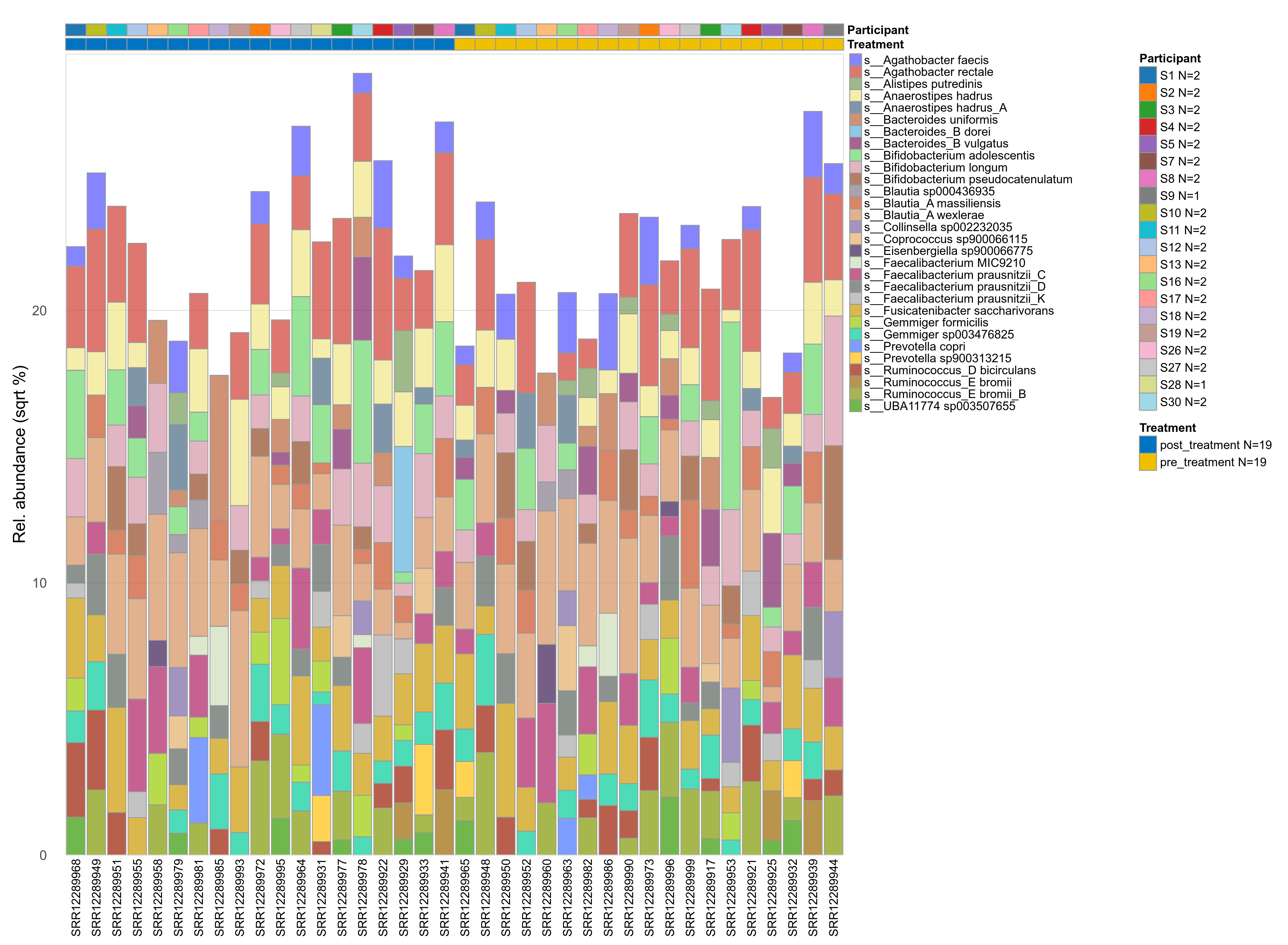
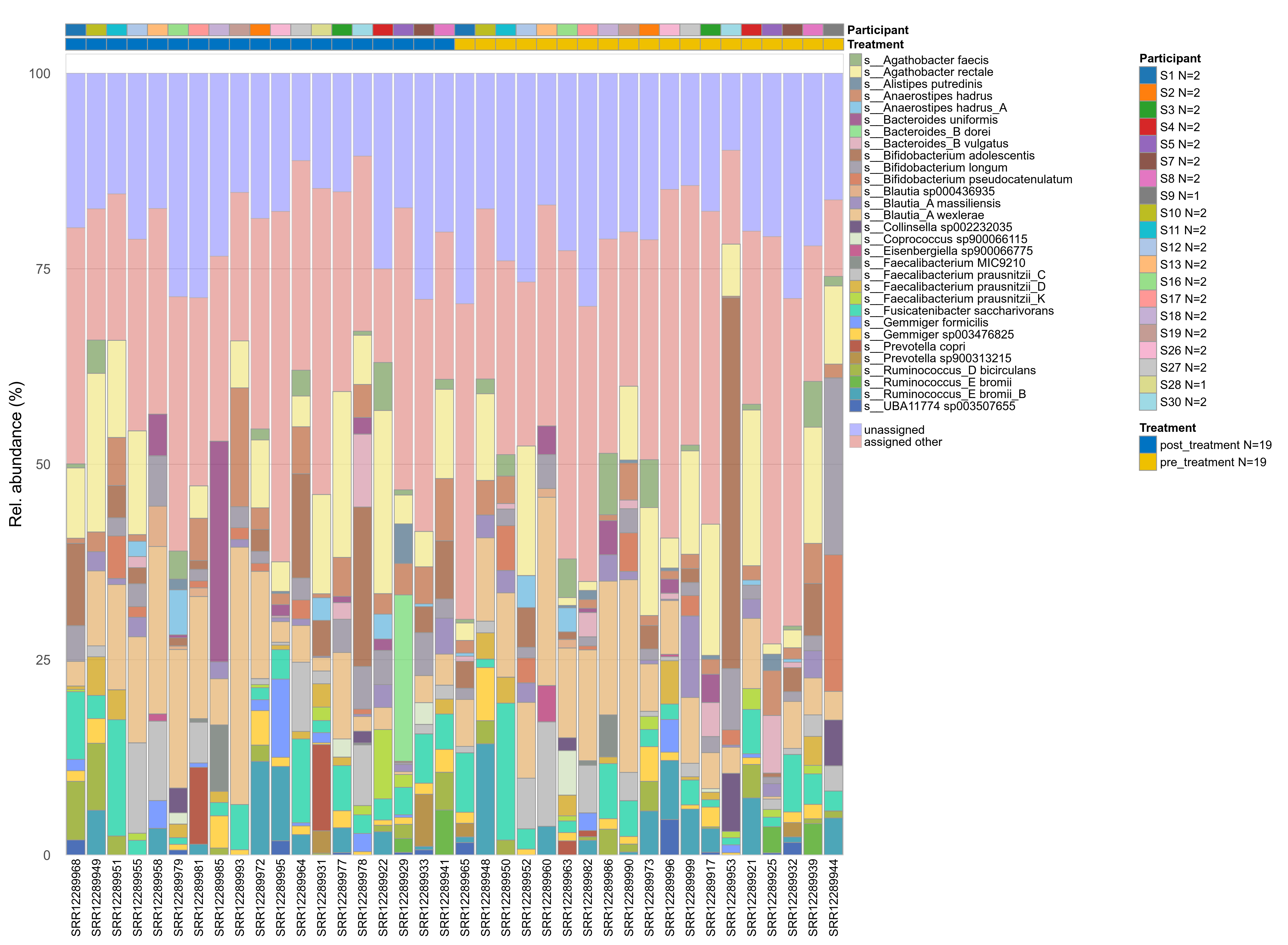
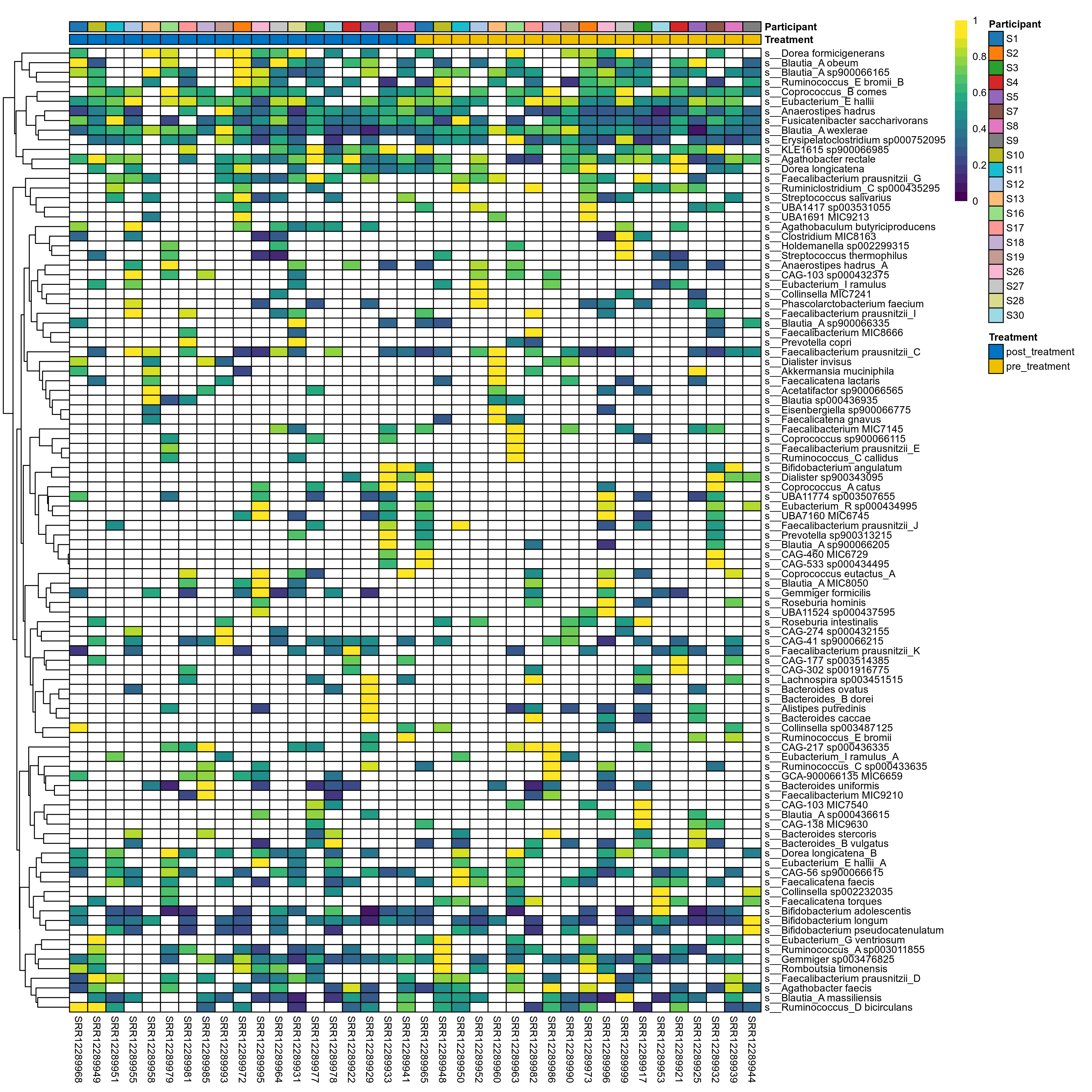
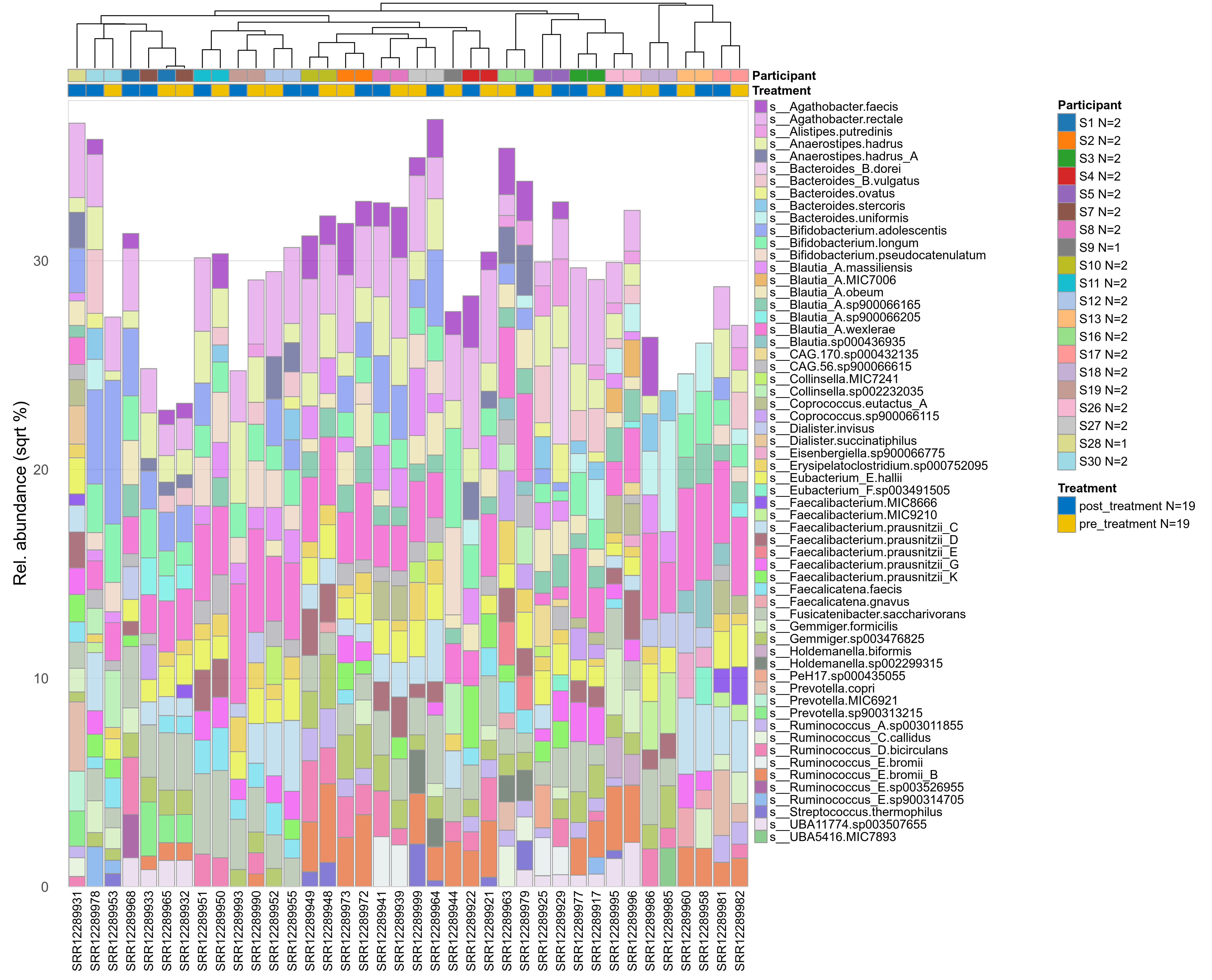
The charts below show the taxonomic composition of the analysed samples using different quantitative visualization techniques. Only the top most abundant species are shown.
Click here to open full-sized image in new window.
Click here to open full-sized image in new window.
Click here to open full-sized image in new window.
Click here to open full-sized image in new window.
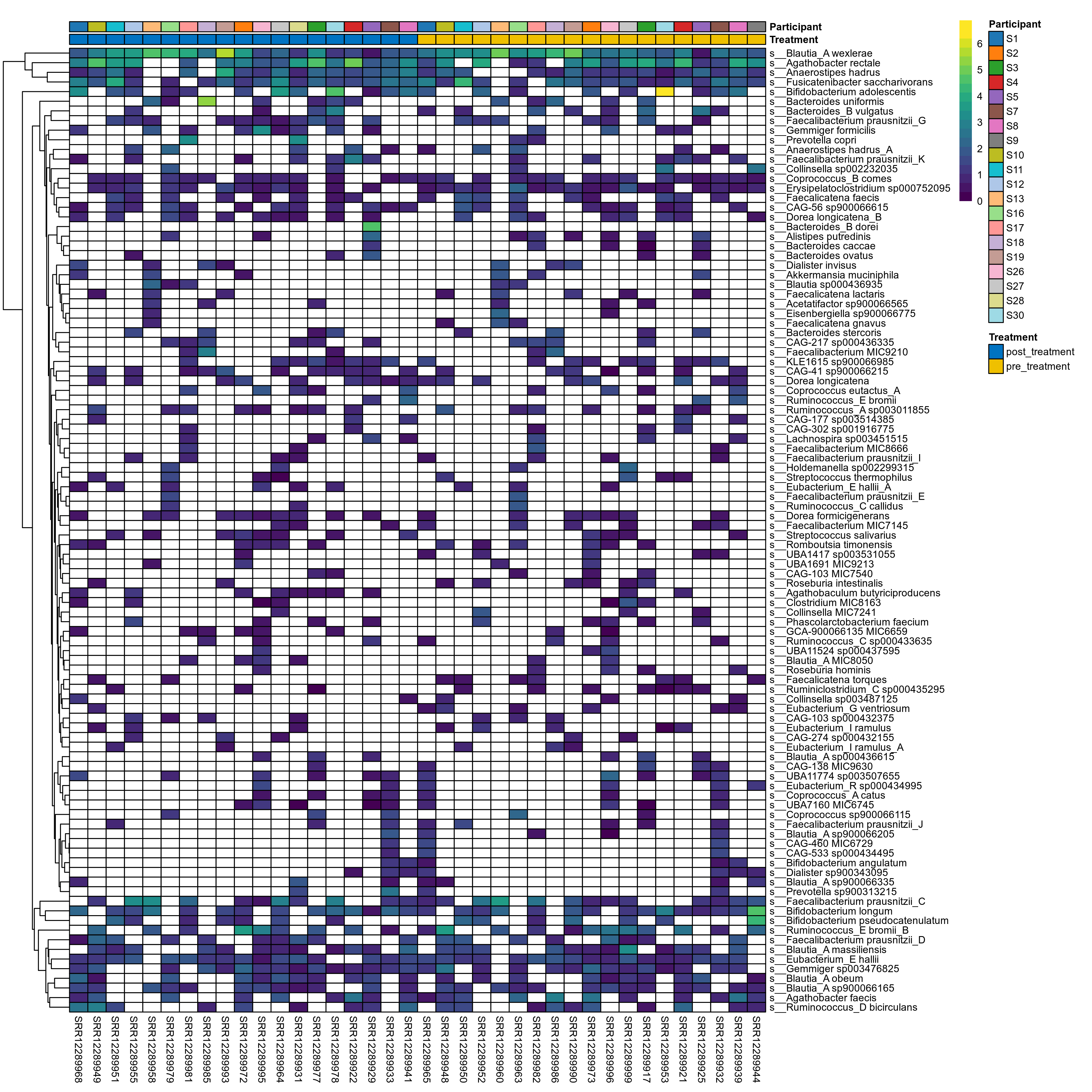
Click here to open full-sized image in new window.
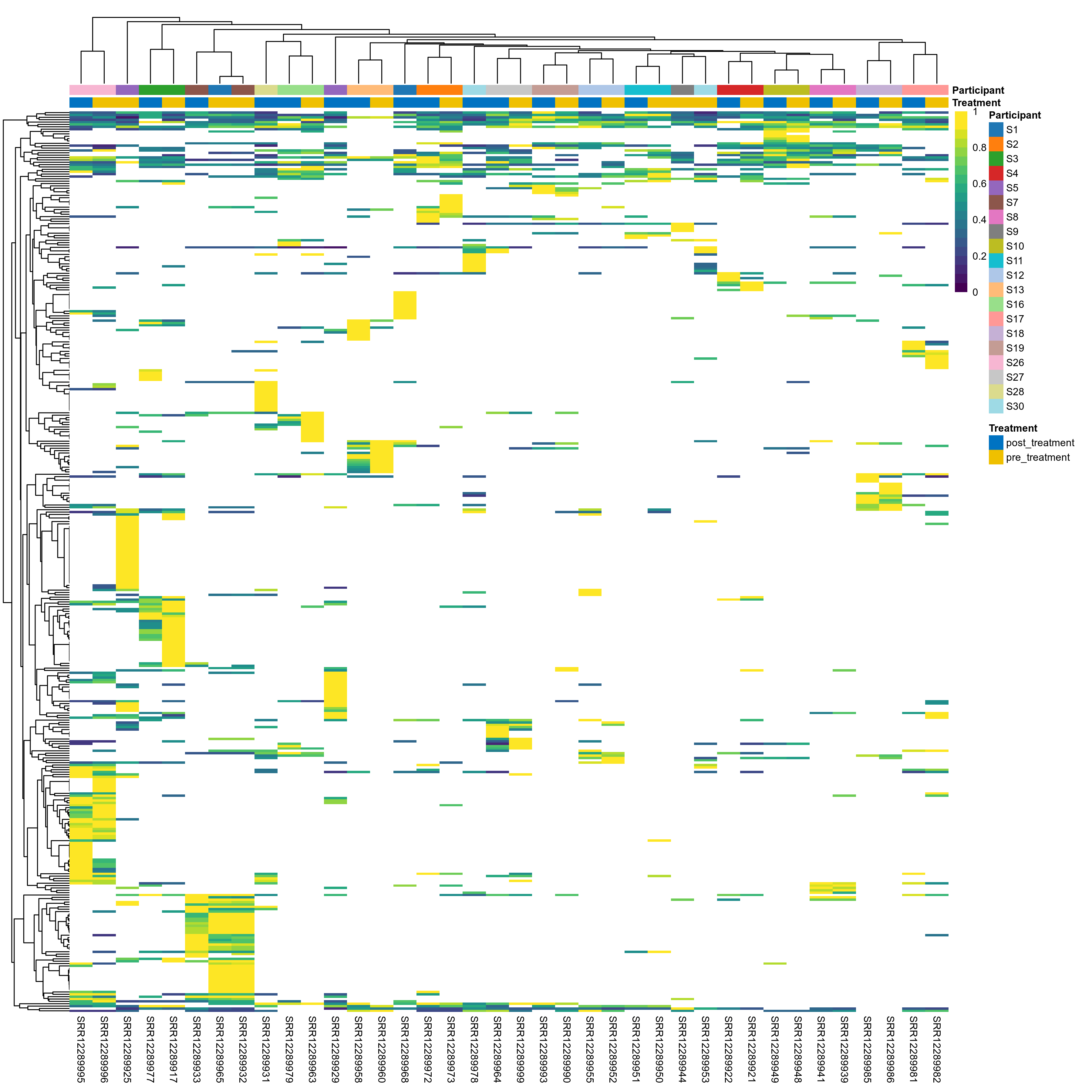
Click here to open full-sized image in new window.
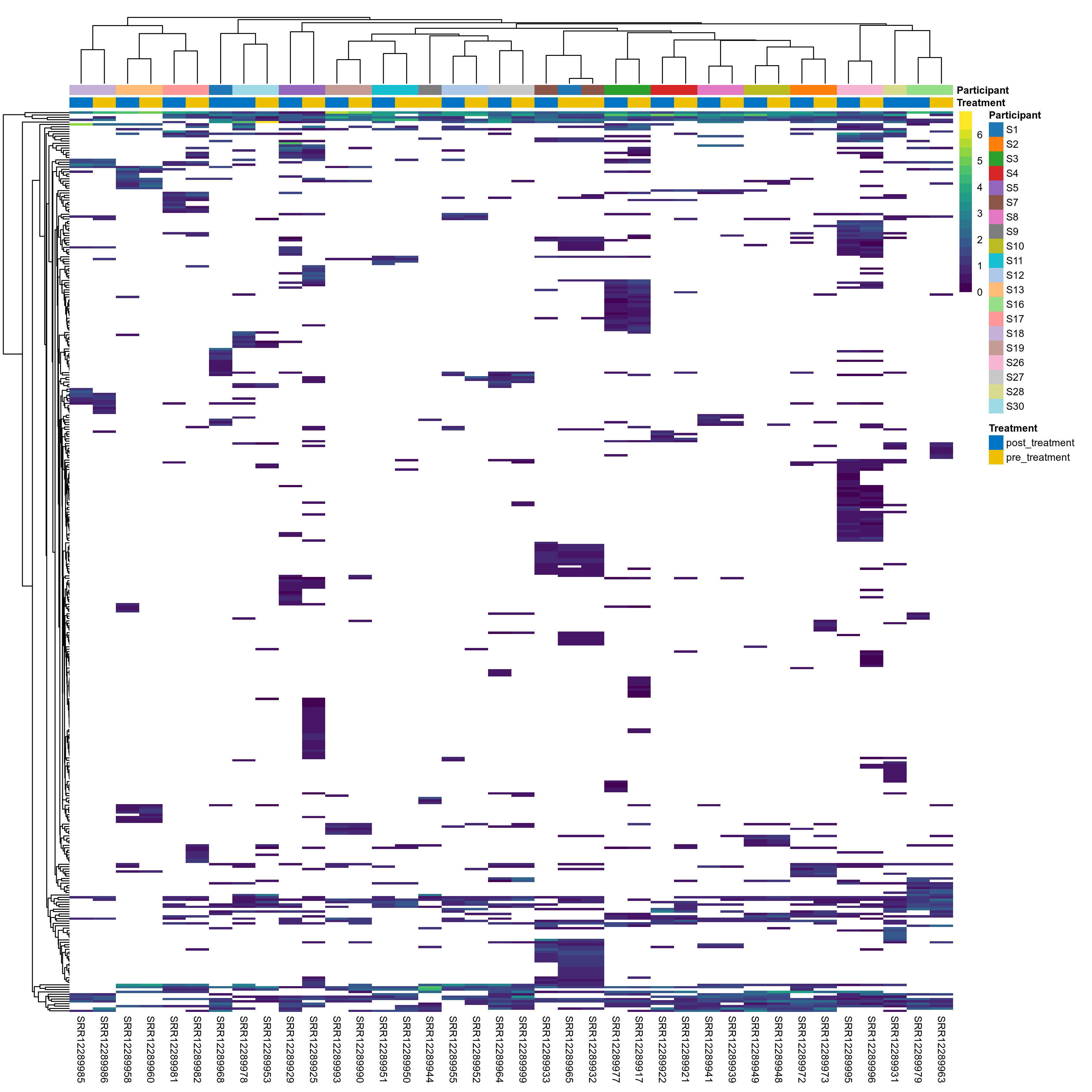
Click here to open full-sized image in new window.
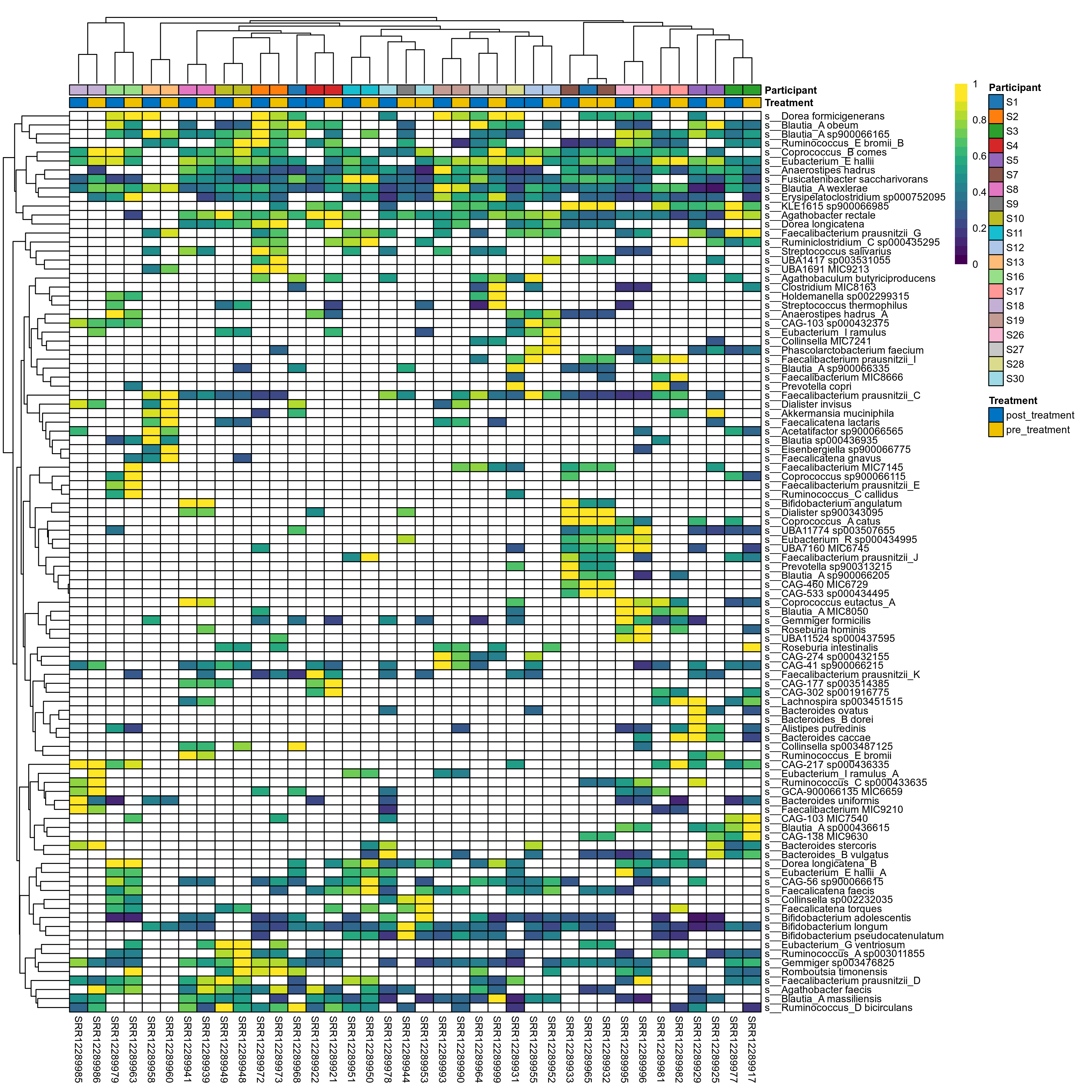
Click here to open full-sized image in new window.
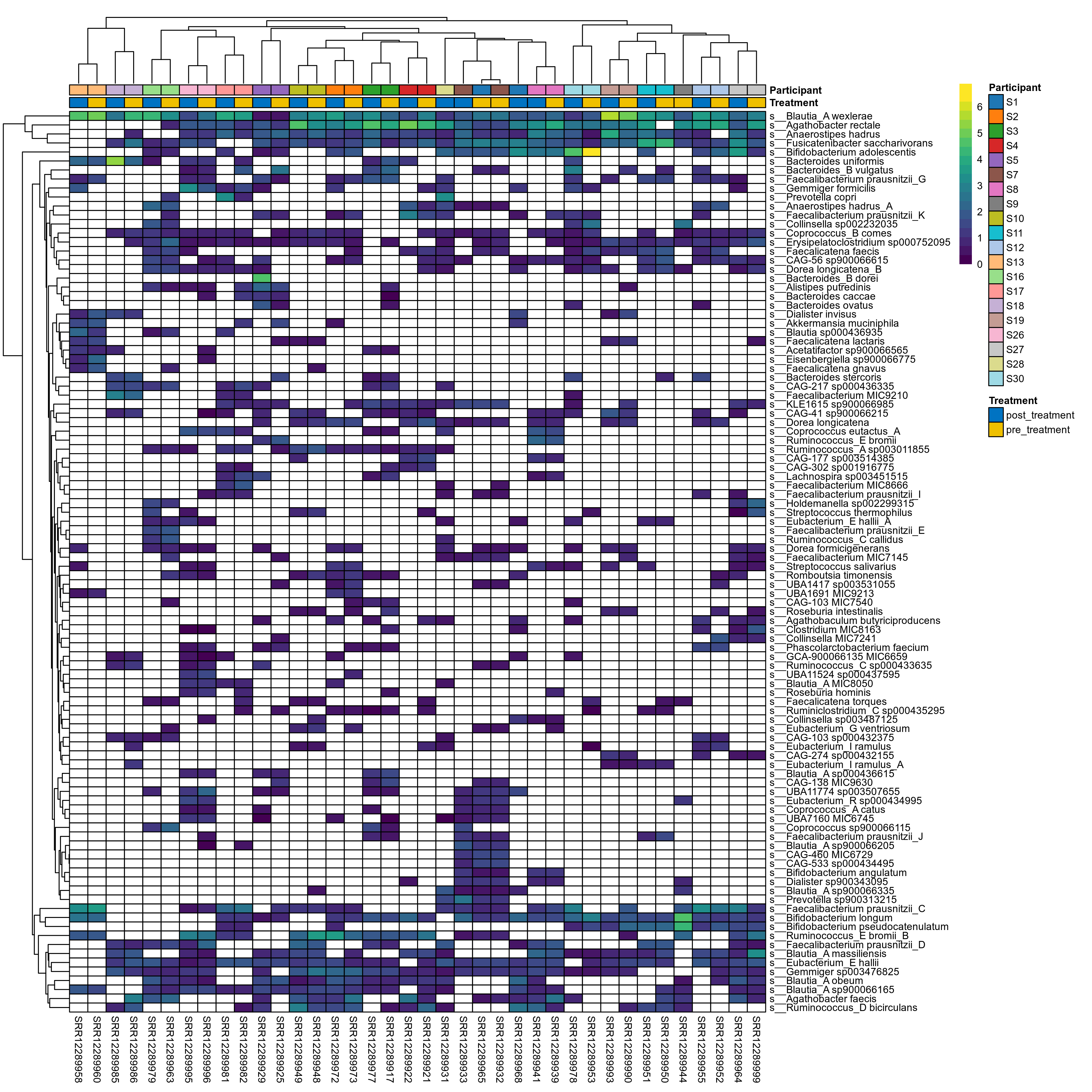
Click here to open full-sized image in new window.
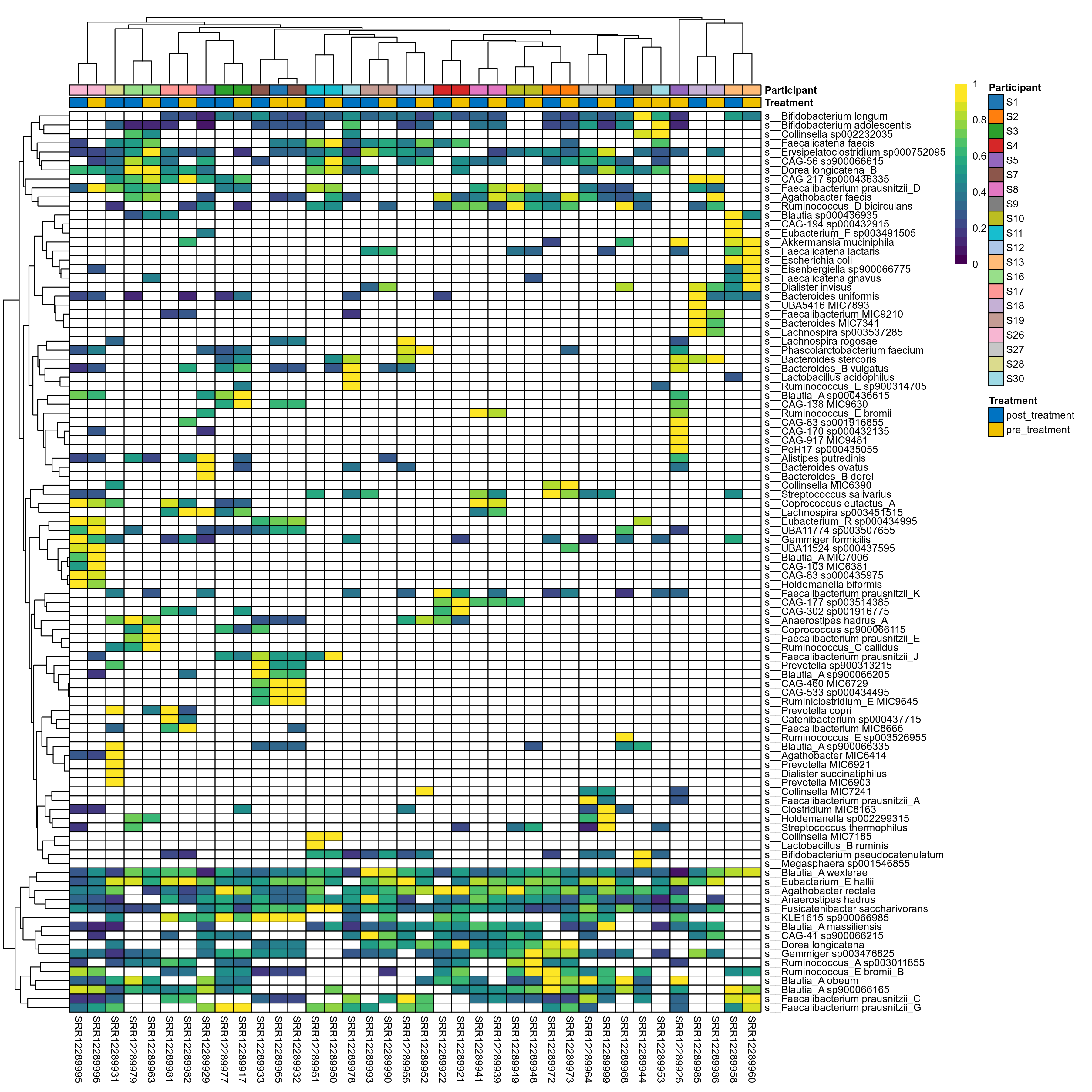
Click here to open full-sized image in new window.
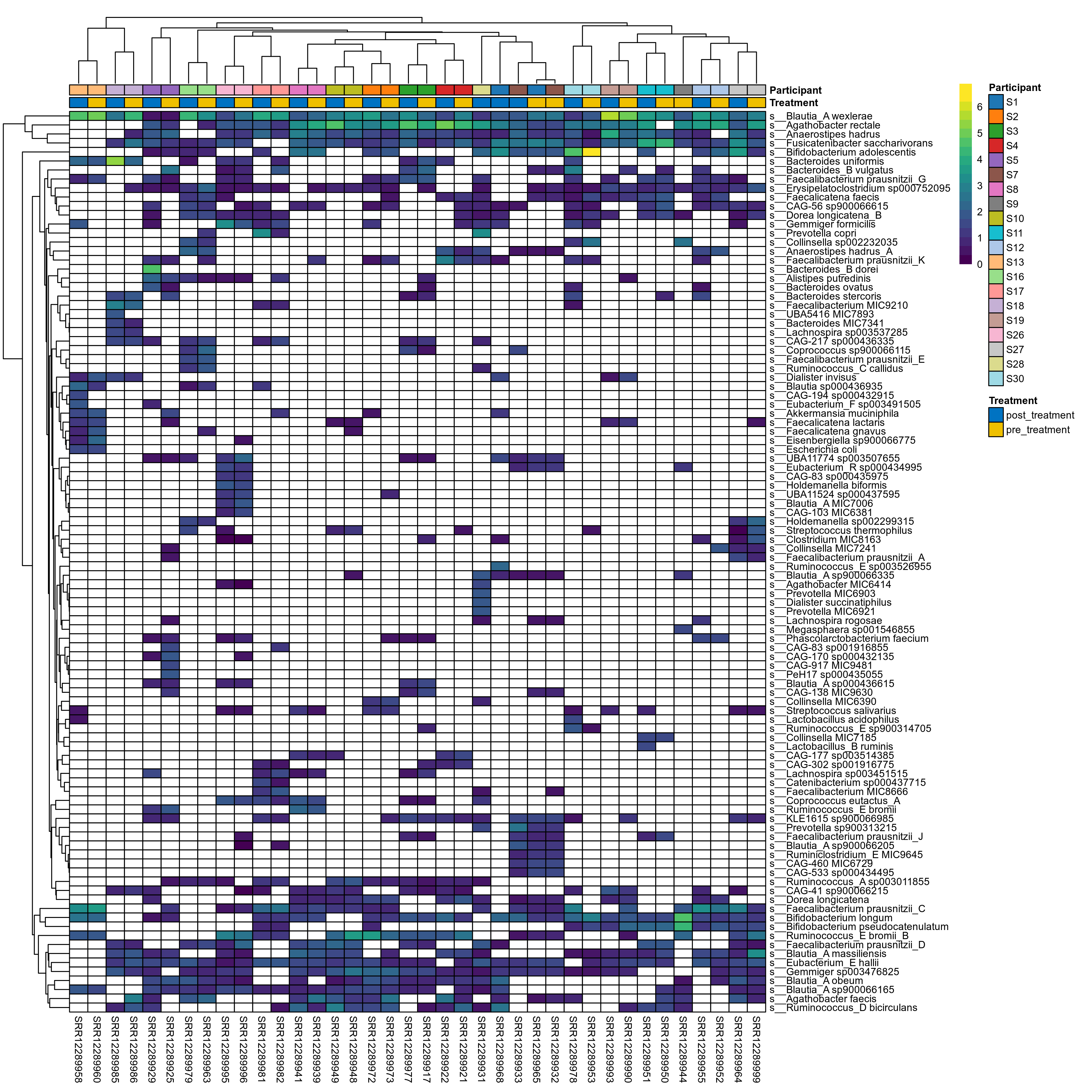
Click here to open full-sized image in new window.
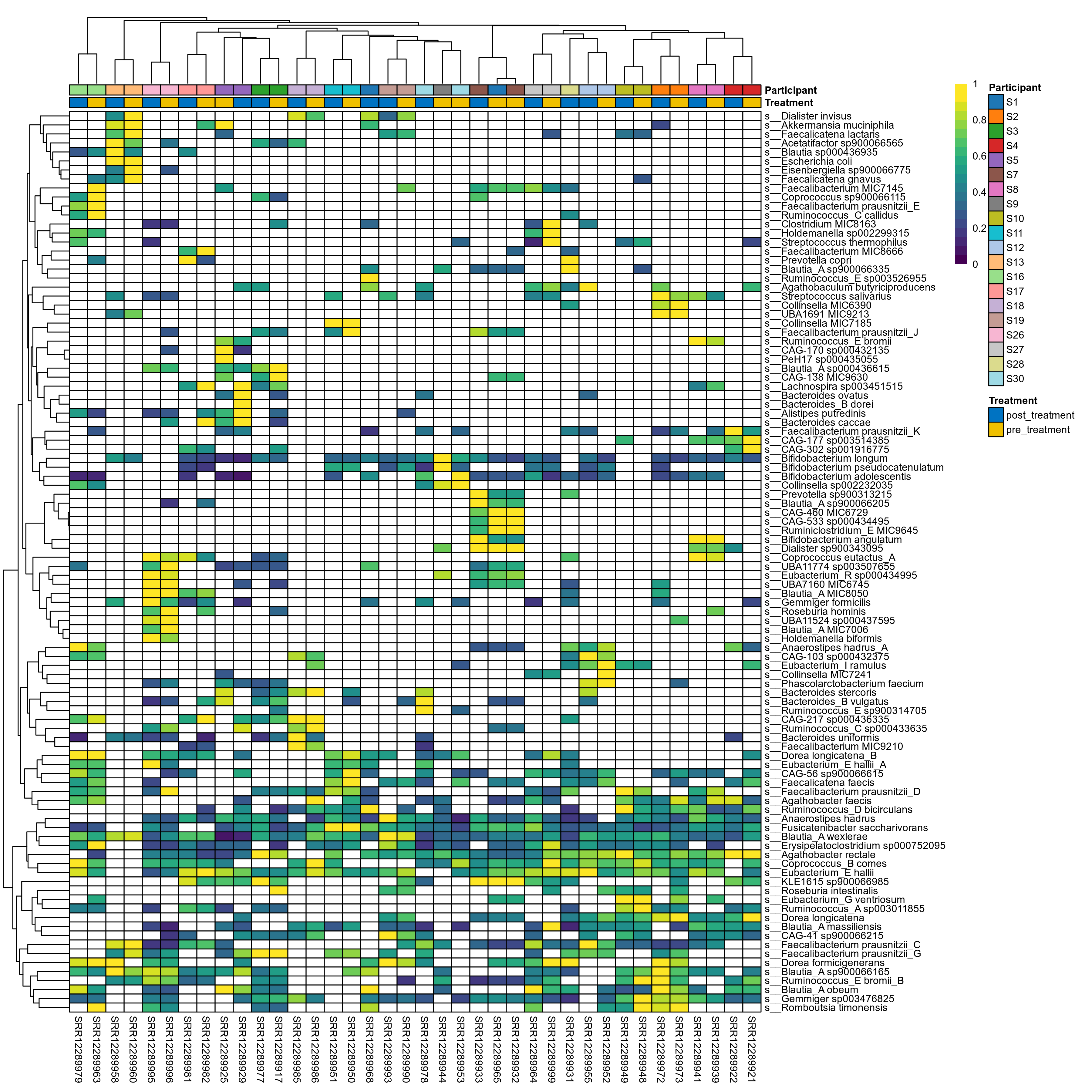
Click here to open full-sized image in new window.
Click here to open full-sized image in new window.
Click here to open interactive barchart in new window.
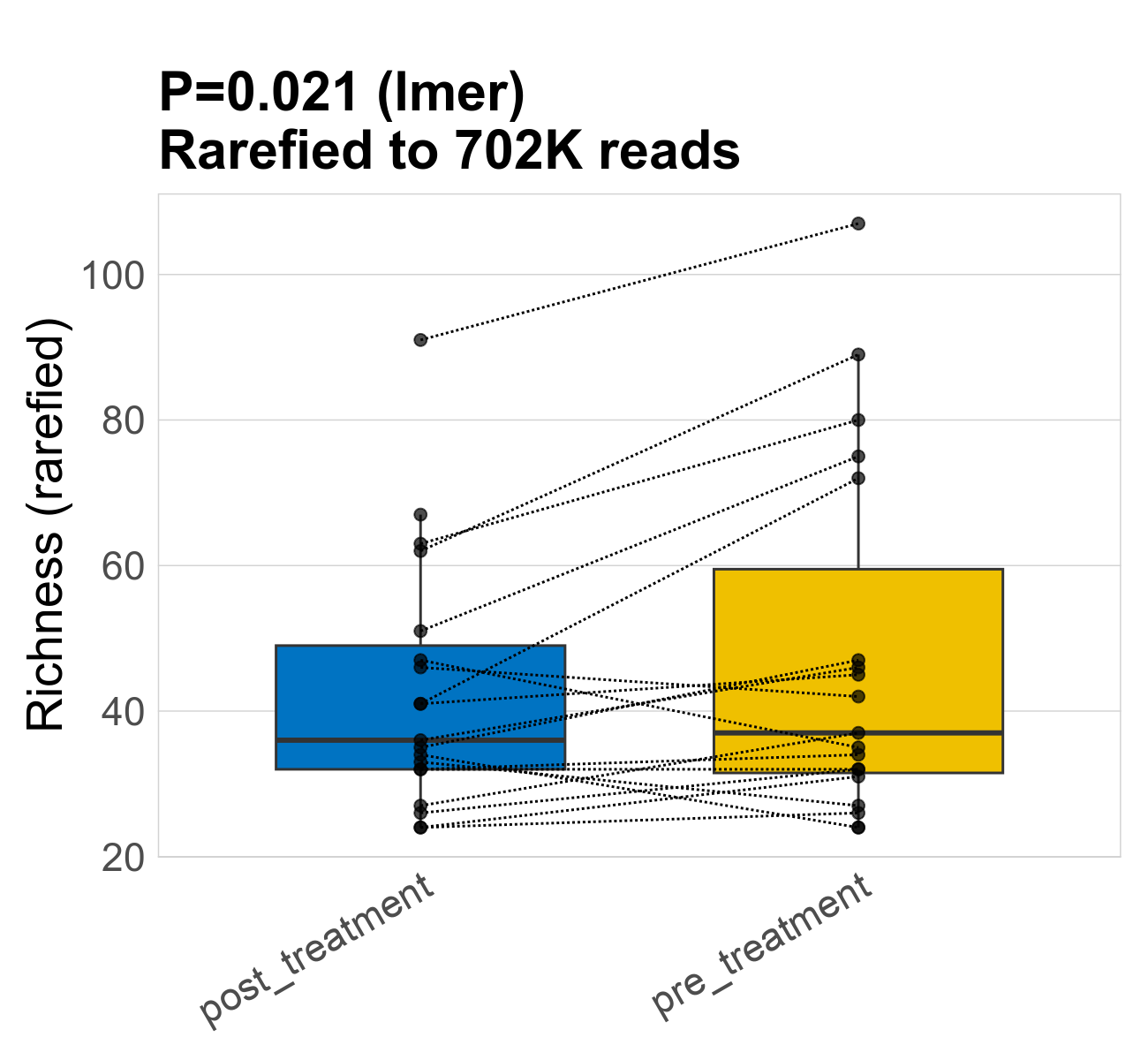
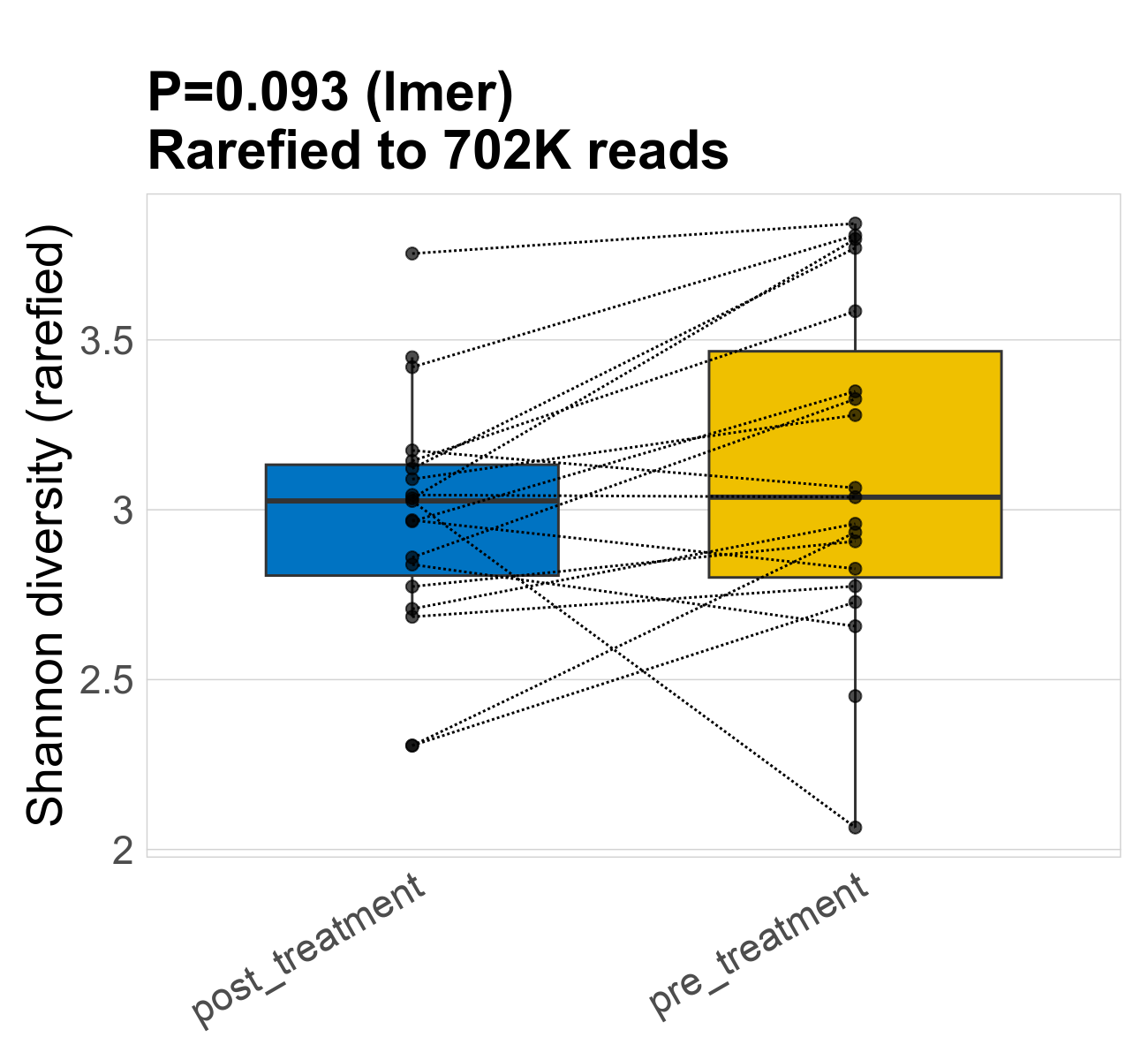
This page provides an overview of the microbial alpha diversity of the analysed samples. Alpha diversity is measured by the Shannon index and species richnes. Richness simply quantifies the total number of species present in each sample. Shannon index additionally accounts for relative abundance and evenness of the species present and quantifies the entropy of microbial communties. Barcharts and boxplots present the mean diversity in each study group.
| Index | rarefiedTo | P lmer condition effect | Mean Pos | Mean Abundance | Median Abundance | Mean Treatmentpost_treatment | Median Treatmentpost_treatment | SD Treatmentpost_treatment | Mean Treatmentpre_treatment | Median Treatmentpre_treatment | SD Treatmentpre_treatment | Fold Change Log2(Treatmentpre_treatment/Treatmentpost_treatment) | Positive samples | Positive Treatmentpost_treatment | Positive Treatmentpre_treatment | Positive_Treatmentpost_treatment_percent | Positive_Treatmentpre_treatment_percent |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Shannon | 702268 | 0.093 | 3 | 3 | 3 | 3 | 3 | 0.35 | 3.1 | 3 | 0.5 | 0.047 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| Richness | 702268 | 0.021 | 45 | 45 | 36 | 43 | 36 | 18 | 48 | 37 | 25 | 0.16 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
Taxonomic profiles were analyzed using supervised and unsupervised multivariate methods. Profiles were ordinated using the unsupervised methods Principal Coordinates Analysis (PCoA), Non-Metric Multidimensional Scaling (NMDS) and Principal Component Analysis (PCA). PCoA and NMDS are related to PCA, but take dissimlarity matrices as input. PCoA and NMDS both attempt to represent the pairwise dissimlarities between samples in low dimensional space as close as possible. NMDS is a rank-based approach and therefore less effected by outliers.
The supervised methods Adonis and Redundancy analysis (RDA) were used to assess if variance in microbial community composition can be attributed to the study condition. NMDS, PCoA and Adonis were run on Bray-Curtis dissimilarities. A short introduction of the used methods can be found at the GUide to STatistical Analysis in Microbial Ecology (GUSTA ME). Sparse Partial Least Square Discriminant Analysis (sPLS-DA) from the MixMc package was additionaly used to extract features associated with the study condition.
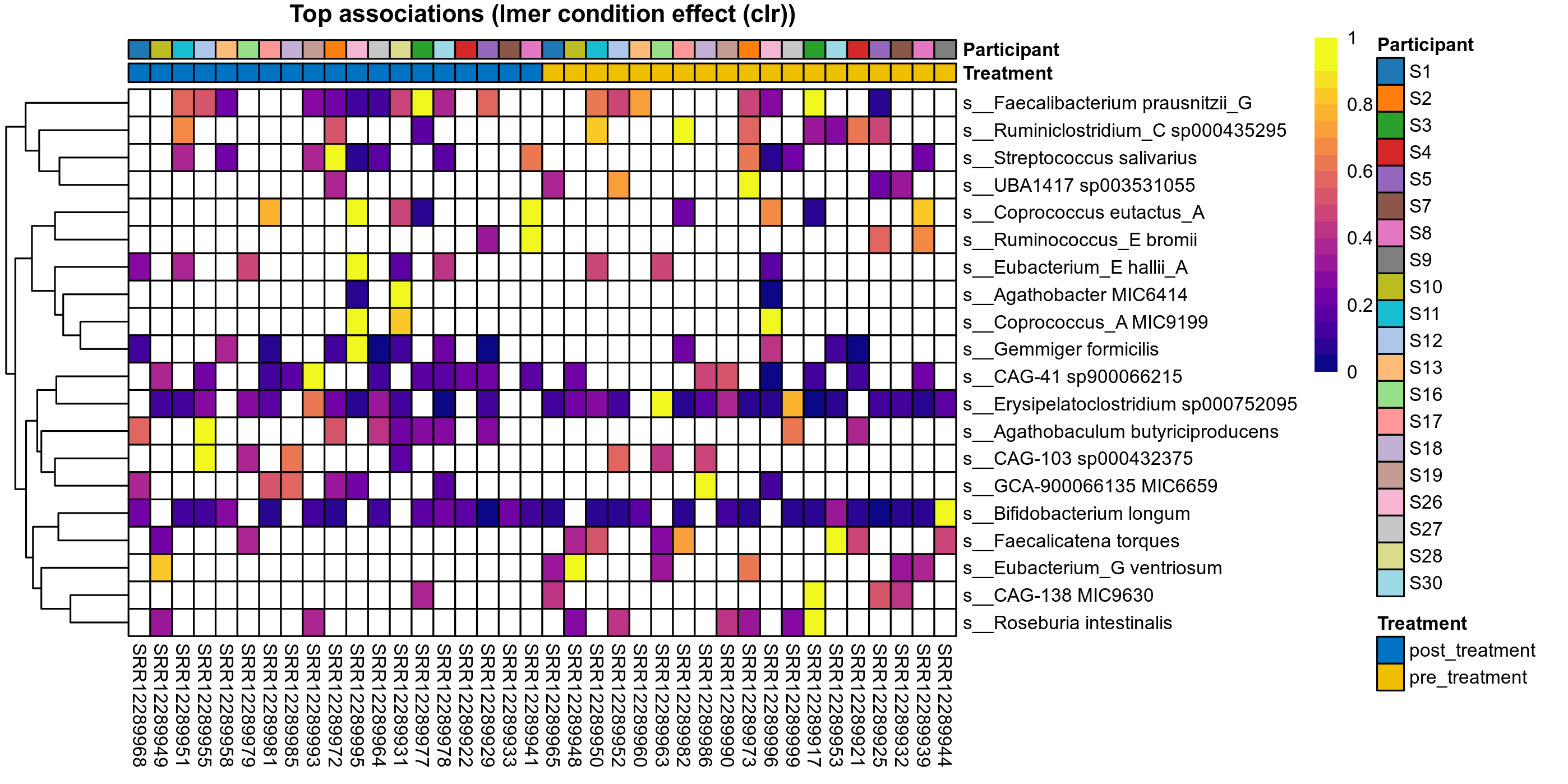
Differentially abundant species were identified by ANOVA or LMER (linear mixed effect regression) of clr transformed relative abudances, Fisher's exact test and/or ALDEx2 (on species read counts). Fisher's exact test is used to test for differences in the detection rate, i.e. number of samples in which each species has been detected.
LMER is used for repeated measures data, using random effects to control for correlation between samples from the same subject. Fixed effects are included for treatment groups, time, and treatment over time, where appropriate. The LMER P values correspond to a nested model test of the significance of including the corresponding fixed effect.
ALDEx2 uses subsampling (Bayesian sampling) to estimate the underlying technical variation. For each subsample instance, center log-ratio transformed data is statistically compared across study groups and computed P values are corrected for multiple testing using the Benjamini–Hochberg procedure. The expected P value (mean P value) is reported, which are those that would likely have been observed if the same samples had been run multiple times. The expected values are reported for both the distribution of P values and for the distribution of Benjamini–Hochberg corrected values.
| Taxon | GTDB taxonomy | P lmer condition effect (sqrt) | FDR lmer condition effect (sqrt) | Pbonf lmer condition effect (sqrt) | P lmer condition effect (clr) | FDR lmer condition effect (clr) | Pbonf lmer condition effect (clr) | Mean Pos | Mean Abundance | Median Abundance | Mean Treatmentpost_treatment | Median Treatmentpost_treatment | SD Treatmentpost_treatment | Mean Treatmentpre_treatment | Median Treatmentpre_treatment | SD Treatmentpre_treatment | Fold Change Log2(Treatmentpre_treatment/Treatmentpost_treatment) | Positive samples | Positive Treatmentpost_treatment | Positive Treatmentpre_treatment | Positive_Treatmentpost_treatment_percent | Positive_Treatmentpre_treatment_percent |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| s__Acetatifactor sp900066365 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Acetatifactor | 0.7 | 0.84 | 1 | 0.76 | 0.86 | 1 | 0.3 | 0.032 | 0 | 0.042 | 0 | 0.13 | 0.021 | 0 | 0.07 | -1 | 4 / 38 (11%) | 2 / 19 (11%) | 2 / 19 (11%) | 0.105 | 0.105 |
| s__Acetatifactor sp900066565 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Acetatifactor | 0.3 | 0.64 | 1 | 0.57 | 0.74 | 1 | 0.69 | 0.11 | 0 | 0.15 | 0 | 0.42 | 0.065 | 0 | 0.2 | -1.2 | 6 / 38 (16%) | 3 / 19 (16%) | 3 / 19 (16%) | 0.158 | 0.158 |
| s__Adlercreutzia equolifaciens | d__Bacteria; p__Actinobacteriota; c__Coriobacteriia; o__Coriobacteriales; f__Eggerthellaceae; g__Adlercreutzia | 0.82 | 0.94 | 1 | 0.8 | 0.88 | 1 | 0.22 | 0.023 | 0 | 0.029 | 0 | 0.099 | 0.017 | 0 | 0.054 | -0.77 | 4 / 38 (11%) | 2 / 19 (11%) | 2 / 19 (11%) | 0.105 | 0.105 |
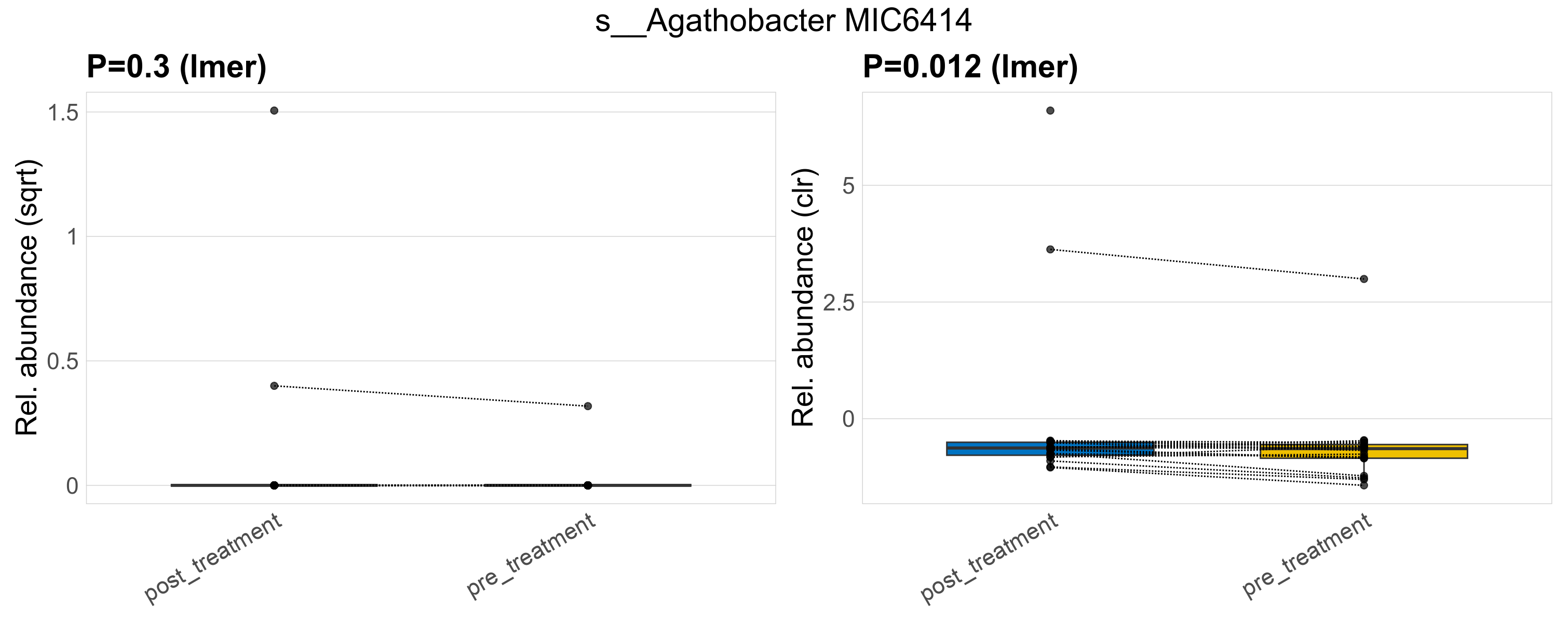
| s__Agathobacter MIC6414 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Agathobacter | 0.3 | 0.64 | 1 | 0.012 | 0.45 | 1 | 0.84 | 0.067 | 0 | 0.13 | 0 | 0.52 | 0.0053 | 0 | 0.023 | -4.6 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
| s__Agathobacter faecis | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Agathobacter | 0.37 | 0.66 | 1 | 0.51 | 0.71 | 1 | 2.7 | 1.4 | 0.49 | 1.1 | 0 | 1.8 | 1.7 | 0.51 | 2.5 | 0.63 | 20 / 38 (53%) | 9 / 19 (47%) | 11 / 19 (58%) | 0.474 | 0.579 |
| s__Agathobacter rectale | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Agathobacter | 0.35 | 0.66 | 1 | 0.56 | 0.73 | 1 | 9.7 | 8.1 | 6.5 | 8.7 | 6.3 | 7.1 | 7.6 | 6.7 | 6.8 | -0.2 | 32 / 38 (84%) | 16 / 19 (84%) | 16 / 19 (84%) | 0.842 | 0.842 |
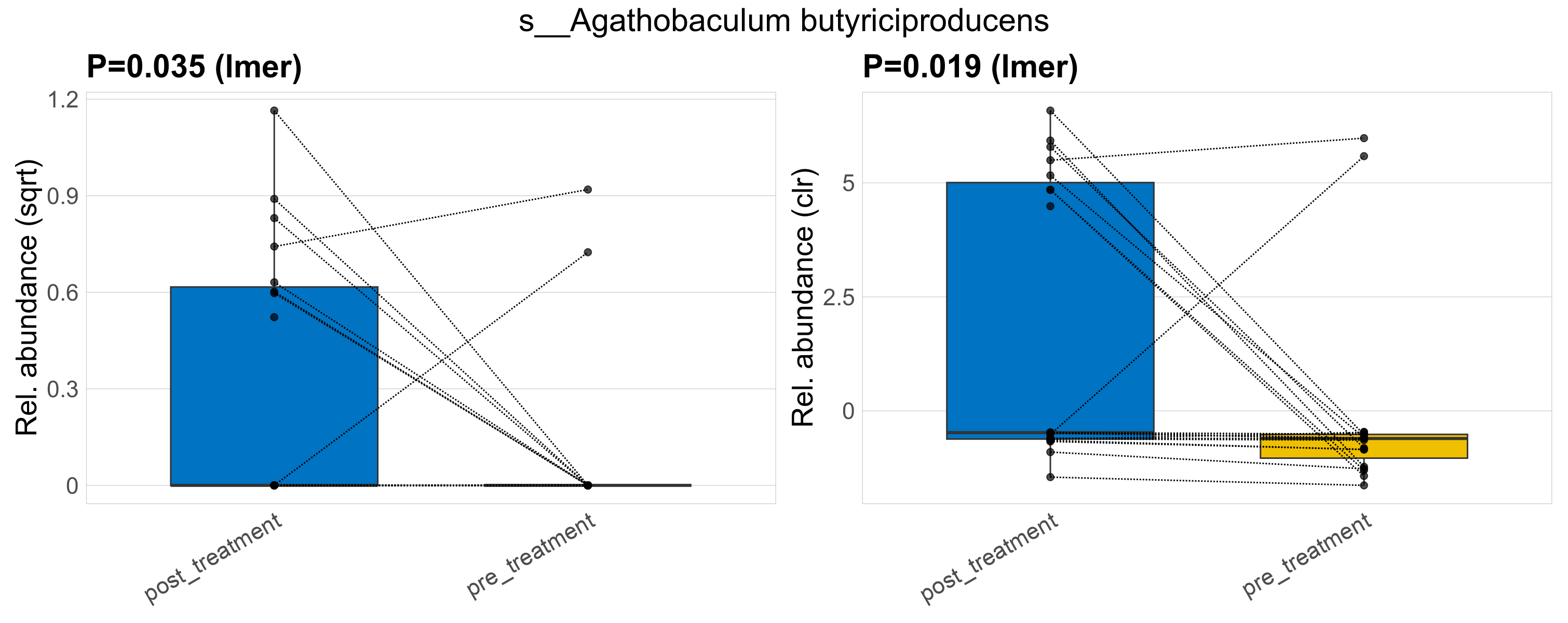
| s__Agathobaculum butyriciproducens | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Butyricicoccaceae; g__Agathobaculum | 0.035 | 0.52 | 1 | 0.019 | 0.45 | 1 | 0.61 | 0.16 | 0 | 0.25 | 0 | 0.38 | 0.072 | 0 | 0.22 | -1.8 | 10 / 38 (26%) | 8 / 19 (42%) | 2 / 19 (11%) | 0.421 | 0.105 |
| s__Akkermansia muciniphila | d__Bacteria; p__Verrucomicrobiota; c__Verrucomicrobiae; o__Verrucomicrobiales; f__Akkermansiaceae; g__Akkermansia | 0.54 | 0.77 | 1 | 0.92 | 0.95 | 1 | 1.8 | 0.28 | 0 | 0.2 | 0 | 0.59 | 0.37 | 0 | 0.95 | 0.89 | 6 / 38 (16%) | 3 / 19 (16%) | 3 / 19 (16%) | 0.158 | 0.158 |
| s__Alistipes finegoldii | d__Bacteria; p__Bacteroidota; c__Bacteroidia; o__Bacteroidales; f__Rikenellaceae; g__Alistipes | 0.077 | 0.59 | 1 | 0.17 | 0.6 | 1 | 0.61 | 0.064 | 0 | 0.034 | 0 | 0.15 | 0.094 | 0 | 0.3 | 1.5 | 4 / 38 (11%) | 1 / 19 (5.3%) | 3 / 19 (16%) | 0.0526 | 0.158 |
| s__Alistipes onderdonkii | d__Bacteria; p__Bacteroidota; c__Bacteroidia; o__Bacteroidales; f__Rikenellaceae; g__Alistipes | 0.45 | 0.7 | 1 | 0.29 | 0.62 | 1 | 0.94 | 0.074 | 0 | 0.11 | 0 | 0.4 | 0.04 | 0 | 0.17 | -1.5 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
| s__Alistipes putredinis | d__Bacteria; p__Bacteroidota; c__Bacteroidia; o__Bacteroidales; f__Rikenellaceae; g__Alistipes | 0.55 | 0.77 | 1 | 0.15 | 0.6 | 1 | 1.3 | 0.3 | 0 | 0.35 | 0 | 1.2 | 0.25 | 0 | 0.53 | -0.49 | 9 / 38 (24%) | 3 / 19 (16%) | 6 / 19 (32%) | 0.158 | 0.316 |
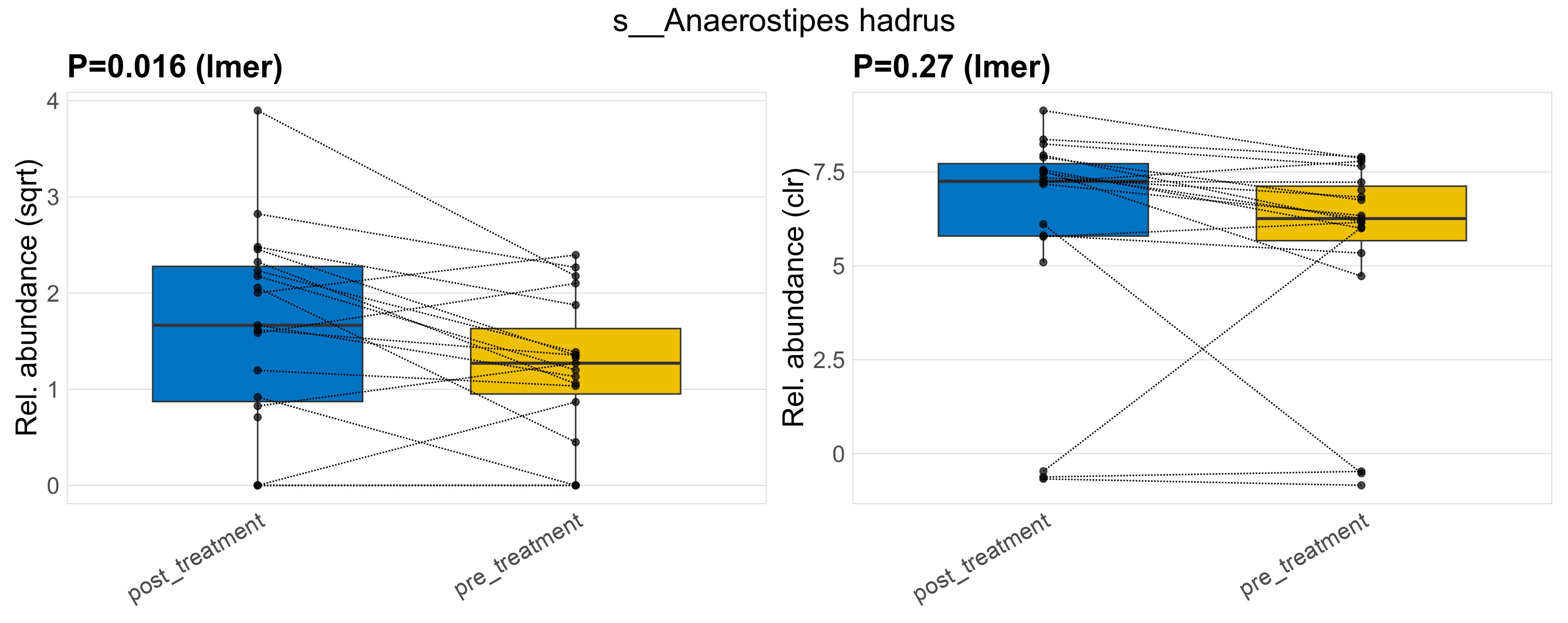
| s__Anaerostipes hadrus | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Anaerostipes | 0.016 | 0.52 | 1 | 0.27 | 0.62 | 1 | 3.4 | 2.9 | 1.8 | 3.7 | 2.8 | 3.7 | 2 | 1.6 | 1.8 | -0.89 | 32 / 38 (84%) | 16 / 19 (84%) | 16 / 19 (84%) | 0.842 | 0.842 |
| s__Anaerostipes hadrus_A | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Anaerostipes | 0.72 | 0.86 | 1 | 0.74 | 0.85 | 1 | 2.3 | 0.6 | 0 | 0.75 | 0 | 1.6 | 0.46 | 0 | 1.1 | -0.71 | 10 / 38 (26%) | 5 / 19 (26%) | 5 / 19 (26%) | 0.263 | 0.263 |
| s__Bacteroides caccae | d__Bacteria; p__Bacteroidota; c__Bacteroidia; o__Bacteroidales; f__Bacteroidaceae; g__Bacteroides | 0.26 | 0.62 | 1 | 0.13 | 0.6 | 1 | 0.88 | 0.12 | 0 | 0.092 | 0 | 0.4 | 0.14 | 0 | 0.36 | 0.61 | 5 / 38 (13%) | 1 / 19 (5.3%) | 4 / 19 (21%) | 0.0526 | 0.211 |
| s__Bacteroides ovatus | d__Bacteria; p__Bacteroidota; c__Bacteroidia; o__Bacteroidales; f__Bacteroidaceae; g__Bacteroides | 0.19 | 0.62 | 1 | 0.3 | 0.62 | 1 | 0.99 | 0.13 | 0 | 0.22 | 0 | 0.78 | 0.036 | 0 | 0.11 | -2.6 | 5 / 38 (13%) | 3 / 19 (16%) | 2 / 19 (11%) | 0.158 | 0.105 |
| s__Bacteroides stercoris | d__Bacteria; p__Bacteroidota; c__Bacteroidia; o__Bacteroidales; f__Bacteroidaceae; g__Bacteroides | 0.99 | 0.99 | 1 | 0.91 | 0.95 | 1 | 1.7 | 0.36 | 0 | 0.36 | 0 | 0.8 | 0.36 | 0 | 0.89 | 0 | 8 / 38 (21%) | 4 / 19 (21%) | 4 / 19 (21%) | 0.211 | 0.211 |
| s__Bacteroides uniformis | d__Bacteria; p__Bacteroidota; c__Bacteroidia; o__Bacteroidales; f__Bacteroidaceae; g__Bacteroides | 0.2 | 0.62 | 1 | 0.21 | 0.6 | 1 | 4.5 | 1.4 | 0 | 2.1 | 0 | 6.5 | 0.74 | 0 | 1.5 | -1.5 | 12 / 38 (32%) | 7 / 19 (37%) | 5 / 19 (26%) | 0.368 | 0.263 |
| s__Bacteroides_B massiliensis | d__Bacteria; p__Bacteroidota; c__Bacteroidia; o__Bacteroidales; f__Bacteroidaceae; g__Bacteroides_B | 0.34 | 0.66 | 1 | 0.21 | 0.6 | 1 | 0.44 | 0.059 | 0 | 0.044 | 0 | 0.19 | 0.073 | 0 | 0.18 | 0.73 | 5 / 38 (13%) | 1 / 19 (5.3%) | 4 / 19 (21%) | 0.0526 | 0.211 |
| s__Bacteroides_B vulgatus | d__Bacteria; p__Bacteroidota; c__Bacteroidia; o__Bacteroidales; f__Bacteroidaceae; g__Bacteroides_B | 0.32 | 0.66 | 1 | 0.17 | 0.6 | 1 | 2.7 | 0.84 | 0 | 0.69 | 0 | 2.2 | 0.99 | 0 | 1.9 | 0.52 | 12 / 38 (32%) | 4 / 19 (21%) | 8 / 19 (42%) | 0.211 | 0.421 |
| s__Barnesiella intestinihominis | d__Bacteria; p__Bacteroidota; c__Bacteroidia; o__Bacteroidales; f__Barnesiellaceae; g__Barnesiella | 0.25 | 0.62 | 1 | 0.4 | 0.68 | 1 | 0.31 | 0.025 | 0 | 0.0066 | 0 | 0.029 | 0.043 | 0 | 0.14 | 2.7 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| s__Bifidobacterium MIC6680 | d__Bacteria; p__Actinobacteriota; c__Actinobacteria; o__Actinomycetales; f__Bifidobacteriaceae; g__Bifidobacterium | 0.61 | 0.8 | 1 | 0.28 | 0.62 | 1 | 1.2 | 0.095 | 0 | 0.11 | 0 | 0.37 | 0.082 | 0 | 0.36 | -0.42 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
| s__Bifidobacterium MIC7686 | d__Bacteria; p__Actinobacteriota; c__Actinobacteria; o__Actinomycetales; f__Bifidobacteriaceae; g__Bifidobacterium | 0.62 | 0.81 | 1 | 0.28 | 0.62 | 1 | 0.64 | 0.05 | 0 | 0.056 | 0 | 0.19 | 0.045 | 0 | 0.19 | -0.32 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
| s__Bifidobacterium adolescentis | d__Bacteria; p__Actinobacteriota; c__Actinobacteria; o__Actinomycetales; f__Bifidobacteriaceae; g__Bifidobacterium | 0.36 | 0.66 | 1 | 0.081 | 0.6 | 1 | 6.8 | 3.8 | 0.74 | 3.7 | 1.1 | 5.6 | 3.8 | 0 | 11 | 0.038 | 21 / 38 (55%) | 12 / 19 (63%) | 9 / 19 (47%) | 0.632 | 0.474 |
| s__Bifidobacterium angulatum | d__Bacteria; p__Actinobacteriota; c__Actinobacteria; o__Actinomycetales; f__Bifidobacteriaceae; g__Bifidobacterium | 0.92 | 0.97 | 1 | 0.63 | 0.79 | 1 | 1.1 | 0.15 | 0 | 0.16 | 0 | 0.48 | 0.13 | 0 | 0.4 | -0.3 | 5 / 38 (13%) | 2 / 19 (11%) | 3 / 19 (16%) | 0.105 | 0.158 |
| s__Bifidobacterium bifidum | d__Bacteria; p__Actinobacteriota; c__Actinobacteria; o__Actinomycetales; f__Bifidobacteriaceae; g__Bifidobacterium | 0.59 | 0.78 | 1 | 0.48 | 0.71 | 1 | 0.43 | 0.056 | 0 | 0.063 | 0 | 0.15 | 0.049 | 0 | 0.16 | -0.36 | 5 / 38 (13%) | 3 / 19 (16%) | 2 / 19 (11%) | 0.158 | 0.105 |
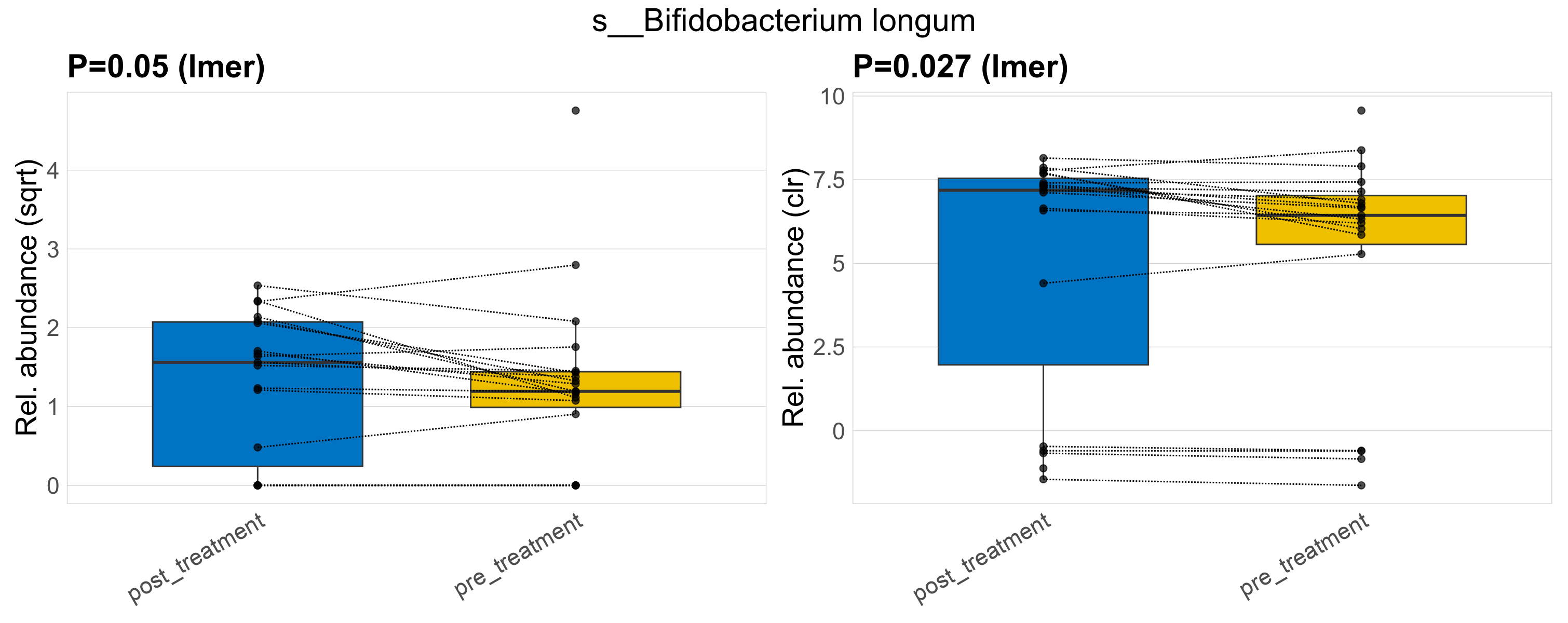
| s__Bifidobacterium longum | d__Bacteria; p__Actinobacteriota; c__Actinobacteria; o__Actinomycetales; f__Bifidobacteriaceae; g__Bifidobacterium | 0.05 | 0.52 | 1 | 0.027 | 0.45 | 1 | 3.5 | 2.7 | 1.7 | 2.5 | 2.4 | 2.1 | 2.9 | 1.4 | 5.1 | 0.21 | 29 / 38 (76%) | 14 / 19 (74%) | 15 / 19 (79%) | 0.737 | 0.789 |
| s__Bifidobacterium pseudocatenulatum | d__Bacteria; p__Actinobacteriota; c__Actinobacteria; o__Actinomycetales; f__Bifidobacteriaceae; g__Bifidobacterium | 0.43 | 0.69 | 1 | 0.47 | 0.71 | 1 | 3.6 | 1.3 | 0 | 0.7 | 0 | 1.3 | 1.9 | 0 | 4.2 | 1.4 | 14 / 38 (37%) | 7 / 19 (37%) | 7 / 19 (37%) | 0.368 | 0.368 |
| s__Blautia sp000436935 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Blautia | 0.25 | 0.62 | 1 | 0.19 | 0.6 | 1 | 1.8 | 0.24 | 0 | 0.35 | 0 | 1.2 | 0.12 | 0 | 0.35 | -1.5 | 5 / 38 (13%) | 3 / 19 (16%) | 2 / 19 (11%) | 0.158 | 0.105 |
| s__Blautia_A MIC7077 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Blautia_A | 0.45 | 0.7 | 1 | 0.66 | 0.81 | 1 | 0.28 | 0.029 | 0 | 0.029 | 0 | 0.086 | 0.03 | 0 | 0.09 | 0.049 | 4 / 38 (11%) | 2 / 19 (11%) | 2 / 19 (11%) | 0.105 | 0.105 |
| s__Blautia_A MIC8050 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Blautia_A | 0.42 | 0.69 | 1 | 0.16 | 0.6 | 1 | 0.91 | 0.14 | 0 | 0.15 | 0 | 0.36 | 0.13 | 0 | 0.42 | -0.21 | 6 / 38 (16%) | 4 / 19 (21%) | 2 / 19 (11%) | 0.211 | 0.105 |
| s__Blautia_A MIC8343 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Blautia_A | 0.18 | 0.62 | 1 | 0.12 | 0.6 | 1 | 0.18 | 0.023 | 0 | 0.019 | 0 | 0.084 | 0.027 | 0 | 0.057 | 0.51 | 5 / 38 (13%) | 1 / 19 (5.3%) | 4 / 19 (21%) | 0.0526 | 0.211 |
| s__Blautia_A MIC9663 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Blautia_A | 0.33 | 0.66 | 1 | 0.2 | 0.6 | 1 | 0.42 | 0.033 | 0 | 0.046 | 0 | 0.14 | 0.02 | 0 | 0.089 | -1.2 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
| s__Blautia_A massiliensis | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Blautia_A | 0.13 | 0.62 | 1 | 0.29 | 0.62 | 1 | 2.1 | 1.3 | 0.5 | 1 | 0.5 | 1.3 | 1.7 | 0.5 | 2.5 | 0.77 | 24 / 38 (63%) | 12 / 19 (63%) | 12 / 19 (63%) | 0.632 | 0.632 |
| s__Blautia_A obeum | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Blautia_A | 0.14 | 0.62 | 1 | 0.32 | 0.64 | 1 | 1.8 | 1 | 0.4 | 1.3 | 0.53 | 1.6 | 0.74 | 0.36 | 1.1 | -0.81 | 21 / 38 (55%) | 11 / 19 (58%) | 10 / 19 (53%) | 0.579 | 0.526 |
| s__Blautia_A sp000285855 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Blautia_A | 0.44 | 0.7 | 1 | 0.48 | 0.71 | 1 | 0.26 | 0.047 | 0 | 0.06 | 0 | 0.13 | 0.034 | 0 | 0.093 | -0.82 | 7 / 38 (18%) | 4 / 19 (21%) | 3 / 19 (16%) | 0.211 | 0.158 |
| s__Blautia_A sp000436615 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Blautia_A | 0.39 | 0.66 | 1 | 0.13 | 0.6 | 1 | 1.1 | 0.17 | 0 | 0.14 | 0 | 0.36 | 0.19 | 0 | 0.51 | 0.44 | 6 / 38 (16%) | 3 / 19 (16%) | 3 / 19 (16%) | 0.158 | 0.158 |
| s__Blautia_A sp900066145 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Blautia_A | 0.96 | 0.98 | 1 | 0.2 | 0.6 | 1 | 0.84 | 0.066 | 0 | 0.049 | 0 | 0.18 | 0.083 | 0 | 0.36 | 0.76 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
| s__Blautia_A sp900066165 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Blautia_A | 0.91 | 0.97 | 1 | 0.77 | 0.86 | 1 | 1.5 | 0.96 | 0.69 | 1 | 0.58 | 1.2 | 0.88 | 0.78 | 0.8 | -0.18 | 25 / 38 (66%) | 12 / 19 (63%) | 13 / 19 (68%) | 0.632 | 0.684 |
| s__Blautia_A sp900066205 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Blautia_A | 0.3 | 0.64 | 1 | 0.12 | 0.6 | 1 | 1.3 | 0.17 | 0 | 0.16 | 0 | 0.71 | 0.17 | 0 | 0.43 | 0.087 | 5 / 38 (13%) | 1 / 19 (5.3%) | 4 / 19 (21%) | 0.0526 | 0.211 |
| s__Blautia_A sp900066335 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Blautia_A | 0.67 | 0.83 | 1 | 0.62 | 0.79 | 1 | 0.87 | 0.16 | 0 | 0.22 | 0 | 0.71 | 0.11 | 0 | 0.26 | -1 | 7 / 38 (18%) | 3 / 19 (16%) | 4 / 19 (21%) | 0.158 | 0.211 |
| s__Blautia_A sp900066355 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Blautia_A | 0.61 | 0.8 | 1 | 0.68 | 0.83 | 1 | 0.23 | 0.03 | 0 | 0.023 | 0 | 0.073 | 0.036 | 0 | 0.092 | 0.65 | 5 / 38 (13%) | 2 / 19 (11%) | 3 / 19 (16%) | 0.105 | 0.158 |
| s__Blautia_A sp900120195 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Blautia_A | 0.46 | 0.71 | 1 | 0.24 | 0.62 | 1 | 0.21 | 0.017 | 0 | 0.021 | 0 | 0.069 | 0.012 | 0 | 0.054 | -0.81 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
| s__Blautia_A wexlerae | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Blautia_A | 0.55 | 0.77 | 1 | 0.73 | 0.85 | 1 | 9.5 | 9.5 | 7.7 | 9.5 | 5.9 | 8.4 | 9.6 | 8.4 | 6.6 | 0.015 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| s__CAG-103 MIC7540 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Oscillospiraceae; g__CAG-103 | 0.34 | 0.66 | 1 | 0.59 | 0.75 | 1 | 0.56 | 0.074 | 0 | 0.049 | 0 | 0.16 | 0.098 | 0 | 0.26 | 1 | 5 / 38 (13%) | 2 / 19 (11%) | 3 / 19 (16%) | 0.105 | 0.158 |
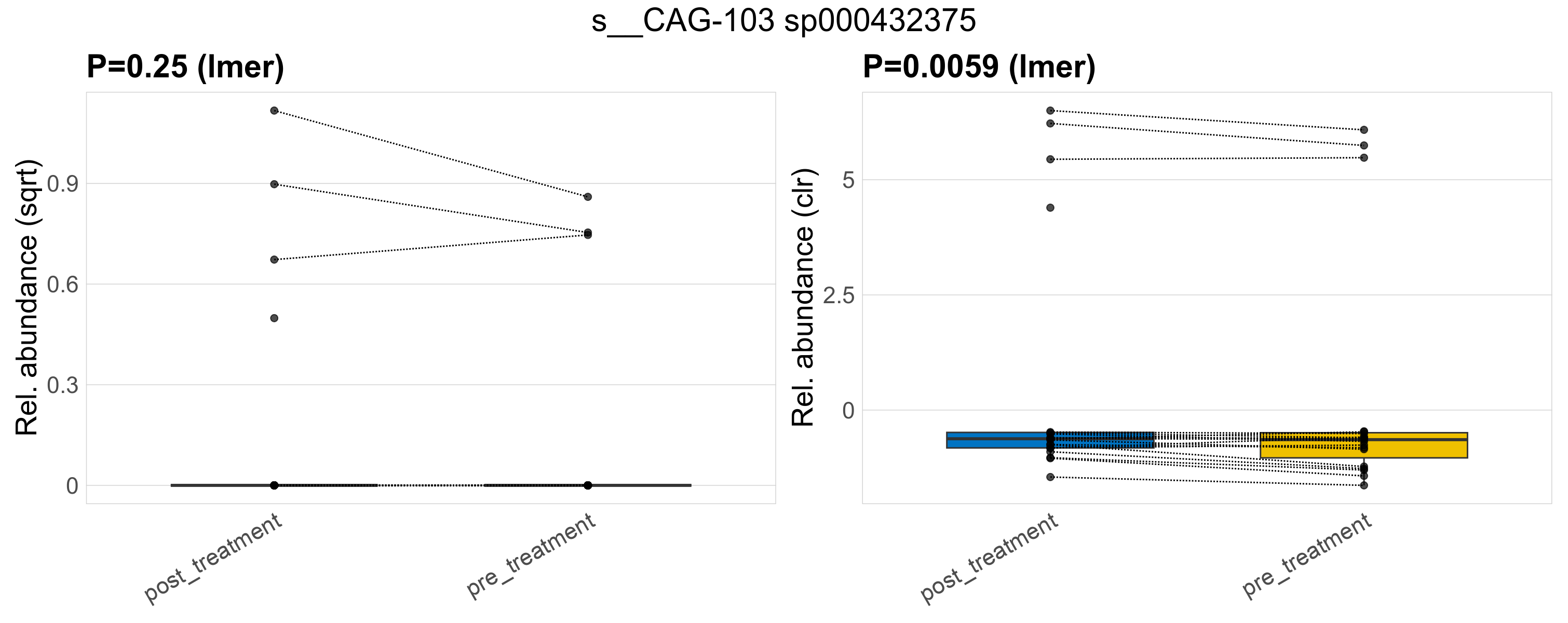
| s__CAG-103 sp000432375 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Oscillospiraceae; g__CAG-103 | 0.25 | 0.62 | 1 | 0.0059 | 0.45 | 1 | 0.66 | 0.12 | 0 | 0.15 | 0 | 0.34 | 0.098 | 0 | 0.24 | -0.61 | 7 / 38 (18%) | 4 / 19 (21%) | 3 / 19 (16%) | 0.211 | 0.158 |
| s__CAG-110 sp000435995 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Oscillospiraceae; g__CAG-110 | 0.26 | 0.62 | 1 | 0.21 | 0.6 | 1 | 0.18 | 0.014 | 0 | 0.025 | 0 | 0.085 | 0.0037 | 0 | 0.016 | -2.8 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
| s__CAG-110 sp003525905 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Oscillospiraceae; g__CAG-110 | 0.31 | 0.65 | 1 | 0.22 | 0.6 | 1 | 0.37 | 0.029 | 0 | 0.036 | 0 | 0.11 | 0.022 | 0 | 0.097 | -0.71 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
| s__CAG-127 sp900319515 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__CAG-127 | 0.96 | 0.98 | 1 | 0.76 | 0.85 | 1 | 0.54 | 0.071 | 0 | 0.089 | 0 | 0.34 | 0.054 | 0 | 0.13 | -0.72 | 5 / 38 (13%) | 2 / 19 (11%) | 3 / 19 (16%) | 0.105 | 0.158 |
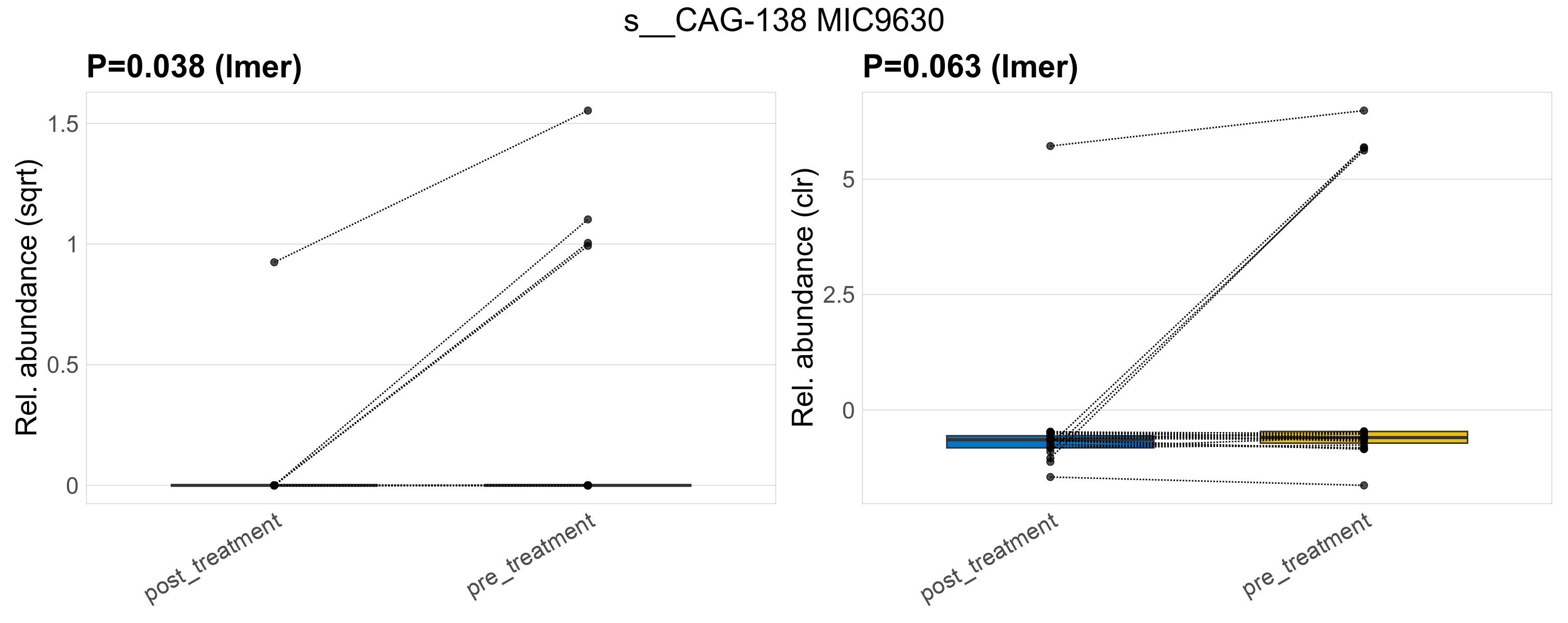
| s__CAG-138 MIC9630 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Christensenellales; f__CAG-138; g__CAG-138 MIC9630 | 0.038 | 0.52 | 1 | 0.063 | 0.54 | 1 | 1.3 | 0.17 | 0 | 0.045 | 0 | 0.2 | 0.3 | 0 | 0.65 | 2.7 | 5 / 38 (13%) | 1 / 19 (5.3%) | 4 / 19 (21%) | 0.0526 | 0.211 |
| s__CAG-1427 sp000435675 | d__Bacteria; p__Actinobacteriota; c__Coriobacteriia; o__Coriobacteriales; f__Eggerthellaceae; g__CAG-1427 | 0.89 | 0.97 | 1 | 0.82 | 0.89 | 1 | 0.21 | 0.022 | 0 | 0.025 | 0 | 0.08 | 0.019 | 0 | 0.058 | -0.4 | 4 / 38 (11%) | 2 / 19 (11%) | 2 / 19 (11%) | 0.105 | 0.105 |
| s__CAG-1427 sp000436075 | d__Bacteria; p__Actinobacteriota; c__Coriobacteriia; o__Coriobacteriales; f__Eggerthellaceae; g__CAG-1427 | 0.17 | 0.62 | 1 | 0.19 | 0.6 | 1 | 0.46 | 0.049 | 0 | 0.025 | 0 | 0.11 | 0.072 | 0 | 0.17 | 1.5 | 4 / 38 (11%) | 1 / 19 (5.3%) | 3 / 19 (16%) | 0.0526 | 0.158 |
| s__CAG-170 sp000432135 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Oscillospiraceae; g__CAG-170 | 0.19 | 0.62 | 1 | 0.24 | 0.61 | 1 | 1.5 | 0.12 | 0 | 0.0063 | 0 | 0.027 | 0.23 | 0 | 0.9 | 5.2 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| s__CAG-177 sp003514385 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Acutalibacteraceae; g__CAG-177 | 0.57 | 0.78 | 1 | 0.22 | 0.61 | 1 | 1.3 | 0.17 | 0 | 0.16 | 0 | 0.39 | 0.17 | 0 | 0.55 | 0.087 | 5 / 38 (13%) | 3 / 19 (16%) | 2 / 19 (11%) | 0.158 | 0.105 |
| s__CAG-217 sp000436335 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Acutalibacteraceae; g__CAG-217 | 0.89 | 0.97 | 1 | 0.3 | 0.62 | 1 | 1.6 | 0.42 | 0 | 0.38 | 0 | 0.71 | 0.45 | 0 | 0.94 | 0.24 | 10 / 38 (26%) | 6 / 19 (32%) | 4 / 19 (21%) | 0.316 | 0.211 |
| s__CAG-245 sp000435175 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__TANB77; f__CAG-508; g__CAG-245 | 0.27 | 0.62 | 1 | 0.33 | 0.64 | 1 | 0.48 | 0.05 | 0 | 0.02 | 0 | 0.085 | 0.081 | 0 | 0.24 | 2 | 4 / 38 (11%) | 1 / 19 (5.3%) | 3 / 19 (16%) | 0.0526 | 0.158 |
| s__CAG-274 MIC6942 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__CAG-274; g__CAG-274 | 0.15 | 0.62 | 1 | 0.11 | 0.6 | 1 | 0.33 | 0.026 | 0 | 0.048 | 0 | 0.17 | 0.0046 | 0 | 0.02 | -3.4 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
| s__CAG-274 sp000432155 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__CAG-274; g__CAG-274 | 0.21 | 0.62 | 1 | 0.17 | 0.6 | 1 | 0.6 | 0.079 | 0 | 0.12 | 0 | 0.32 | 0.043 | 0 | 0.14 | -1.5 | 5 / 38 (13%) | 3 / 19 (16%) | 2 / 19 (11%) | 0.158 | 0.105 |
| s__CAG-302 sp001916775 | d__Bacteria; p__Firmicutes; c__Bacilli; o__RF39; f__CAG-302; g__CAG-302 | 0.28 | 0.62 | 1 | 0.45 | 0.7 | 1 | 0.83 | 0.11 | 0 | 0.077 | 0 | 0.23 | 0.14 | 0 | 0.43 | 0.86 | 5 / 38 (13%) | 2 / 19 (11%) | 3 / 19 (16%) | 0.105 | 0.158 |
| s__CAG-313 sp000433035 | d__Bacteria; p__Firmicutes; c__Bacilli; o__ML615J-28; f__CAG-313; g__CAG-313 | 0.17 | 0.62 | 1 | 0.42 | 0.68 | 1 | 0.46 | 0.037 | 0 | 0.023 | 0 | 0.1 | 0.05 | 0 | 0.18 | 1.1 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| s__CAG-354 sp001915925 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__TANB77; f__CAG-508; g__CAG-354 | 0.92 | 0.97 | 1 | 0.83 | 0.89 | 1 | 0.41 | 0.054 | 0 | 0.057 | 0 | 0.17 | 0.051 | 0 | 0.14 | -0.16 | 5 / 38 (13%) | 2 / 19 (11%) | 3 / 19 (16%) | 0.105 | 0.158 |
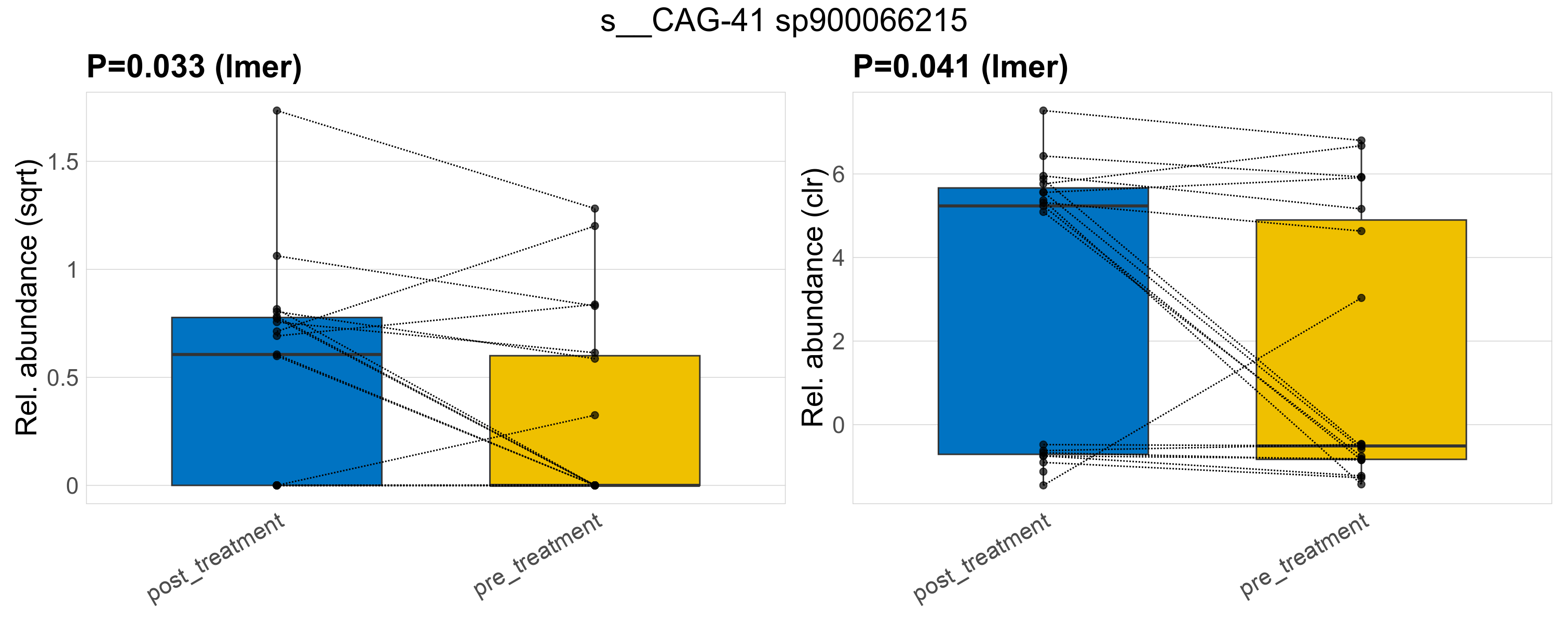
| s__CAG-41 sp900066215 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Monoglobales; f__UBA1381; g__CAG-41 | 0.033 | 0.52 | 1 | 0.041 | 0.45 | 1 | 0.79 | 0.37 | 0 | 0.47 | 0.37 | 0.7 | 0.28 | 0 | 0.5 | -0.75 | 18 / 38 (47%) | 11 / 19 (58%) | 7 / 19 (37%) | 0.579 | 0.368 |
| s__CAG-417 sp000432835 | d__Bacteria; p__Firmicutes; c__Bacilli; o__RF39; f__CAG-611; g__CAG-417 | 0.68 | 0.84 | 1 | 0.3 | 0.62 | 1 | 1.2 | 0.092 | 0 | 0.091 | 0 | 0.29 | 0.094 | 0 | 0.41 | 0.047 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
| s__CAG-452 sp000434035 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__TANB77; f__CAG-508; g__CAG-452 | 0.24 | 0.62 | 1 | 0.41 | 0.68 | 1 | 0.29 | 0.023 | 0 | 0.012 | 0 | 0.051 | 0.034 | 0 | 0.1 | 1.5 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| s__CAG-460 MIC6729 | d__Bacteria; p__Firmicutes; c__Bacilli; o__RF39; f__CAG-1000; g__CAG-460 | 0.21 | 0.62 | 1 | 0.36 | 0.66 | 1 | 2 | 0.16 | 0 | 0.069 | 0 | 0.3 | 0.25 | 0 | 0.76 | 1.9 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| s__CAG-485 MIC8260 | d__Bacteria; p__Bacteroidota; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__CAG-485 | 0.7 | 0.84 | 1 | 0.57 | 0.74 | 1 | 0.49 | 0.039 | 0 | 0.043 | 0 | 0.19 | 0.035 | 0 | 0.1 | -0.3 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| s__CAG-533 sp000434495 | d__Bacteria; p__Firmicutes; c__Bacilli; o__RF39; f__CAG-1000; g__CAG-533 | 0.19 | 0.62 | 1 | 0.34 | 0.66 | 1 | 2 | 0.15 | 0 | 0.055 | 0 | 0.24 | 0.25 | 0 | 0.76 | 2.2 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| s__CAG-557 sp000435275 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Acutalibacteraceae; g__CAG-557 | 0.46 | 0.71 | 1 | 0.49 | 0.71 | 1 | 0.63 | 0.049 | 0 | 0.043 | 0 | 0.19 | 0.056 | 0 | 0.17 | 0.38 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| s__CAG-56 sp900066615 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__CAG-56 | 0.51 | 0.75 | 1 | 0.26 | 0.62 | 1 | 0.96 | 0.53 | 0.26 | 0.54 | 0.39 | 0.62 | 0.53 | 0 | 0.88 | -0.027 | 21 / 38 (55%) | 12 / 19 (63%) | 9 / 19 (47%) | 0.632 | 0.474 |
| s__CAG-628 sp003524085 | d__Bacteria; p__Firmicutes; c__Bacilli; o__RF39; f__CAG-1000; g__CAG-628 | 0.22 | 0.62 | 1 | 0.39 | 0.68 | 1 | 0.47 | 0.037 | 0 | 0.018 | 0 | 0.08 | 0.056 | 0 | 0.17 | 1.6 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| s__CAG-83 sp000431575 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Oscillospiraceae; g__CAG-83 | 0.37 | 0.66 | 1 | 0.25 | 0.62 | 1 | 0.53 | 0.042 | 0 | 0.06 | 0 | 0.2 | 0.023 | 0 | 0.1 | -1.4 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
| s__CAG-83 sp000435555 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Oscillospiraceae; g__CAG-83 | 0.22 | 0.62 | 1 | 0.37 | 0.66 | 1 | 1 | 0.083 | 0 | 0.035 | 0 | 0.15 | 0.13 | 0 | 0.39 | 1.9 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| s__CAG-83 sp003487665 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Oscillospiraceae; g__CAG-83 | 0.24 | 0.62 | 1 | 0.4 | 0.68 | 1 | 0.45 | 0.035 | 0 | 0.018 | 0 | 0.078 | 0.053 | 0 | 0.16 | 1.6 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| s__CAG-877 sp000433455 | d__Bacteria; p__Firmicutes; c__Bacilli; o__RF39; f__CAG-611; g__CAG-877 | 0.32 | 0.66 | 1 | 0.25 | 0.62 | 1 | 0.41 | 0.043 | 0 | 0.03 | 0 | 0.095 | 0.056 | 0 | 0.2 | 0.9 | 4 / 38 (11%) | 2 / 19 (11%) | 2 / 19 (11%) | 0.105 | 0.105 |
| s__Clostridium MIC8163 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Clostridiales; f__Clostridiaceae; g__Clostridium | 0.65 | 0.82 | 1 | 0.69 | 0.84 | 1 | 0.71 | 0.13 | 0 | 0.059 | 0 | 0.14 | 0.2 | 0 | 0.71 | 1.8 | 7 / 38 (18%) | 4 / 19 (21%) | 3 / 19 (16%) | 0.211 | 0.158 |
| s__Collinsella MIC6390 | d__Bacteria; p__Actinobacteriota; c__Coriobacteriia; o__Coriobacteriales; f__Coriobacteriaceae; g__Collinsella | 0.33 | 0.66 | 1 | 0.065 | 0.54 | 1 | 1.5 | 0.12 | 0 | 0.11 | 0 | 0.38 | 0.12 | 0 | 0.53 | 0.13 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
| s__Collinsella MIC7086 | d__Bacteria; p__Actinobacteriota; c__Coriobacteriia; o__Coriobacteriales; f__Coriobacteriaceae; g__Collinsella | 0.39 | 0.66 | 1 | 0.46 | 0.71 | 1 | 0.7 | 0.055 | 0 | 0.043 | 0 | 0.19 | 0.068 | 0 | 0.2 | 0.66 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| s__Collinsella MIC7241 | d__Bacteria; p__Actinobacteriota; c__Coriobacteriia; o__Coriobacteriales; f__Coriobacteriaceae; g__Collinsella | 0.19 | 0.62 | 1 | 0.2 | 0.6 | 1 | 1.3 | 0.14 | 0 | 0.038 | 0 | 0.17 | 0.24 | 0 | 0.78 | 2.7 | 4 / 38 (11%) | 1 / 19 (5.3%) | 3 / 19 (16%) | 0.0526 | 0.158 |
| s__Collinsella MIC7504 | d__Bacteria; p__Actinobacteriota; c__Coriobacteriia; o__Coriobacteriales; f__Coriobacteriaceae; g__Collinsella | 0.12 | 0.62 | 1 | 0.2 | 0.6 | 1 | 0.75 | 0.079 | 0 | 0.049 | 0 | 0.21 | 0.11 | 0 | 0.31 | 1.2 | 4 / 38 (11%) | 1 / 19 (5.3%) | 3 / 19 (16%) | 0.0526 | 0.158 |
| s__Collinsella aerofaciens_F | d__Bacteria; p__Actinobacteriota; c__Coriobacteriia; o__Coriobacteriales; f__Coriobacteriaceae; g__Collinsella | 0.2 | 0.62 | 1 | 0.2 | 0.6 | 1 | 0.81 | 0.064 | 0 | 0.11 | 0 | 0.33 | 0.019 | 0 | 0.084 | -2.5 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
| s__Collinsella sp002232035 | d__Bacteria; p__Actinobacteriota; c__Coriobacteriia; o__Coriobacteriales; f__Coriobacteriaceae; g__Collinsella | 0.41 | 0.67 | 1 | 0.66 | 0.81 | 1 | 3.9 | 0.52 | 0 | 0.25 | 0 | 0.79 | 0.79 | 0 | 2.1 | 1.7 | 5 / 38 (13%) | 2 / 19 (11%) | 3 / 19 (16%) | 0.105 | 0.158 |
| s__Collinsella sp003487125 | d__Bacteria; p__Actinobacteriota; c__Coriobacteriia; o__Coriobacteriales; f__Coriobacteriaceae; g__Collinsella | 0.91 | 0.97 | 1 | 0.78 | 0.87 | 1 | 0.58 | 0.077 | 0 | 0.085 | 0 | 0.28 | 0.069 | 0 | 0.19 | -0.3 | 5 / 38 (13%) | 2 / 19 (11%) | 3 / 19 (16%) | 0.105 | 0.158 |
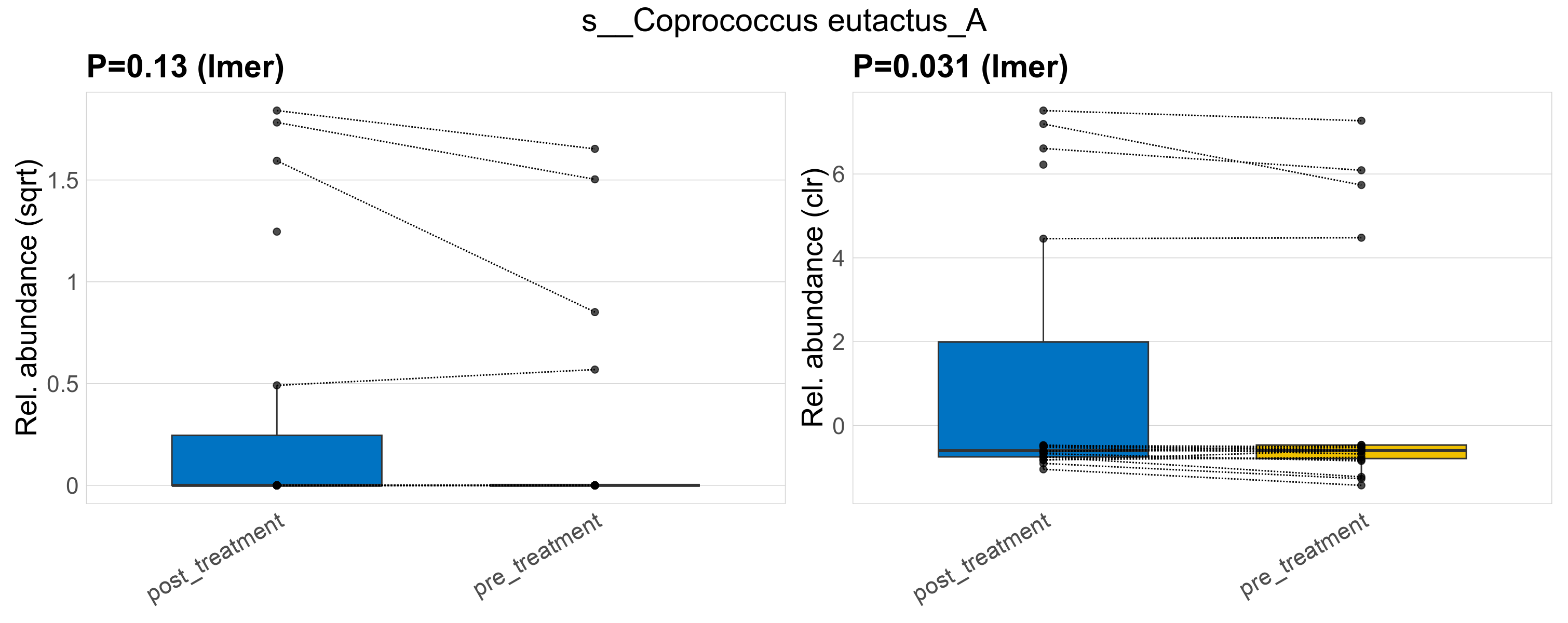
| s__Coprococcus eutactus_A | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Coprococcus | 0.13 | 0.62 | 1 | 0.031 | 0.45 | 1 | 1.9 | 0.45 | 0 | 0.57 | 0 | 1.2 | 0.32 | 0 | 0.79 | -0.83 | 9 / 38 (24%) | 5 / 19 (26%) | 4 / 19 (21%) | 0.263 | 0.211 |
| s__Coprococcus sp900066115 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Coprococcus | 0.55 | 0.77 | 1 | 0.24 | 0.61 | 1 | 2.5 | 0.33 | 0 | 0.35 | 0 | 0.85 | 0.32 | 0 | 1.3 | -0.13 | 5 / 38 (13%) | 3 / 19 (16%) | 2 / 19 (11%) | 0.158 | 0.105 |
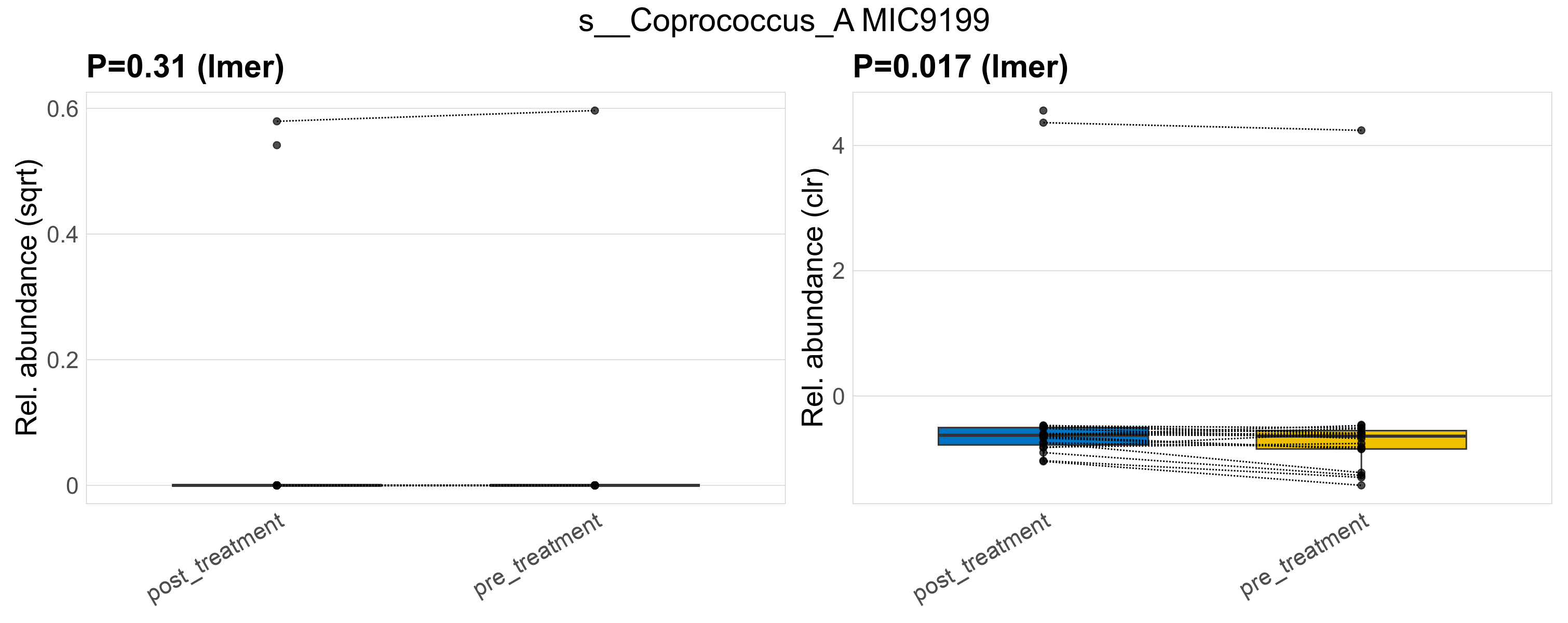
| s__Coprococcus_A MIC9199 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Coprococcus_A | 0.31 | 0.65 | 1 | 0.017 | 0.45 | 1 | 0.33 | 0.026 | 0 | 0.033 | 0 | 0.099 | 0.019 | 0 | 0.082 | -0.8 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
| s__Coprococcus_A catus | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Coprococcus_A | 0.68 | 0.84 | 1 | 0.48 | 0.71 | 1 | 0.42 | 0.078 | 0 | 0.081 | 0 | 0.18 | 0.074 | 0 | 0.2 | -0.13 | 7 / 38 (18%) | 4 / 19 (21%) | 3 / 19 (16%) | 0.211 | 0.158 |
| s__Coprococcus_B comes | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Coprococcus_B | 0.097 | 0.62 | 1 | 0.24 | 0.62 | 1 | 0.76 | 0.56 | 0.59 | 0.46 | 0.52 | 0.42 | 0.65 | 0.63 | 0.5 | 0.5 | 28 / 38 (74%) | 13 / 19 (68%) | 15 / 19 (79%) | 0.684 | 0.789 |
| s__Dialister invisus | d__Bacteria; p__Firmicutes_C; c__Negativicutes; o__Veillonellales; f__Dialisteraceae; g__Dialister | 0.95 | 0.98 | 1 | 0.41 | 0.68 | 1 | 2 | 0.37 | 0 | 0.34 | 0 | 0.82 | 0.4 | 0 | 1 | 0.23 | 7 / 38 (18%) | 4 / 19 (21%) | 3 / 19 (16%) | 0.211 | 0.158 |
| s__Dialister sp900343095 | d__Bacteria; p__Firmicutes_C; c__Negativicutes; o__Veillonellales; f__Dialisteraceae; g__Dialister | 0.55 | 0.77 | 1 | 0.86 | 0.91 | 1 | 1 | 0.19 | 0 | 0.15 | 0 | 0.41 | 0.24 | 0 | 0.49 | 0.68 | 7 / 38 (18%) | 3 / 19 (16%) | 4 / 19 (21%) | 0.158 | 0.211 |
| s__Dorea formicigenerans | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Dorea | 0.25 | 0.62 | 1 | 0.16 | 0.6 | 1 | 0.65 | 0.29 | 0 | 0.34 | 0.23 | 0.4 | 0.24 | 0 | 0.37 | -0.5 | 17 / 38 (45%) | 10 / 19 (53%) | 7 / 19 (37%) | 0.526 | 0.368 |
| s__Dorea longicatena | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Dorea | 0.23 | 0.62 | 1 | 0.97 | 0.97 | 1 | 1.2 | 0.52 | 0 | 0.43 | 0 | 0.59 | 0.62 | 0 | 0.95 | 0.53 | 17 / 38 (45%) | 9 / 19 (47%) | 8 / 19 (42%) | 0.474 | 0.421 |
| s__Dorea longicatena_B | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Dorea | 0.99 | 0.99 | 1 | 0.41 | 0.68 | 1 | 1 | 0.49 | 0 | 0.45 | 0.29 | 0.66 | 0.52 | 0 | 0.75 | 0.21 | 18 / 38 (47%) | 10 / 19 (53%) | 8 / 19 (42%) | 0.526 | 0.421 |
| s__Dorea sp000433215 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Dorea | 0.078 | 0.59 | 1 | 0.14 | 0.6 | 1 | 0.2 | 0.021 | 0 | 0.0049 | 0 | 0.022 | 0.038 | 0 | 0.092 | 3 | 4 / 38 (11%) | 1 / 19 (5.3%) | 3 / 19 (16%) | 0.0526 | 0.158 |
| s__Dorea sp900066555 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Dorea | 0.28 | 0.63 | 1 | 0.27 | 0.62 | 1 | 0.18 | 0.019 | 0 | 0.012 | 0 | 0.051 | 0.025 | 0 | 0.068 | 1.1 | 4 / 38 (11%) | 1 / 19 (5.3%) | 3 / 19 (16%) | 0.0526 | 0.158 |
| s__Duncaniella MIC6738 | d__Bacteria; p__Bacteroidota; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__Duncaniella | 0.66 | 0.83 | 1 | 0.56 | 0.73 | 1 | 0.49 | 0.039 | 0 | 0.041 | 0 | 0.18 | 0.036 | 0 | 0.11 | -0.19 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| s__ER4 sp900317525 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Oscillospiraceae; g__ER4 | 0.94 | 0.98 | 1 | 0.36 | 0.66 | 1 | 0.51 | 0.067 | 0 | 0.057 | 0 | 0.15 | 0.077 | 0 | 0.24 | 0.43 | 5 / 38 (13%) | 3 / 19 (16%) | 2 / 19 (11%) | 0.158 | 0.105 |
| s__Eisenbergiella sp900066775 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Eisenbergiella | 0.17 | 0.62 | 1 | 0.35 | 0.66 | 1 | 2 | 0.15 | 0 | 0.048 | 0 | 0.21 | 0.26 | 0 | 1.1 | 2.4 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| s__Erysipelatoclostridium ramosum | d__Bacteria; p__Firmicutes; c__Bacilli; o__Erysipelotrichales; f__Erysipelatoclostridiaceae; g__Erysipelatoclostridium | 0.15 | 0.62 | 1 | 0.37 | 0.66 | 1 | 0.37 | 0.029 | 0 | 0.012 | 0 | 0.051 | 0.047 | 0 | 0.17 | 2 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
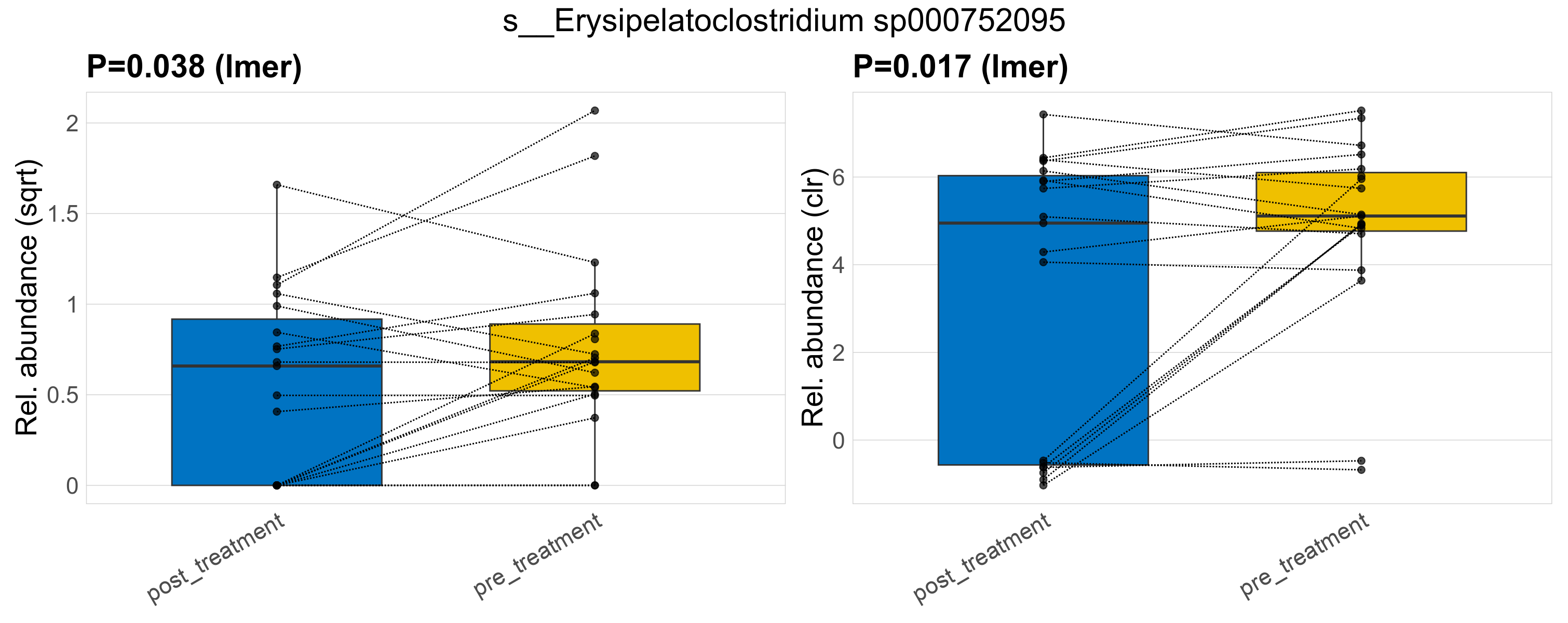
| s__Erysipelatoclostridium sp000752095 | d__Bacteria; p__Firmicutes; c__Bacilli; o__Erysipelotrichales; f__Erysipelatoclostridiaceae; g__Erysipelatoclostridium | 0.038 | 0.52 | 1 | 0.017 | 0.45 | 1 | 0.92 | 0.7 | 0.46 | 0.56 | 0.43 | 0.7 | 0.84 | 0.47 | 1.1 | 0.58 | 29 / 38 (76%) | 12 / 19 (63%) | 17 / 19 (89%) | 0.632 | 0.895 |
| s__Eubacterium_E hallii | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Eubacterium_E | 0.44 | 0.69 | 1 | 0.35 | 0.66 | 1 | 2 | 1.8 | 1.7 | 1.8 | 1.7 | 1.2 | 1.7 | 1.4 | 1.1 | -0.082 | 33 / 38 (87%) | 16 / 19 (84%) | 17 / 19 (89%) | 0.842 | 0.895 |
| s__Eubacterium_E hallii_A | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Eubacterium_E | 0.062 | 0.52 | 1 | 0.049 | 0.47 | 1 | 0.65 | 0.15 | 0 | 0.22 | 0 | 0.4 | 0.089 | 0 | 0.23 | -1.3 | 9 / 38 (24%) | 6 / 19 (32%) | 3 / 19 (16%) | 0.316 | 0.158 |
| s__Eubacterium_F MIC7104 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Eubacterium_F | 0.051 | 0.52 | 1 | 0.069 | 0.55 | 1 | 0.44 | 0.058 | 0 | 0.015 | 0 | 0.067 | 0.1 | 0 | 0.21 | 2.7 | 5 / 38 (13%) | 1 / 19 (5.3%) | 4 / 19 (21%) | 0.0526 | 0.211 |
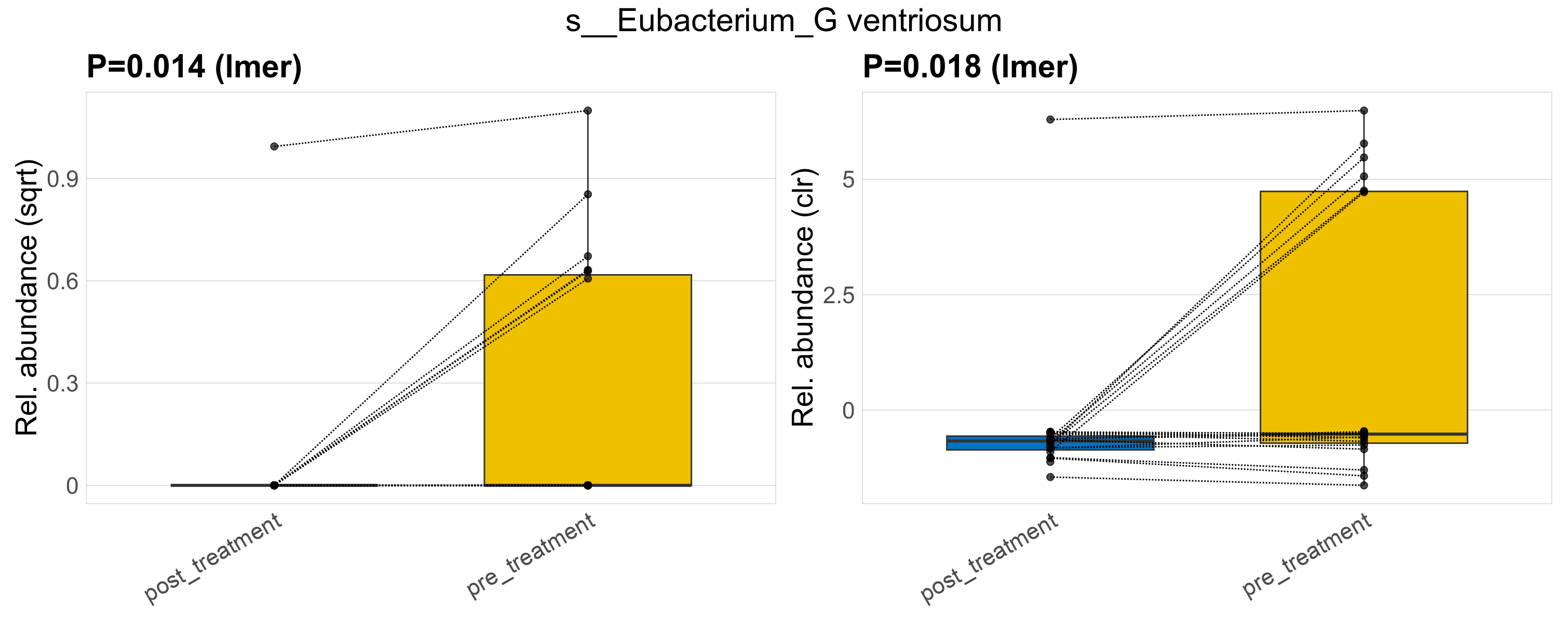
| s__Eubacterium_G ventriosum | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Eubacterium_G | 0.014 | 0.52 | 1 | 0.018 | 0.45 | 1 | 0.65 | 0.12 | 0 | 0.052 | 0 | 0.23 | 0.19 | 0 | 0.33 | 1.9 | 7 / 38 (18%) | 1 / 19 (5.3%) | 6 / 19 (32%) | 0.0526 | 0.316 |
| s__Eubacterium_I ramulus | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Eubacterium_I | 0.094 | 0.62 | 1 | 0.16 | 0.6 | 1 | 0.58 | 0.12 | 0 | 0.078 | 0 | 0.2 | 0.17 | 0 | 0.36 | 1.1 | 8 / 38 (21%) | 3 / 19 (16%) | 5 / 19 (26%) | 0.158 | 0.263 |
| s__Eubacterium_I ramulus_A | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Eubacterium_I | 0.38 | 0.66 | 1 | 0.49 | 0.71 | 1 | 0.82 | 0.11 | 0 | 0.07 | 0 | 0.23 | 0.15 | 0 | 0.42 | 1.1 | 5 / 38 (13%) | 2 / 19 (11%) | 3 / 19 (16%) | 0.105 | 0.158 |
| s__Eubacterium_R sp000434995 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Acutalibacteraceae; g__Eubacterium_R | 0.26 | 0.62 | 1 | 0.33 | 0.64 | 1 | 1.5 | 0.23 | 0 | 0.17 | 0 | 0.55 | 0.3 | 0 | 0.61 | 0.82 | 6 / 38 (16%) | 2 / 19 (11%) | 4 / 19 (21%) | 0.105 | 0.211 |
| s__F23-B02 sp002472405 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Oscillospiraceae; g__F23-B02 | 0.13 | 0.62 | 1 | 0.16 | 0.6 | 1 | 0.73 | 0.077 | 0 | 0.025 | 0 | 0.11 | 0.13 | 0 | 0.32 | 2.4 | 4 / 38 (11%) | 1 / 19 (5.3%) | 3 / 19 (16%) | 0.0526 | 0.158 |
| s__Faecalibacterium MIC7145 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Ruminococcaceae; g__Faecalibacterium | 0.1 | 0.62 | 1 | 0.092 | 0.6 | 1 | 0.6 | 0.14 | 0 | 0.076 | 0 | 0.21 | 0.21 | 0 | 0.38 | 1.5 | 9 / 38 (24%) | 3 / 19 (16%) | 6 / 19 (32%) | 0.158 | 0.316 |
| s__Faecalibacterium MIC7301 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Ruminococcaceae; g__Faecalibacterium | 0.47 | 0.71 | 1 | 0.54 | 0.73 | 1 | 0.23 | 0.018 | 0 | 0.016 | 0 | 0.071 | 0.021 | 0 | 0.062 | 0.39 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| s__Faecalibacterium MIC8666 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Ruminococcaceae; g__Faecalibacterium | 0.18 | 0.62 | 1 | 0.5 | 0.71 | 1 | 1.3 | 0.14 | 0 | 0.083 | 0 | 0.3 | 0.19 | 0 | 0.75 | 1.2 | 4 / 38 (11%) | 2 / 19 (11%) | 2 / 19 (11%) | 0.105 | 0.105 |
| s__Faecalibacterium MIC9210 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Ruminococcaceae; g__Faecalibacterium | 0.18 | 0.62 | 1 | 0.14 | 0.6 | 1 | 3 | 0.4 | 0 | 0.48 | 0 | 1.9 | 0.31 | 0 | 1.2 | -0.63 | 5 / 38 (13%) | 3 / 19 (16%) | 2 / 19 (11%) | 0.158 | 0.105 |
| s__Faecalibacterium prausnitzii_A | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Ruminococcaceae; g__Faecalibacterium | 0.62 | 0.81 | 1 | 0.75 | 0.85 | 1 | 1.2 | 0.093 | 0 | 0.15 | 0 | 0.65 | 0.037 | 0 | 0.12 | -2 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| s__Faecalibacterium prausnitzii_C | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Ruminococcaceae; g__Faecalibacterium | 0.95 | 0.98 | 1 | 0.5 | 0.71 | 1 | 3.9 | 2.5 | 0.8 | 2.7 | 0.75 | 3.9 | 2.3 | 0.83 | 3.3 | -0.23 | 24 / 38 (63%) | 11 / 19 (58%) | 13 / 19 (68%) | 0.579 | 0.684 |
| s__Faecalibacterium prausnitzii_D | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Ruminococcaceae; g__Faecalibacterium | 0.87 | 0.97 | 1 | 0.28 | 0.62 | 1 | 2.3 | 1.1 | 0 | 1 | 0.43 | 1.5 | 1.1 | 0 | 1.7 | 0.14 | 18 / 38 (47%) | 10 / 19 (53%) | 8 / 19 (42%) | 0.526 | 0.421 |
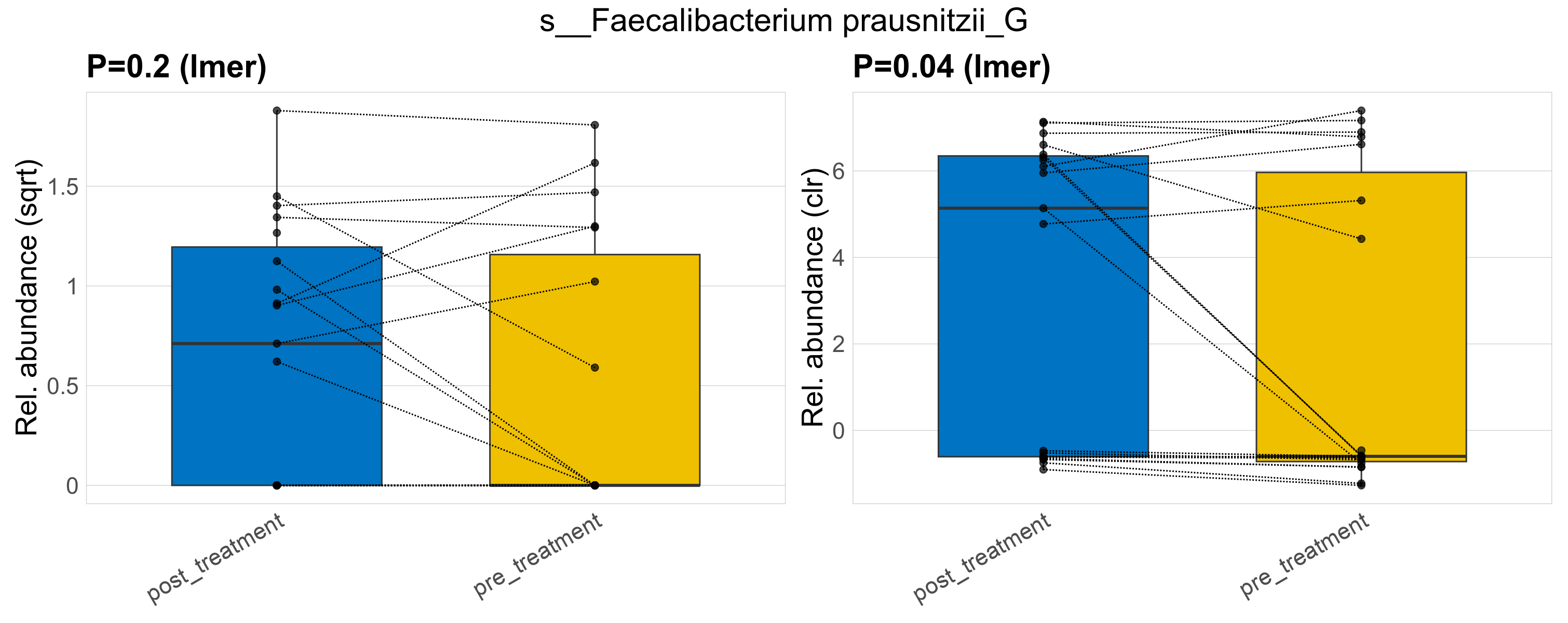
| s__Faecalibacterium prausnitzii_G | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Ruminococcaceae; g__Faecalibacterium | 0.2 | 0.62 | 1 | 0.04 | 0.45 | 1 | 1.6 | 0.75 | 0 | 0.83 | 0.51 | 0.99 | 0.67 | 0 | 1.1 | -0.31 | 18 / 38 (47%) | 11 / 19 (58%) | 7 / 19 (37%) | 0.579 | 0.368 |
| s__Faecalibacterium prausnitzii_I | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Ruminococcaceae; g__Faecalibacterium | 0.99 | 0.99 | 1 | 0.95 | 0.96 | 1 | 0.45 | 0.095 | 0 | 0.1 | 0 | 0.23 | 0.089 | 0 | 0.21 | -0.17 | 8 / 38 (21%) | 4 / 19 (21%) | 4 / 19 (21%) | 0.211 | 0.211 |
| s__Faecalibacterium prausnitzii_J | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Ruminococcaceae; g__Faecalibacterium | 0.29 | 0.64 | 1 | 0.22 | 0.6 | 1 | 0.8 | 0.17 | 0 | 0.13 | 0 | 0.36 | 0.2 | 0 | 0.5 | 0.62 | 8 / 38 (21%) | 3 / 19 (16%) | 5 / 19 (26%) | 0.158 | 0.263 |
| s__Faecalibacterium prausnitzii_K | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Ruminococcaceae; g__Faecalibacterium | 0.59 | 0.78 | 1 | 0.73 | 0.85 | 1 | 1.8 | 0.6 | 0 | 0.79 | 0 | 2 | 0.41 | 0 | 0.74 | -0.95 | 13 / 38 (34%) | 7 / 19 (37%) | 6 / 19 (32%) | 0.368 | 0.316 |
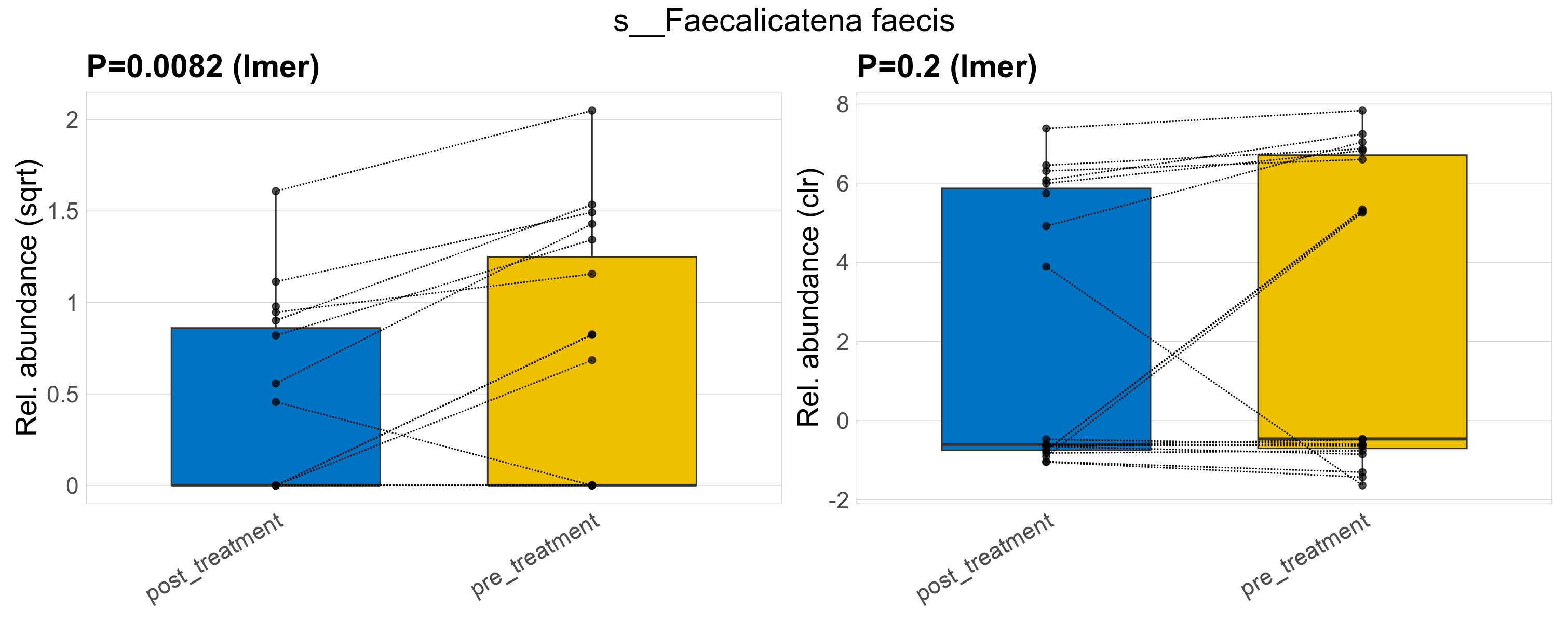
| s__Faecalicatena faecis | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Faecalicatena | 0.0082 | 0.52 | 1 | 0.2 | 0.6 | 1 | 1.4 | 0.62 | 0 | 0.4 | 0 | 0.67 | 0.83 | 0 | 1.2 | 1.1 | 17 / 38 (45%) | 8 / 19 (42%) | 9 / 19 (47%) | 0.421 | 0.474 |
| s__Faecalicatena gnavus | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Faecalicatena | 0.08 | 0.59 | 1 | 0.16 | 0.6 | 1 | 1.3 | 0.13 | 0 | 0.043 | 0 | 0.19 | 0.22 | 0 | 0.79 | 2.4 | 4 / 38 (11%) | 1 / 19 (5.3%) | 3 / 19 (16%) | 0.0526 | 0.158 |
| s__Faecalicatena lactaris | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Faecalicatena | 0.06 | 0.52 | 1 | 0.18 | 0.6 | 1 | 0.93 | 0.2 | 0 | 0.13 | 0 | 0.36 | 0.26 | 0 | 0.7 | 1 | 8 / 38 (21%) | 3 / 19 (16%) | 5 / 19 (26%) | 0.158 | 0.263 |
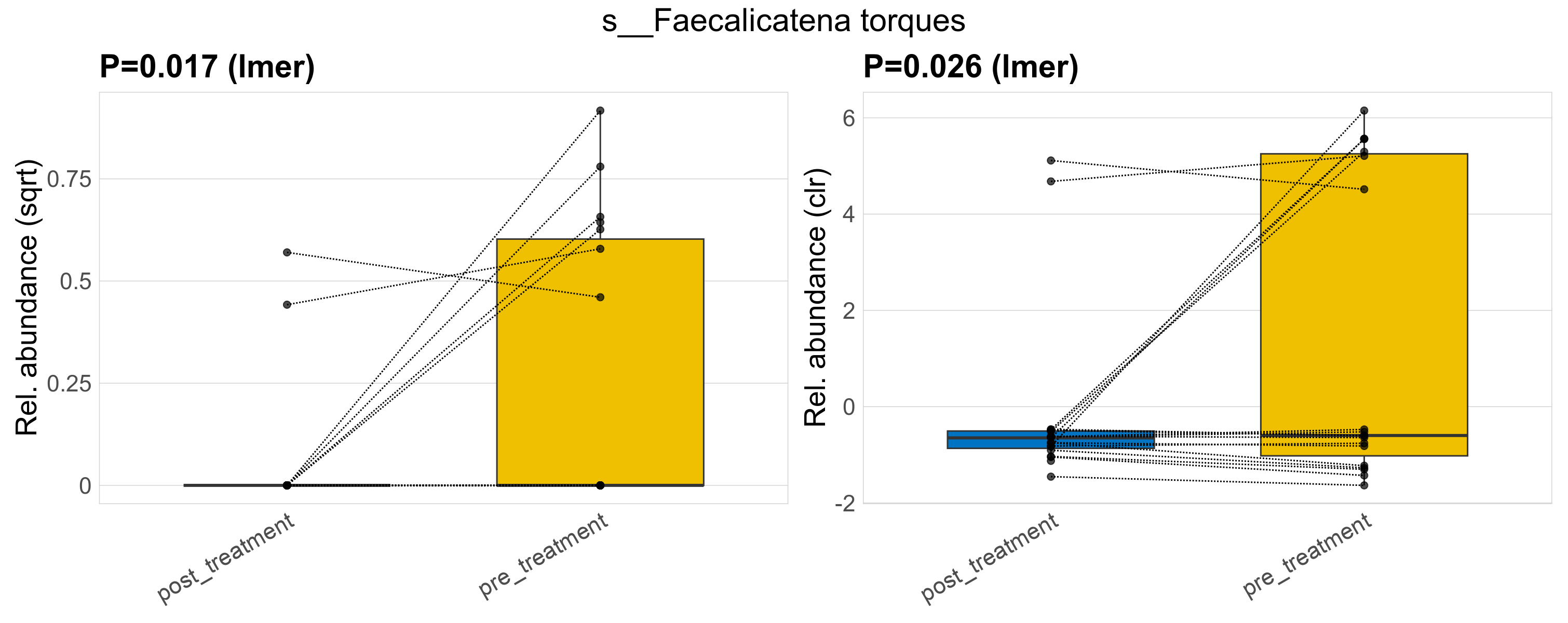
| s__Faecalicatena torques | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Faecalicatena | 0.017 | 0.52 | 1 | 0.026 | 0.45 | 1 | 0.42 | 0.099 | 0 | 0.027 | 0 | 0.085 | 0.17 | 0 | 0.26 | 2.7 | 9 / 38 (24%) | 2 / 19 (11%) | 7 / 19 (37%) | 0.105 | 0.368 |
| s__Fusicatenibacter saccharivorans | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Fusicatenibacter | 0.37 | 0.66 | 1 | 0.12 | 0.6 | 1 | 4.5 | 4 | 2.7 | 4.2 | 3 | 3.8 | 3.8 | 2.6 | 4.1 | -0.14 | 34 / 38 (89%) | 17 / 19 (89%) | 17 / 19 (89%) | 0.895 | 0.895 |
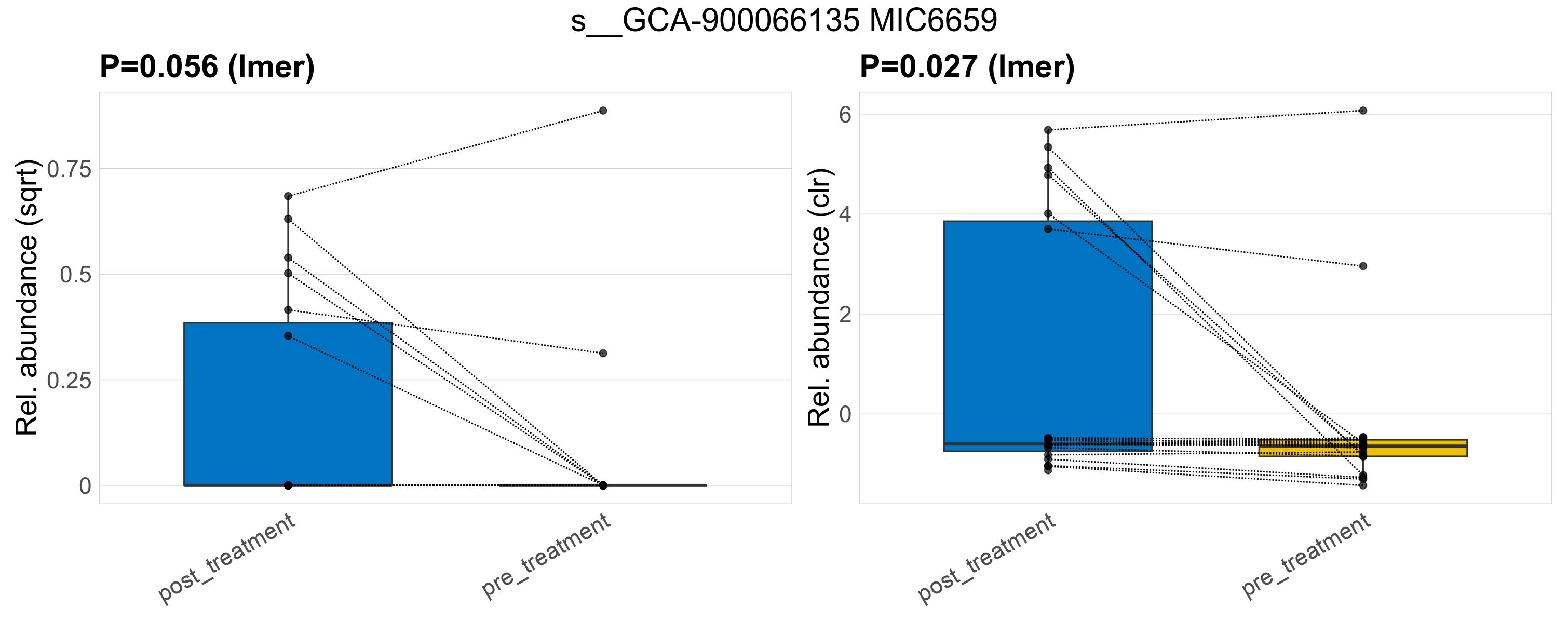
| s__GCA-900066135 MIC6659 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__GCA-900066135 | 0.056 | 0.52 | 1 | 0.027 | 0.45 | 1 | 0.32 | 0.068 | 0 | 0.09 | 0 | 0.15 | 0.047 | 0 | 0.18 | -0.94 | 8 / 38 (21%) | 6 / 19 (32%) | 2 / 19 (11%) | 0.316 | 0.105 |
| s__Gemmiger MIC9530 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Ruminococcaceae; g__Gemmiger | 0.69 | 0.84 | 1 | 0.72 | 0.85 | 1 | 0.37 | 0.059 | 0 | 0.064 | 0 | 0.17 | 0.054 | 0 | 0.13 | -0.25 | 6 / 38 (16%) | 3 / 19 (16%) | 3 / 19 (16%) | 0.158 | 0.158 |
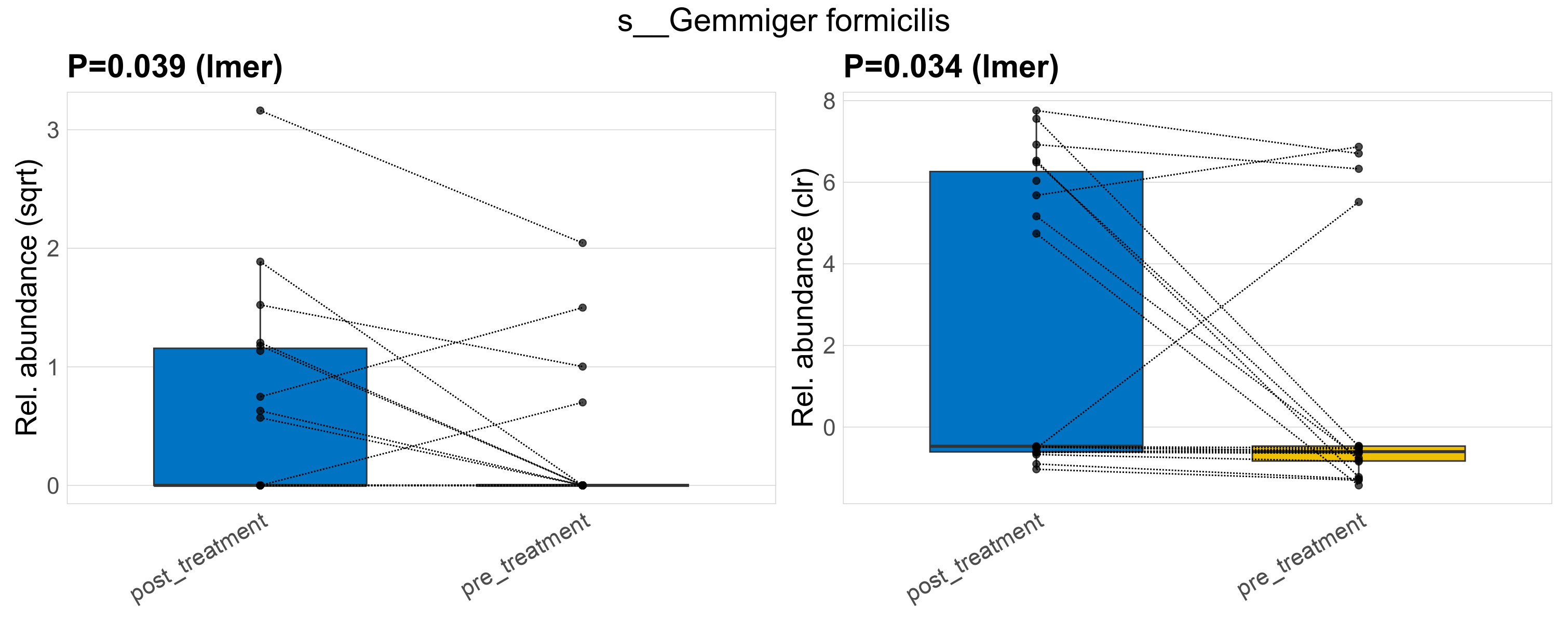
| s__Gemmiger formicilis | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Ruminococcaceae; g__Gemmiger | 0.039 | 0.52 | 1 | 0.034 | 0.45 | 1 | 2.2 | 0.77 | 0 | 1.1 | 0 | 2.4 | 0.42 | 0 | 1.1 | -1.4 | 13 / 38 (34%) | 9 / 19 (47%) | 4 / 19 (21%) | 0.474 | 0.211 |
| s__Gemmiger sp003476825 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Ruminococcaceae; g__Gemmiger | 0.8 | 0.94 | 1 | 0.66 | 0.81 | 1 | 1.8 | 1.3 | 0.94 | 1.3 | 0.89 | 1.4 | 1.3 | 0.98 | 1.7 | 0 | 29 / 38 (76%) | 15 / 19 (79%) | 14 / 19 (74%) | 0.789 | 0.737 |
| s__Holdemanella sp002299315 | d__Bacteria; p__Firmicutes; c__Bacilli; o__Erysipelotrichales; f__Erysipelotrichaceae; g__Holdemanella | 0.48 | 0.71 | 1 | 0.28 | 0.62 | 1 | 2.5 | 0.26 | 0 | 0.21 | 0 | 0.62 | 0.31 | 0 | 1 | 0.56 | 4 / 38 (11%) | 2 / 19 (11%) | 2 / 19 (11%) | 0.105 | 0.105 |
| s__Intestinibacter bartlettii | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Peptostreptococcales; f__Peptostreptococcaceae; g__Intestinibacter | 0.87 | 0.97 | 1 | 0.85 | 0.9 | 1 | 0.27 | 0.05 | 0 | 0.049 | 0 | 0.12 | 0.05 | 0 | 0.12 | 0.029 | 7 / 38 (18%) | 3 / 19 (16%) | 4 / 19 (21%) | 0.158 | 0.211 |
| s__KLE1615 sp900066985 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__KLE1615 | 0.42 | 0.69 | 1 | 0.45 | 0.7 | 1 | 1.2 | 0.55 | 0 | 0.52 | 0 | 0.76 | 0.58 | 0 | 0.74 | 0.16 | 17 / 38 (45%) | 8 / 19 (42%) | 9 / 19 (47%) | 0.421 | 0.474 |
| s__Lachnospira eligens_B | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Lachnospira | 0.2 | 0.62 | 1 | 0.29 | 0.62 | 1 | 0.37 | 0.039 | 0 | 0.011 | 0 | 0.047 | 0.067 | 0 | 0.17 | 2.6 | 4 / 38 (11%) | 1 / 19 (5.3%) | 3 / 19 (16%) | 0.0526 | 0.158 |
| s__Lachnospira rogosae | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Lachnospira | 0.85 | 0.95 | 1 | 0.84 | 0.9 | 1 | 0.57 | 0.075 | 0 | 0.11 | 0 | 0.43 | 0.04 | 0 | 0.1 | -1.5 | 5 / 38 (13%) | 2 / 19 (11%) | 3 / 19 (16%) | 0.105 | 0.158 |
| s__Lachnospira sp000437735 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Lachnospira | 0.2 | 0.62 | 1 | 0.15 | 0.6 | 1 | 0.49 | 0.039 | 0 | 0.065 | 0 | 0.2 | 0.013 | 0 | 0.057 | -2.3 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
| s__Lachnospira sp003451515 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Lachnospira | 0.93 | 0.98 | 1 | 0.45 | 0.7 | 1 | 1.2 | 0.21 | 0 | 0.18 | 0 | 0.48 | 0.24 | 0 | 0.62 | 0.42 | 7 / 38 (18%) | 4 / 19 (21%) | 3 / 19 (16%) | 0.211 | 0.158 |
| s__Lactobacillus_B salivarius | d__Bacteria; p__Firmicutes; c__Bacilli; o__Lactobacillales; f__Lactobacillaceae; g__Lactobacillus_B | 0.15 | 0.62 | 1 | 0.13 | 0.6 | 1 | 0.81 | 0.064 | 0 | 0.11 | 0 | 0.34 | 0.022 | 0 | 0.098 | -2.3 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
| s__Megamonas funiformis | d__Bacteria; p__Firmicutes_C; c__Negativicutes; o__Selenomonadales; f__Selenomonadaceae; g__Megamonas | 0.83 | 0.94 | 1 | 0.97 | 0.97 | 1 | 0.46 | 0.049 | 0 | 0.063 | 0 | 0.19 | 0.035 | 0 | 0.13 | -0.85 | 4 / 38 (11%) | 2 / 19 (11%) | 2 / 19 (11%) | 0.105 | 0.105 |
| s__Methanobrevibacter_A smithii | d__Archaea; p__Euryarchaeota; c__Methanobacteria; o__Methanobacteriales; f__Methanobacteriaceae; g__Methanobrevibacter_A | 0.18 | 0.62 | 1 | 0.36 | 0.66 | 1 | 1 | 0.083 | 0 | 0.025 | 0 | 0.11 | 0.14 | 0 | 0.42 | 2.5 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| s__Monoglobus pectinilyticus | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Monoglobales; f__Monoglobaceae; g__Monoglobus | 0.062 | 0.52 | 1 | 0.09 | 0.6 | 1 | 0.2 | 0.038 | 0 | 0.0091 | 0 | 0.027 | 0.066 | 0 | 0.13 | 2.9 | 7 / 38 (18%) | 2 / 19 (11%) | 5 / 19 (26%) | 0.105 | 0.263 |
| s__NK3B98 sp003150485 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Oscillospiraceae; g__NK3B98 | 0.4 | 0.67 | 1 | 0.52 | 0.72 | 1 | 0.2 | 0.016 | 0 | 0.012 | 0 | 0.054 | 0.019 | 0 | 0.057 | 0.66 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| s__Oxalobacter formigenes_A | d__Bacteria; p__Proteobacteria; c__Gammaproteobacteria; o__Burkholderiales; f__Burkholderiaceae; g__Oxalobacter | 0.23 | 0.62 | 1 | 0.18 | 0.6 | 1 | 0.11 | 0.0083 | 0 | 0.011 | 0 | 0.036 | 0.0051 | 0 | 0.022 | -1.1 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
| s__Parabacteroides distasonis | d__Bacteria; p__Bacteroidota; c__Bacteroidia; o__Bacteroidales; f__Tannerellaceae; g__Parabacteroides | 0.18 | 0.62 | 1 | 0.23 | 0.61 | 1 | 0.6 | 0.063 | 0 | 0.057 | 0 | 0.25 | 0.07 | 0 | 0.24 | 0.3 | 4 / 38 (11%) | 1 / 19 (5.3%) | 3 / 19 (16%) | 0.0526 | 0.158 |
| s__Parabacteroides merdae | d__Bacteria; p__Bacteroidota; c__Bacteroidia; o__Bacteroidales; f__Tannerellaceae; g__Parabacteroides | 0.39 | 0.66 | 1 | 0.48 | 0.71 | 1 | 0.27 | 0.05 | 0 | 0.04 | 0 | 0.12 | 0.061 | 0 | 0.14 | 0.61 | 7 / 38 (18%) | 3 / 19 (16%) | 4 / 19 (21%) | 0.158 | 0.211 |
| s__Parasutterella excrementihominis | d__Bacteria; p__Proteobacteria; c__Gammaproteobacteria; o__Burkholderiales; f__Burkholderiaceae; g__Parasutterella | 0.26 | 0.62 | 1 | 0.17 | 0.6 | 1 | 0.46 | 0.036 | 0 | 0.059 | 0 | 0.19 | 0.013 | 0 | 0.055 | -2.2 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
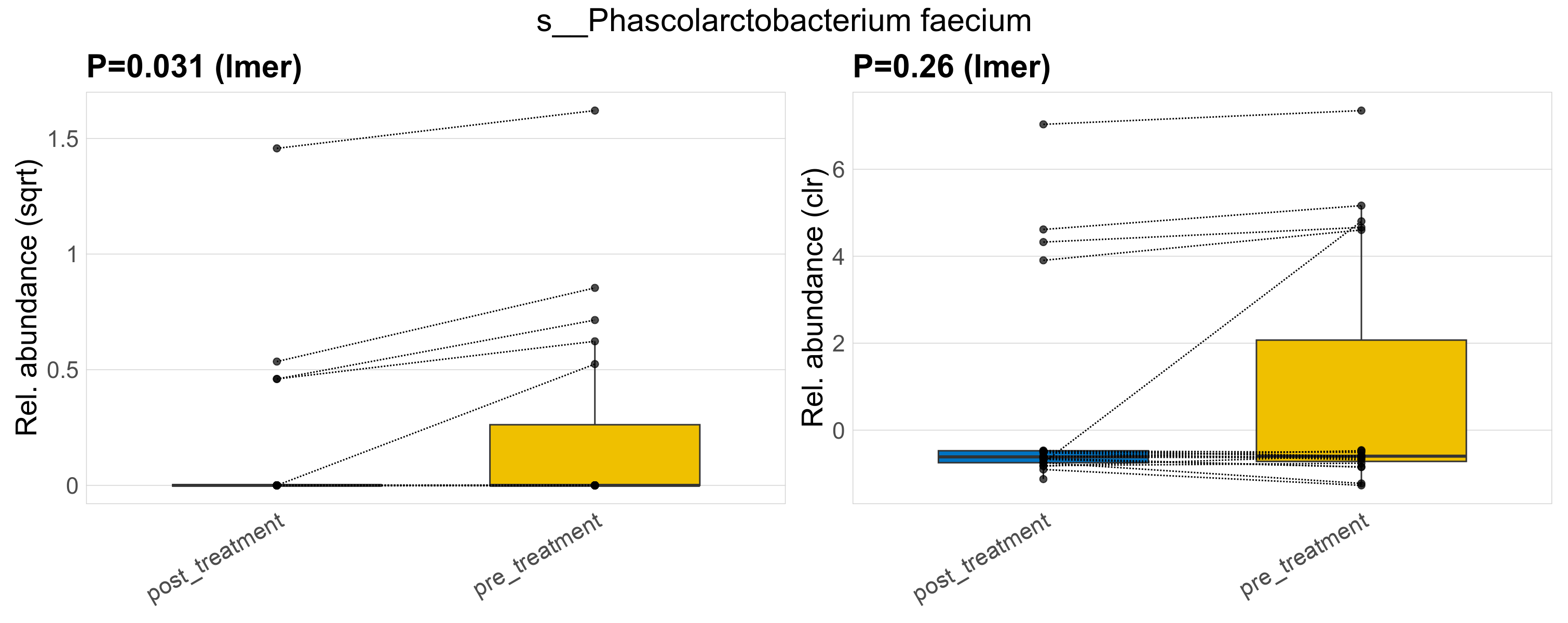
| s__Phascolarctobacterium faecium | d__Bacteria; p__Firmicutes_C; c__Negativicutes; o__Acidaminococcales; f__Acidaminococcaceae; g__Phascolarctobacterium | 0.031 | 0.52 | 1 | 0.26 | 0.62 | 1 | 0.82 | 0.19 | 0 | 0.15 | 0 | 0.49 | 0.24 | 0 | 0.62 | 0.68 | 9 / 38 (24%) | 4 / 19 (21%) | 5 / 19 (26%) | 0.211 | 0.263 |
| s__Prevotella copri | d__Bacteria; p__Bacteroidota; c__Bacteroidia; o__Bacteroidales; f__Bacteroidaceae; g__Prevotella | 0.48 | 0.71 | 1 | 0.92 | 0.95 | 1 | 5.9 | 0.62 | 0 | 1.1 | 0 | 3.3 | 0.14 | 0 | 0.45 | -3 | 4 / 38 (11%) | 2 / 19 (11%) | 2 / 19 (11%) | 0.105 | 0.105 |
| s__Prevotella sp000436915 | d__Bacteria; p__Bacteroidota; c__Bacteroidia; o__Bacteroidales; f__Bacteroidaceae; g__Prevotella | 0.59 | 0.78 | 1 | 0.54 | 0.73 | 1 | 0.45 | 0.035 | 0 | 0.036 | 0 | 0.16 | 0.035 | 0 | 0.1 | -0.041 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| s__Prevotella sp900313215 | d__Bacteria; p__Bacteroidota; c__Bacteroidia; o__Bacteroidales; f__Bacteroidaceae; g__Prevotella | 0.84 | 0.94 | 1 | 0.75 | 0.85 | 1 | 3.3 | 0.35 | 0 | 0.5 | 0 | 1.6 | 0.19 | 0 | 0.57 | -1.4 | 4 / 38 (11%) | 2 / 19 (11%) | 2 / 19 (11%) | 0.105 | 0.105 |
| s__QAMH01 MIC6543 | d__Bacteria; p__Actinobacteriota; c__Coriobacteriia; o__Coriobacteriales; f__QAMH01; g__QAMH01 | 0.27 | 0.62 | 1 | 0.42 | 0.68 | 1 | 0.5 | 0.039 | 0 | 0.022 | 0 | 0.094 | 0.057 | 0 | 0.17 | 1.4 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| s__Romboutsia timonensis | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Peptostreptococcales; f__Peptostreptococcaceae; g__Romboutsia | 0.83 | 0.94 | 1 | 0.93 | 0.95 | 1 | 0.73 | 0.23 | 0 | 0.2 | 0 | 0.35 | 0.26 | 0 | 0.51 | 0.38 | 12 / 38 (32%) | 6 / 19 (32%) | 6 / 19 (32%) | 0.316 | 0.316 |
| s__Roseburia hominis | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Roseburia | 0.05 | 0.52 | 1 | 0.083 | 0.6 | 1 | 0.84 | 0.11 | 0 | 0.042 | 0 | 0.18 | 0.18 | 0 | 0.43 | 2.1 | 5 / 38 (13%) | 1 / 19 (5.3%) | 4 / 19 (21%) | 0.0526 | 0.211 |
| s__Roseburia intestinalis | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Roseburia | 0.041 | 0.52 | 1 | 0.049 | 0.47 | 1 | 0.57 | 0.12 | 0 | 0.048 | 0 | 0.14 | 0.19 | 0 | 0.34 | 2 | 8 / 38 (21%) | 2 / 19 (11%) | 6 / 19 (32%) | 0.105 | 0.316 |
| s__Ruminiclostridium_C MIC9168 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Oscillospiraceae; g__Ruminiclostridium_C | 0.18 | 0.62 | 1 | 0.18 | 0.6 | 1 | 0.3 | 0.023 | 0 | 0.032 | 0 | 0.11 | 0.014 | 0 | 0.062 | -1.2 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
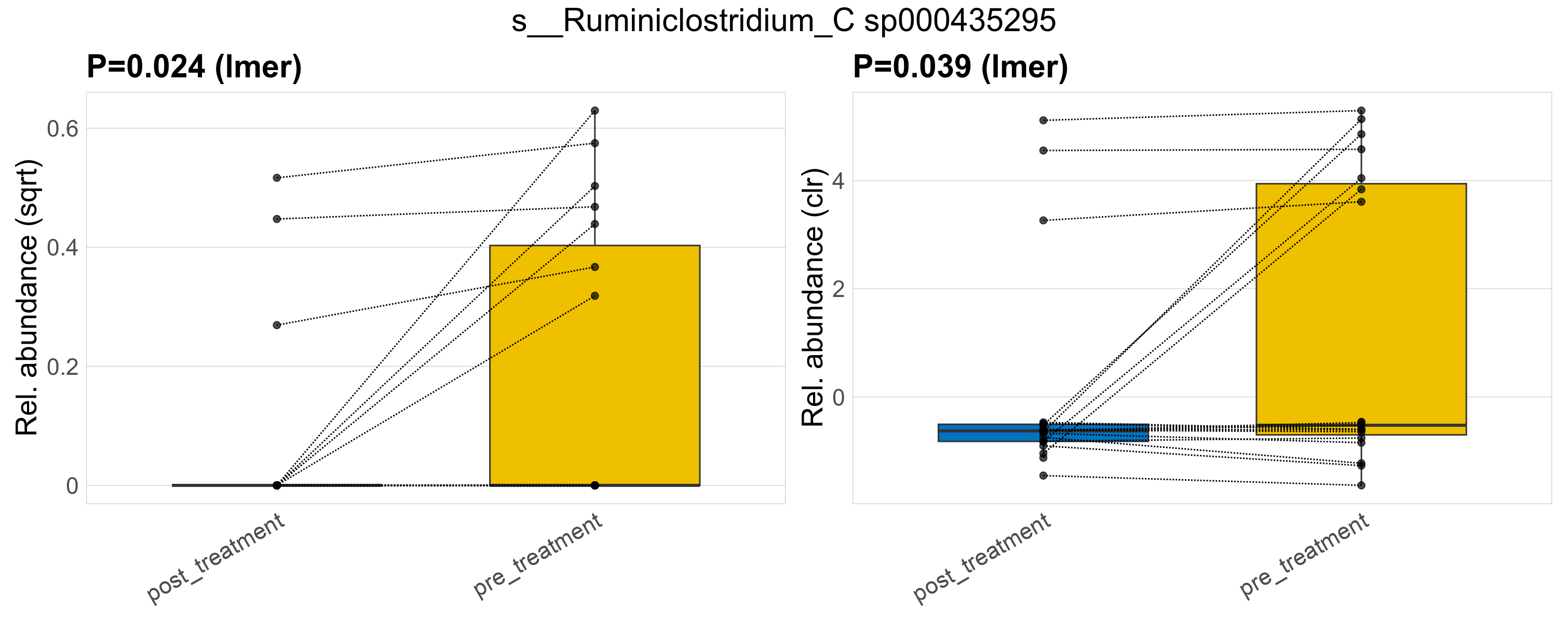
| s__Ruminiclostridium_C sp000435295 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Oscillospiraceae; g__Ruminiclostridium_C | 0.024 | 0.52 | 1 | 0.039 | 0.45 | 1 | 0.22 | 0.057 | 0 | 0.028 | 0 | 0.075 | 0.086 | 0 | 0.13 | 1.6 | 10 / 38 (26%) | 3 / 19 (16%) | 7 / 19 (37%) | 0.158 | 0.368 |
| s__Ruminiclostridium_E MIC9645 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Ruminococcaceae; g__Ruminiclostridium_E | 0.19 | 0.62 | 1 | 0.34 | 0.66 | 1 | 1.5 | 0.12 | 0 | 0.041 | 0 | 0.18 | 0.2 | 0 | 0.59 | 2.3 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| s__Ruminococcus_A sp003011855 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__Ruminococcus_A | 0.8 | 0.94 | 1 | 0.75 | 0.85 | 1 | 0.93 | 0.37 | 0 | 0.36 | 0 | 0.64 | 0.38 | 0 | 0.83 | 0.078 | 15 / 38 (39%) | 8 / 19 (42%) | 7 / 19 (37%) | 0.421 | 0.368 |
| s__Ruminococcus_C callidus | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Ruminococcaceae; g__Ruminococcus_C | 0.38 | 0.66 | 1 | 0.42 | 0.68 | 1 | 2 | 0.16 | 0 | 0.11 | 0 | 0.34 | 0.2 | 0 | 0.87 | 0.86 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
| s__Ruminococcus_C sp000433635 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Ruminococcaceae; g__Ruminococcus_C | 0.75 | 0.89 | 1 | 0.75 | 0.85 | 1 | 0.77 | 0.14 | 0 | 0.13 | 0 | 0.34 | 0.15 | 0 | 0.4 | 0.21 | 7 / 38 (18%) | 3 / 19 (16%) | 4 / 19 (21%) | 0.158 | 0.211 |
| s__Ruminococcus_C sp000980705 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Ruminococcaceae; g__Ruminococcus_C | 0.43 | 0.69 | 1 | 0.45 | 0.7 | 1 | 0.98 | 0.077 | 0 | 0.12 | 0 | 0.36 | 0.036 | 0 | 0.16 | -1.7 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
| s__Ruminococcus_D bicirculans | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Ruminococcaceae; g__Ruminococcus_D | 0.64 | 0.82 | 1 | 0.95 | 0.96 | 1 | 2.6 | 1.3 | 0.11 | 1.5 | 0 | 2.6 | 1 | 0.22 | 1.5 | -0.58 | 19 / 38 (50%) | 9 / 19 (47%) | 10 / 19 (53%) | 0.474 | 0.526 |
| s__Ruminococcus_E bromii | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Acutalibacteraceae; g__Ruminococcus_E | 0.88 | 0.97 | 1 | 0.05 | 0.47 | 1 | 3.7 | 0.39 | 0 | 0.4 | 0 | 1.4 | 0.39 | 0 | 1.2 | -0.037 | 4 / 38 (11%) | 2 / 19 (11%) | 2 / 19 (11%) | 0.105 | 0.105 |
| s__Ruminococcus_E bromii_B | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Acutalibacteraceae; g__Ruminococcus_E | 0.1 | 0.62 | 1 | 0.094 | 0.6 | 1 | 4.6 | 2.6 | 0.57 | 2.2 | 0 | 3.5 | 2.9 | 0.73 | 3.8 | 0.4 | 21 / 38 (55%) | 9 / 19 (47%) | 12 / 19 (63%) | 0.474 | 0.632 |
| s__Ruminococcus_E sp900314705 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Acutalibacteraceae; g__Ruminococcus_E | 0.75 | 0.89 | 1 | 0.71 | 0.85 | 1 | 1.6 | 0.13 | 0 | 0.2 | 0 | 0.85 | 0.056 | 0 | 0.17 | -1.8 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| s__Senegalimassilia anaerobia | d__Bacteria; p__Actinobacteriota; c__Coriobacteriia; o__Coriobacteriales; f__Eggerthellaceae; g__Senegalimassilia | 0.38 | 0.66 | 1 | 0.53 | 0.73 | 1 | 0.31 | 0.025 | 0 | 0.019 | 0 | 0.083 | 0.031 | 0 | 0.092 | 0.71 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
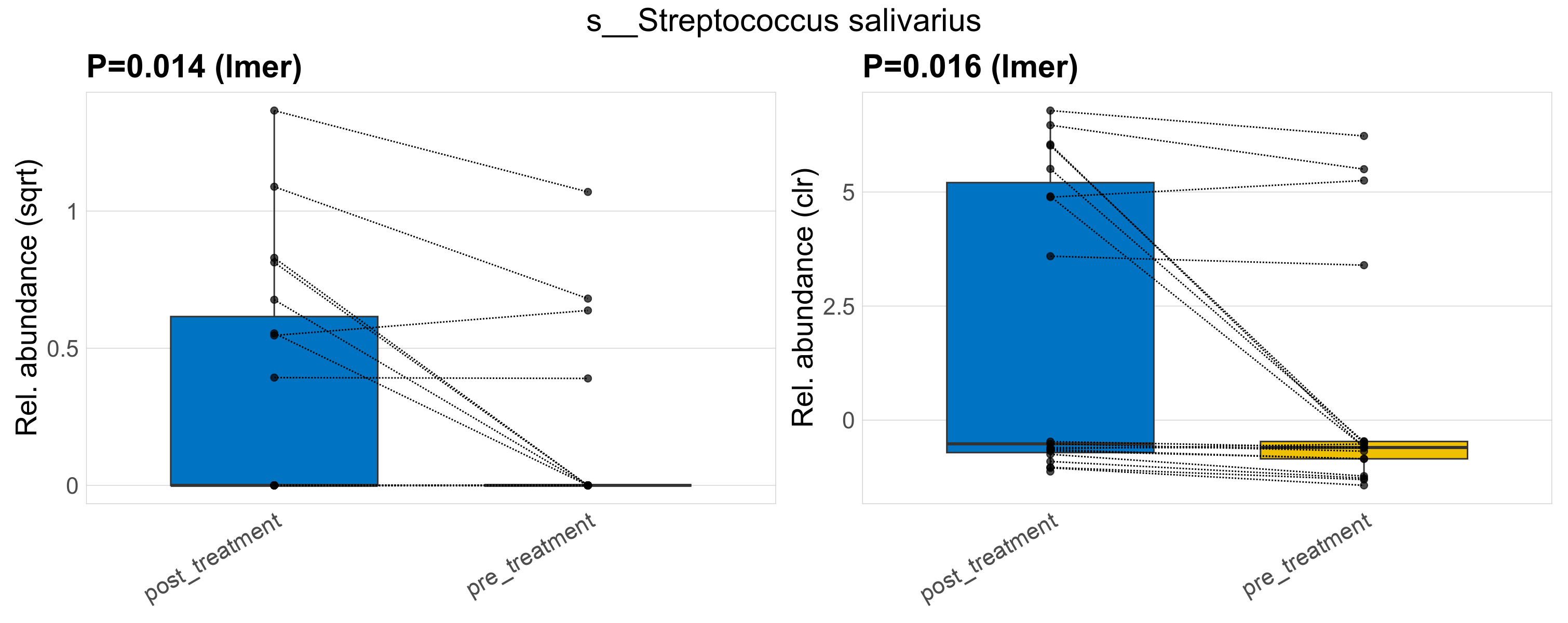
| s__Streptococcus salivarius | d__Bacteria; p__Firmicutes; c__Bacilli; o__Lactobacillales; f__Streptococcaceae; g__Streptococcus | 0.014 | 0.52 | 1 | 0.016 | 0.45 | 1 | 0.65 | 0.2 | 0 | 0.3 | 0 | 0.5 | 0.11 | 0 | 0.29 | -1.4 | 12 / 38 (32%) | 8 / 19 (42%) | 4 / 19 (21%) | 0.421 | 0.211 |
| s__Streptococcus thermophilus | d__Bacteria; p__Firmicutes; c__Bacilli; o__Lactobacillales; f__Streptococcaceae; g__Streptococcus | 0.54 | 0.77 | 1 | 0.88 | 0.92 | 1 | 1.1 | 0.24 | 0 | 0.14 | 0 | 0.46 | 0.33 | 0 | 1 | 1.2 | 8 / 38 (21%) | 4 / 19 (21%) | 4 / 19 (21%) | 0.211 | 0.211 |
| s__TF01-11 sp003149875 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__TF01-11 | 0.37 | 0.66 | 1 | 0.53 | 0.73 | 1 | 0.17 | 0.013 | 0 | 0.0048 | 0 | 0.021 | 0.022 | 0 | 0.065 | 2.2 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| s__TF01-11 sp003524945 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__TF01-11 | 0.27 | 0.62 | 1 | 0.16 | 0.6 | 1 | 0.19 | 0.015 | 0 | 0.025 | 0 | 0.086 | 0.0047 | 0 | 0.021 | -2.4 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
| s__Turicibacter sp001543345 | d__Bacteria; p__Firmicutes; c__Bacilli; o__Haloplasmatales; f__Turicibacteraceae; g__Turicibacter | 0.38 | 0.66 | 1 | 0.52 | 0.72 | 1 | 0.22 | 0.017 | 0 | 0.0079 | 0 | 0.034 | 0.027 | 0 | 0.096 | 1.8 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| s__UBA11524 sp000437595 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Christensenellales; f__CAG-74; g__UBA11524 | 0.23 | 0.62 | 1 | 0.43 | 0.7 | 1 | 1.5 | 0.12 | 0 | 0.082 | 0 | 0.36 | 0.15 | 0 | 0.5 | 0.87 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| s__UBA11774 sp003507655 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__UBA11774 | 0.81 | 0.94 | 1 | 0.31 | 0.62 | 1 | 1.3 | 0.37 | 0 | 0.3 | 0 | 0.6 | 0.44 | 0 | 1.1 | 0.55 | 11 / 38 (29%) | 6 / 19 (32%) | 5 / 19 (26%) | 0.316 | 0.263 |
| s__UBA1191 MIC6696 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Peptostreptococcales; f__Anaerovoracaceae; g__UBA1191 | 0.58 | 0.78 | 1 | 0.55 | 0.73 | 1 | 0.58 | 0.046 | 0 | 0.049 | 0 | 0.21 | 0.042 | 0 | 0.15 | -0.22 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| s__UBA1394 sp900066845 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Ruminococcaceae; g__UBA1394 | 0.65 | 0.82 | 1 | 0.81 | 0.89 | 1 | 0.46 | 0.06 | 0 | 0.082 | 0 | 0.25 | 0.039 | 0 | 0.1 | -1.1 | 5 / 38 (13%) | 2 / 19 (11%) | 3 / 19 (16%) | 0.105 | 0.158 |
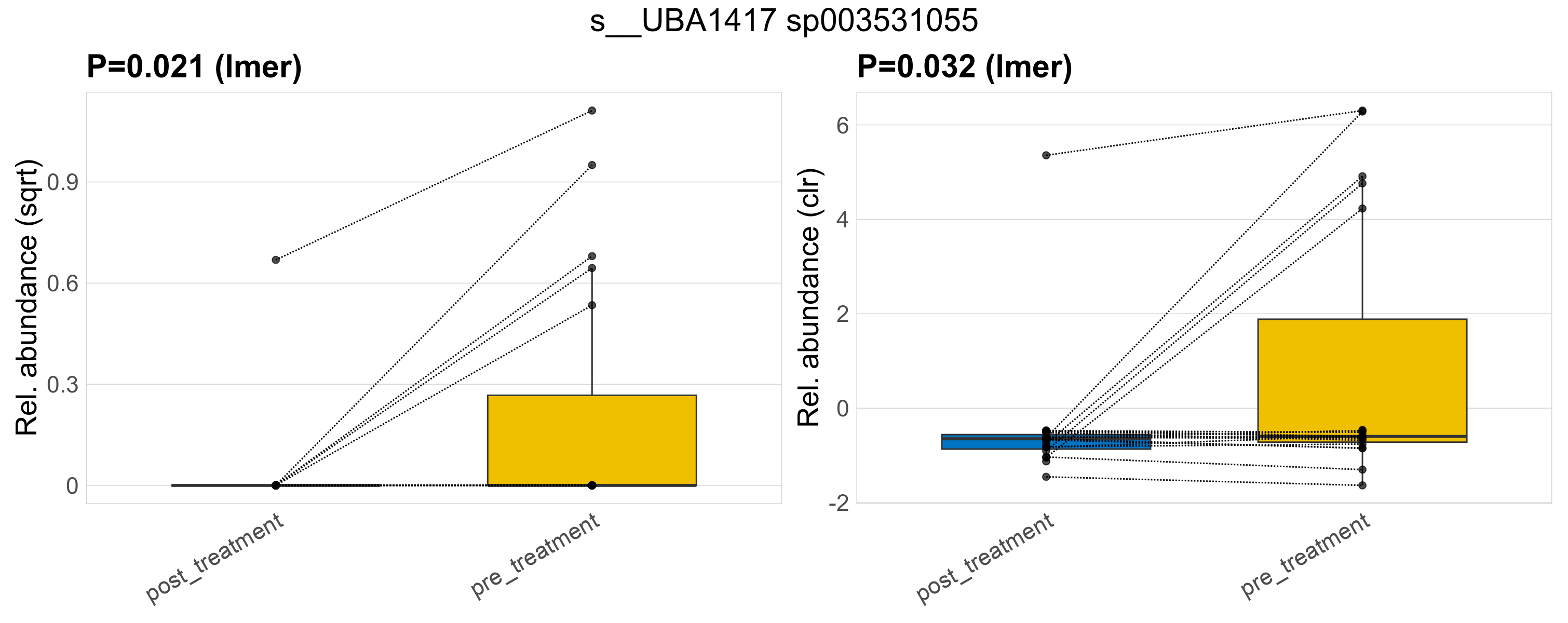
| s__UBA1417 sp003531055 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Acutalibacteraceae; g__UBA1417 | 0.021 | 0.52 | 1 | 0.032 | 0.45 | 1 | 0.62 | 0.099 | 0 | 0.024 | 0 | 0.1 | 0.17 | 0 | 0.35 | 2.8 | 6 / 38 (16%) | 1 / 19 (5.3%) | 5 / 19 (26%) | 0.0526 | 0.263 |
| s__UBA1691 MIC9213 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Acutalibacteraceae; g__UBA1691 | 0.21 | 0.62 | 1 | 0.58 | 0.74 | 1 | 1 | 0.11 | 0 | 0.086 | 0 | 0.31 | 0.13 | 0 | 0.4 | 0.6 | 4 / 38 (11%) | 2 / 19 (11%) | 2 / 19 (11%) | 0.105 | 0.105 |
| s__UBA1777 MIC6732 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Oscillospirales; f__Oscillospiraceae; g__UBA1777 | 0.24 | 0.62 | 1 | 0.39 | 0.68 | 1 | 1.2 | 0.093 | 0 | 0.047 | 0 | 0.2 | 0.14 | 0 | 0.41 | 1.6 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| s__UBA7160 MIC6745 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__UBA7160 | 0.88 | 0.97 | 1 | 0.72 | 0.85 | 1 | 0.51 | 0.12 | 0 | 0.11 | 0 | 0.29 | 0.13 | 0 | 0.34 | 0.24 | 9 / 38 (24%) | 5 / 19 (26%) | 4 / 19 (21%) | 0.263 | 0.211 |
| s__UBA7160 MIC9207 | d__Bacteria; p__Firmicutes_A; c__Clostridia; o__Lachnospirales; f__Lachnospiraceae; g__UBA7160 | 0.53 | 0.77 | 1 | 0.82 | 0.89 | 1 | 0.12 | 0.013 | 0 | 0.012 | 0 | 0.037 | 0.013 | 0 | 0.041 | 0.12 | 4 / 38 (11%) | 2 / 19 (11%) | 2 / 19 (11%) | 0.105 | 0.105 |
Click here to open full-sized image in new window.
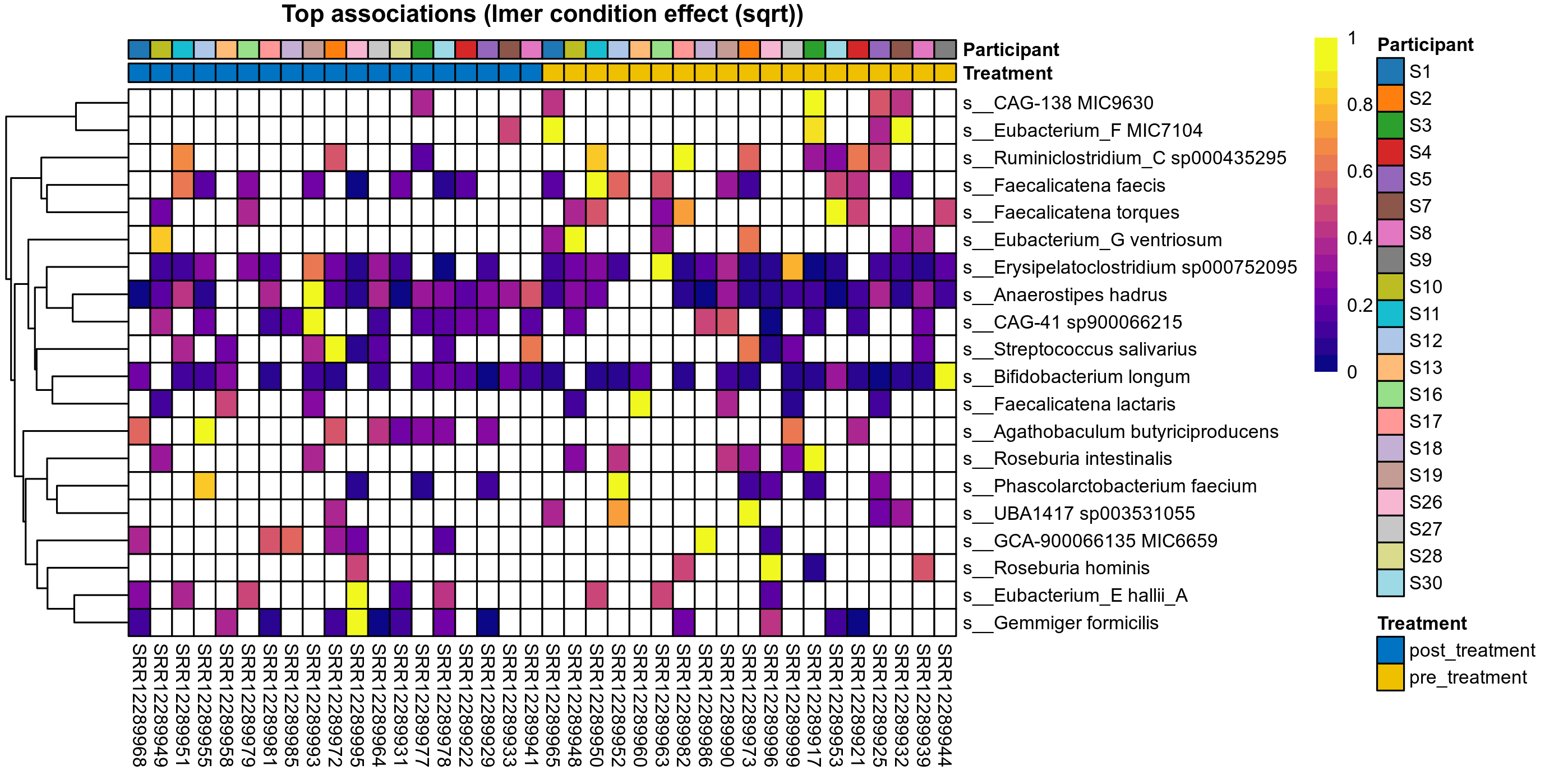
Click here to open full-sized image in new window.
P lmer condition effect (sqrt): Differentially abundant species were identified by linear mixed effect regression without confounders. Random effects: Participant (intercept). Fixed effects: Treatment. Effect tested: Treatment.
P lmer condition effect (clr): Differentially abundant species were identified by linear mixed effect regression without confounders. Random effects: Participant (intercept). Fixed effects: Treatment. Effect tested: Treatment.
The following plots present the distribution of the top most differentially abundant species across all applied statistical analysis. Plots are ordered alphabetically.