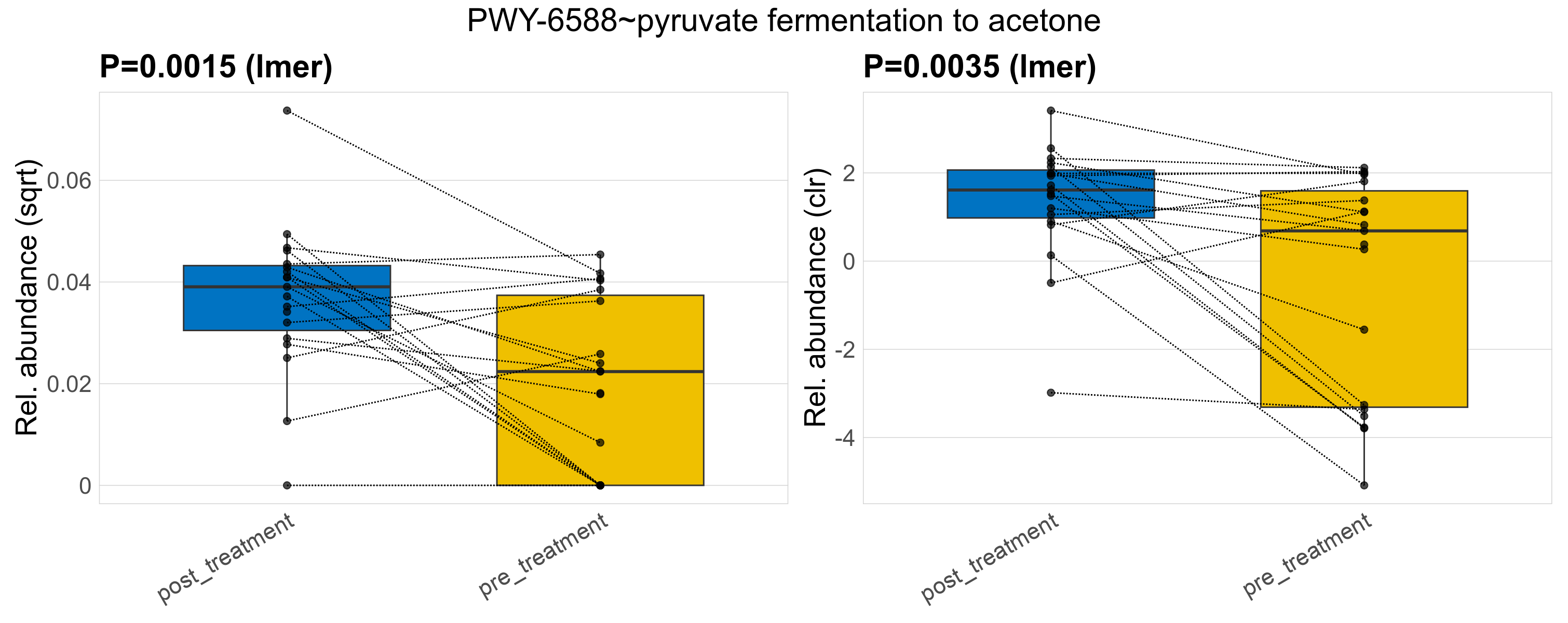
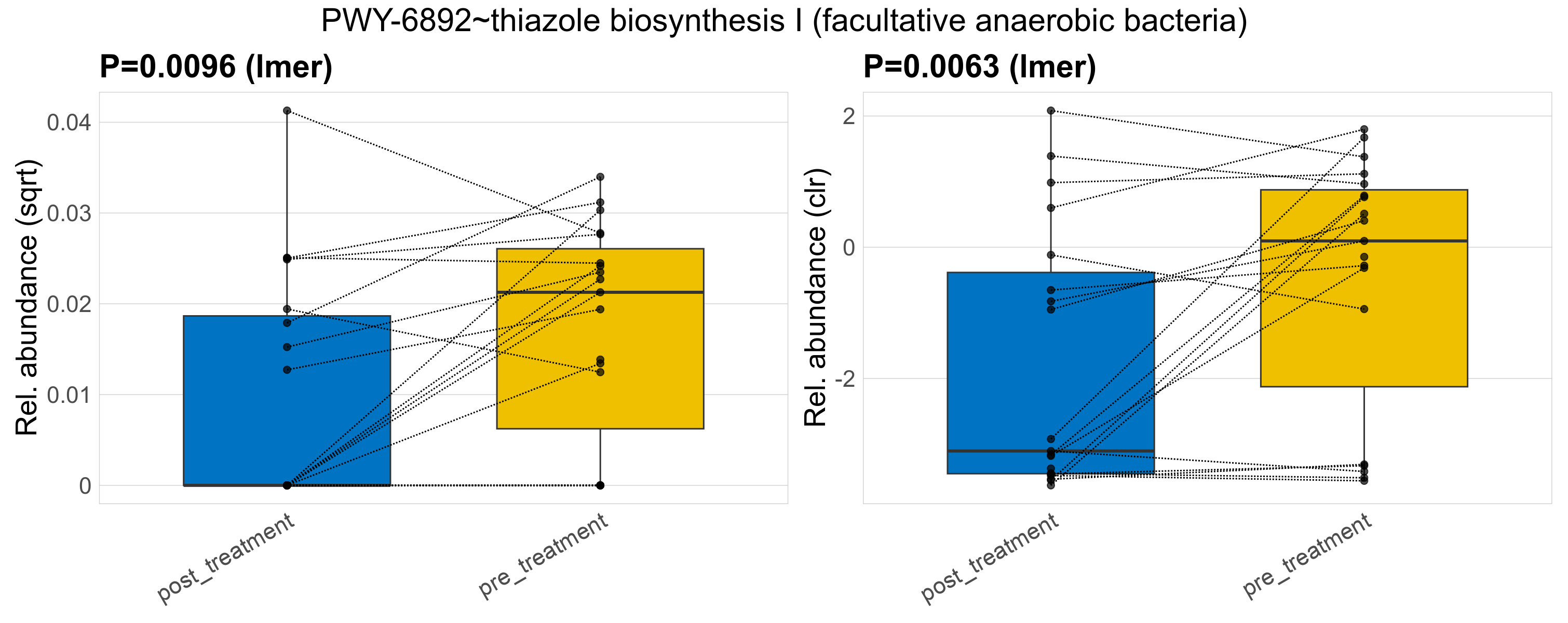
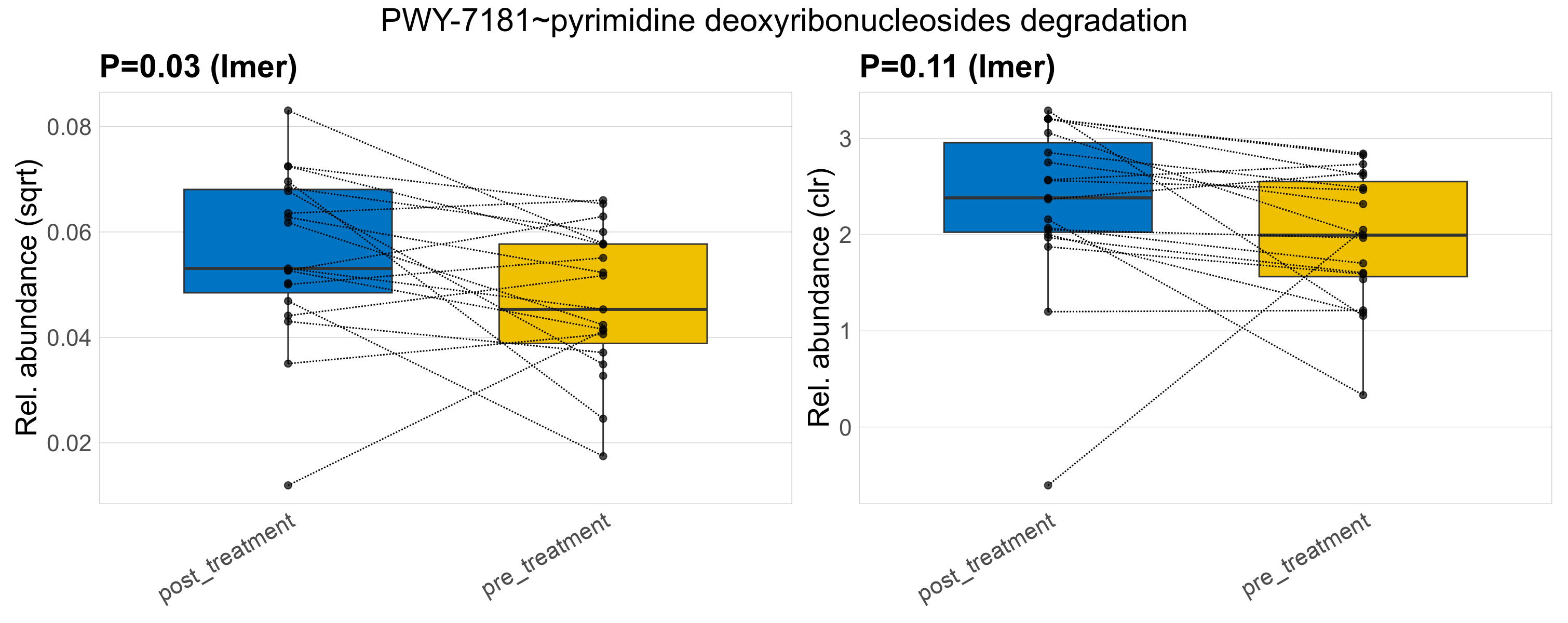
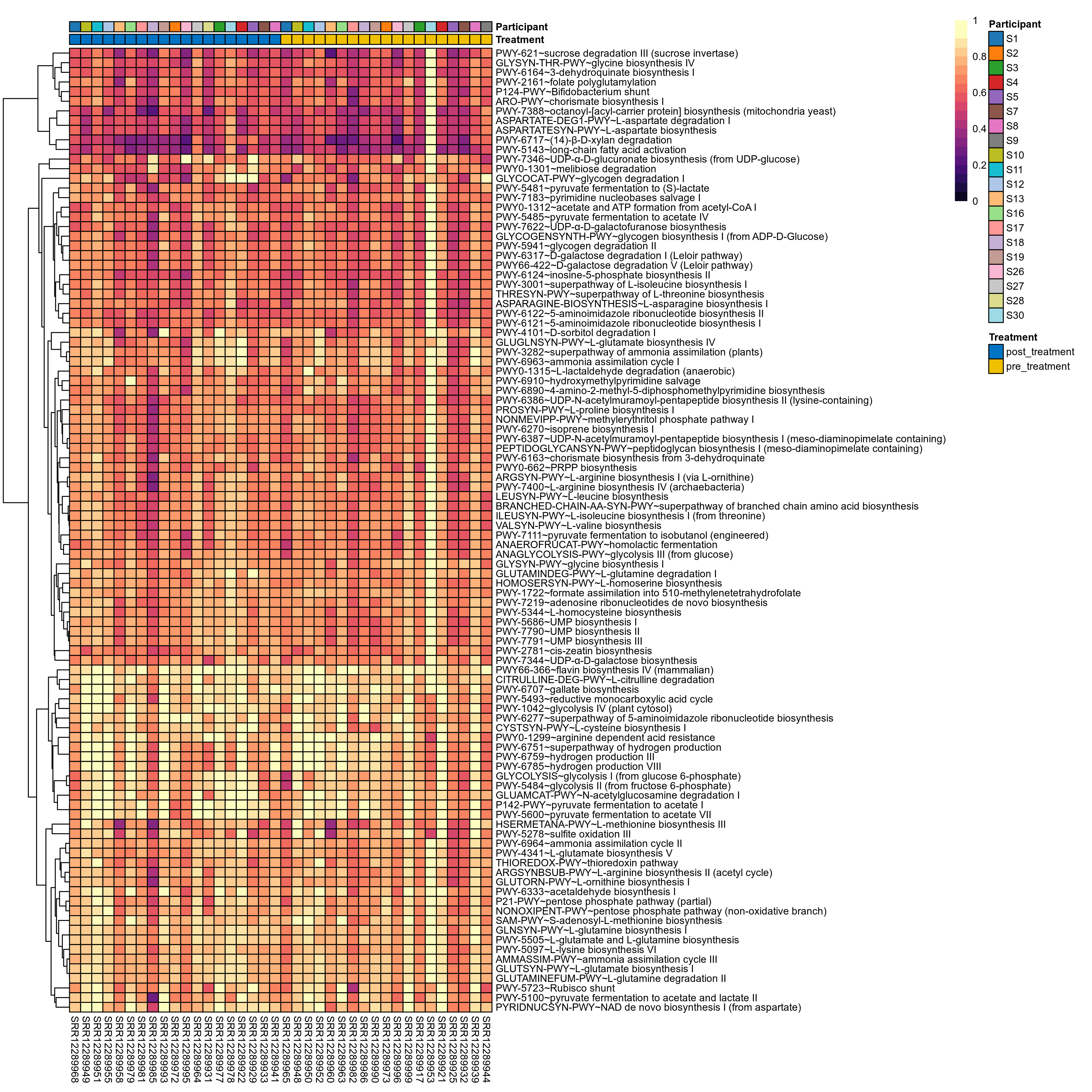
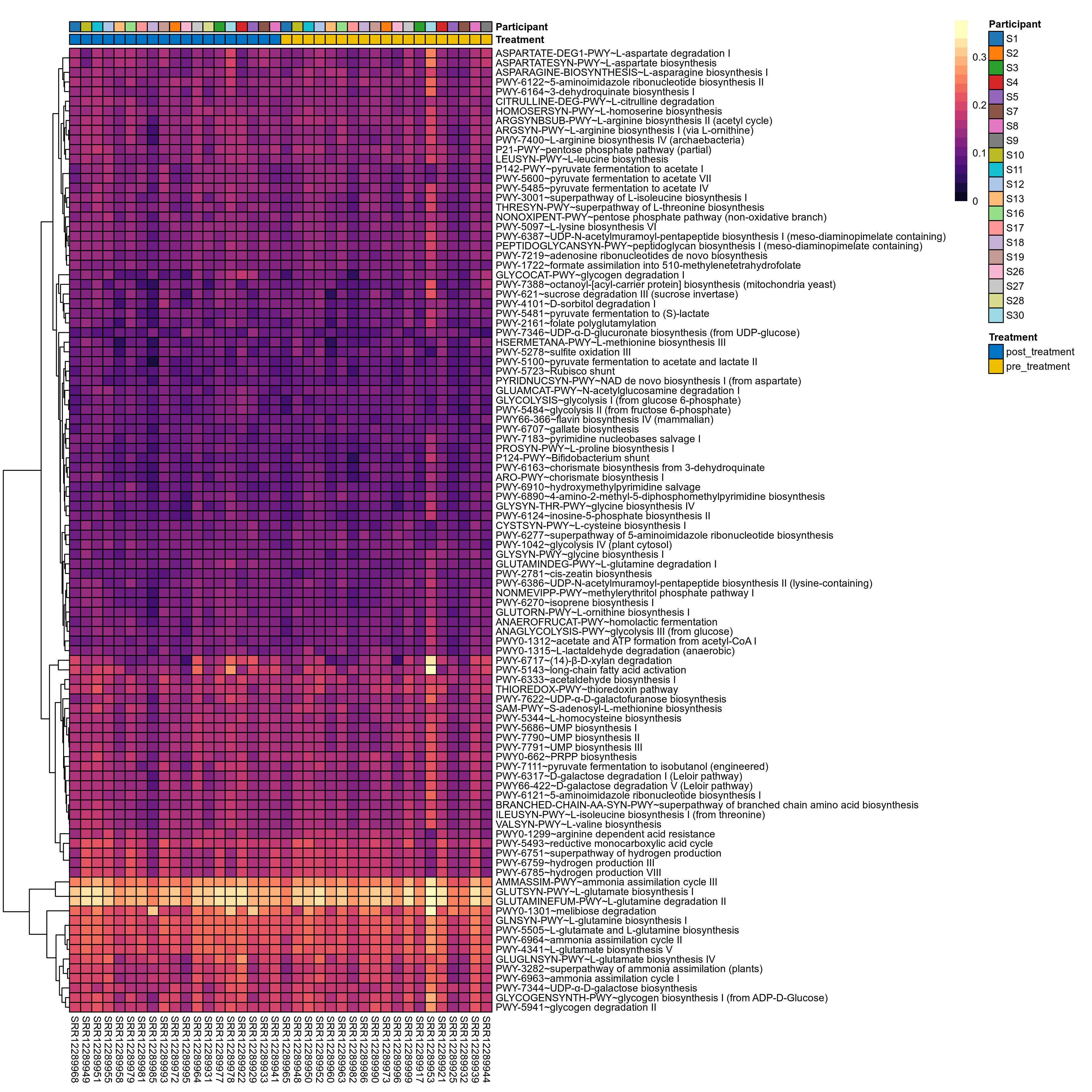
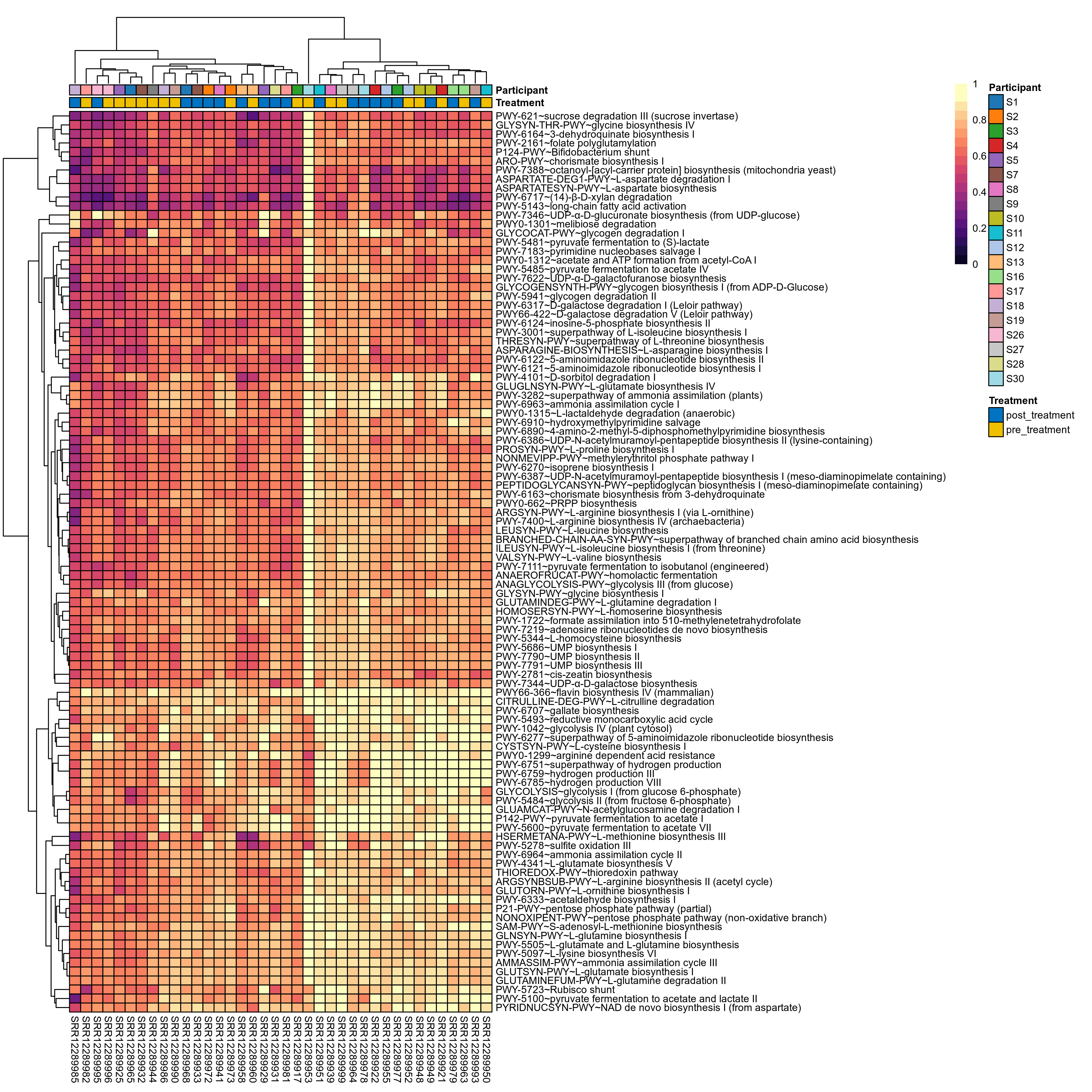
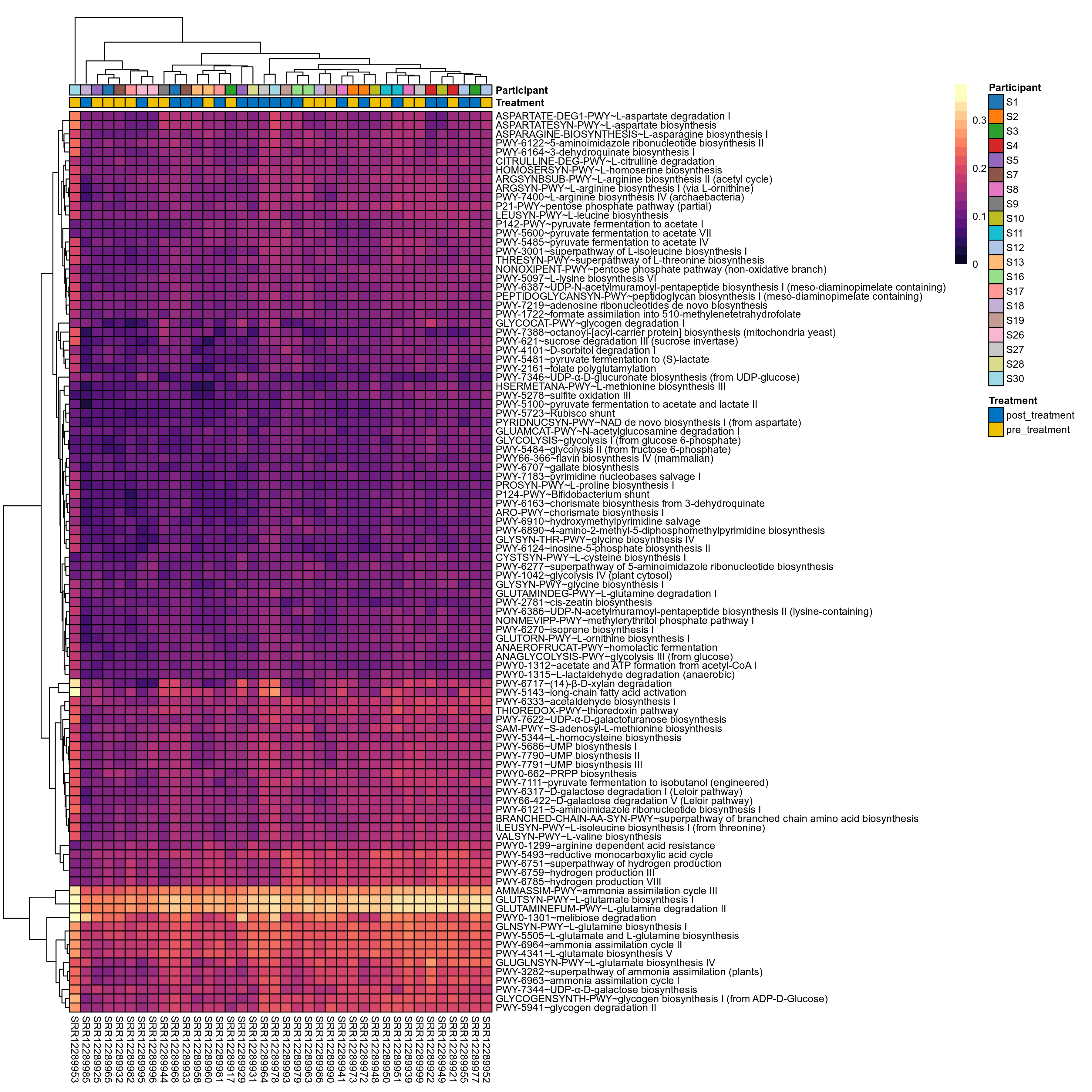
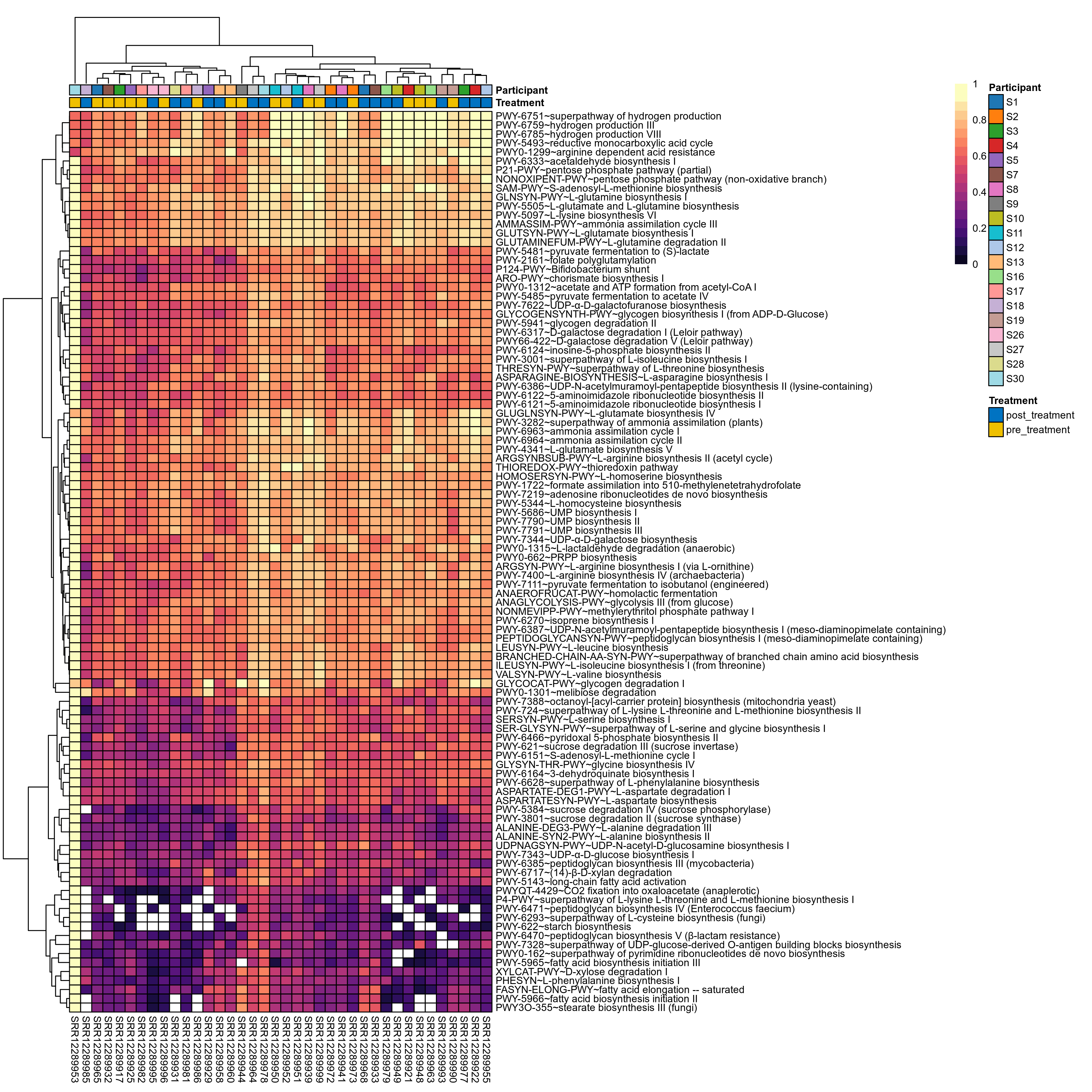
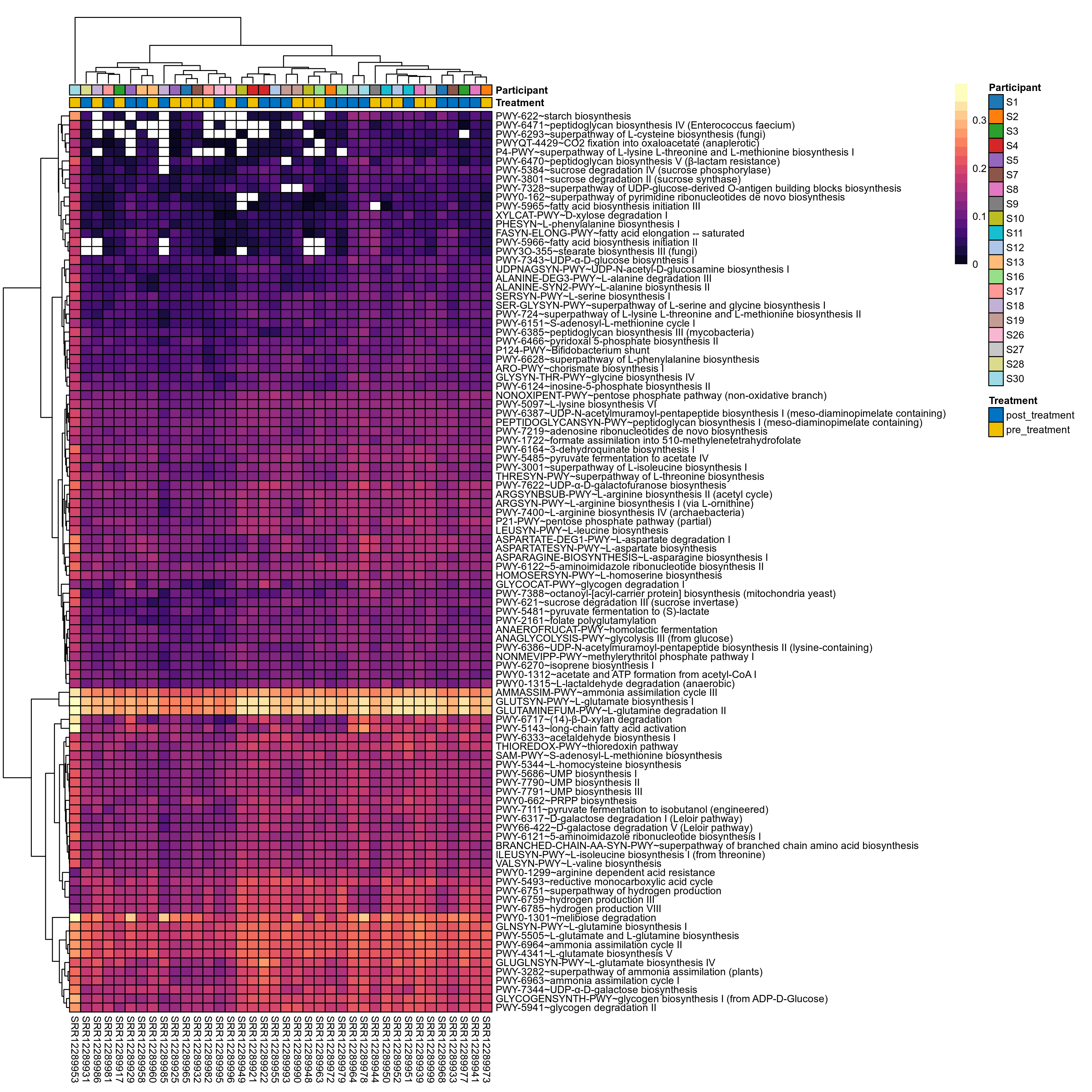
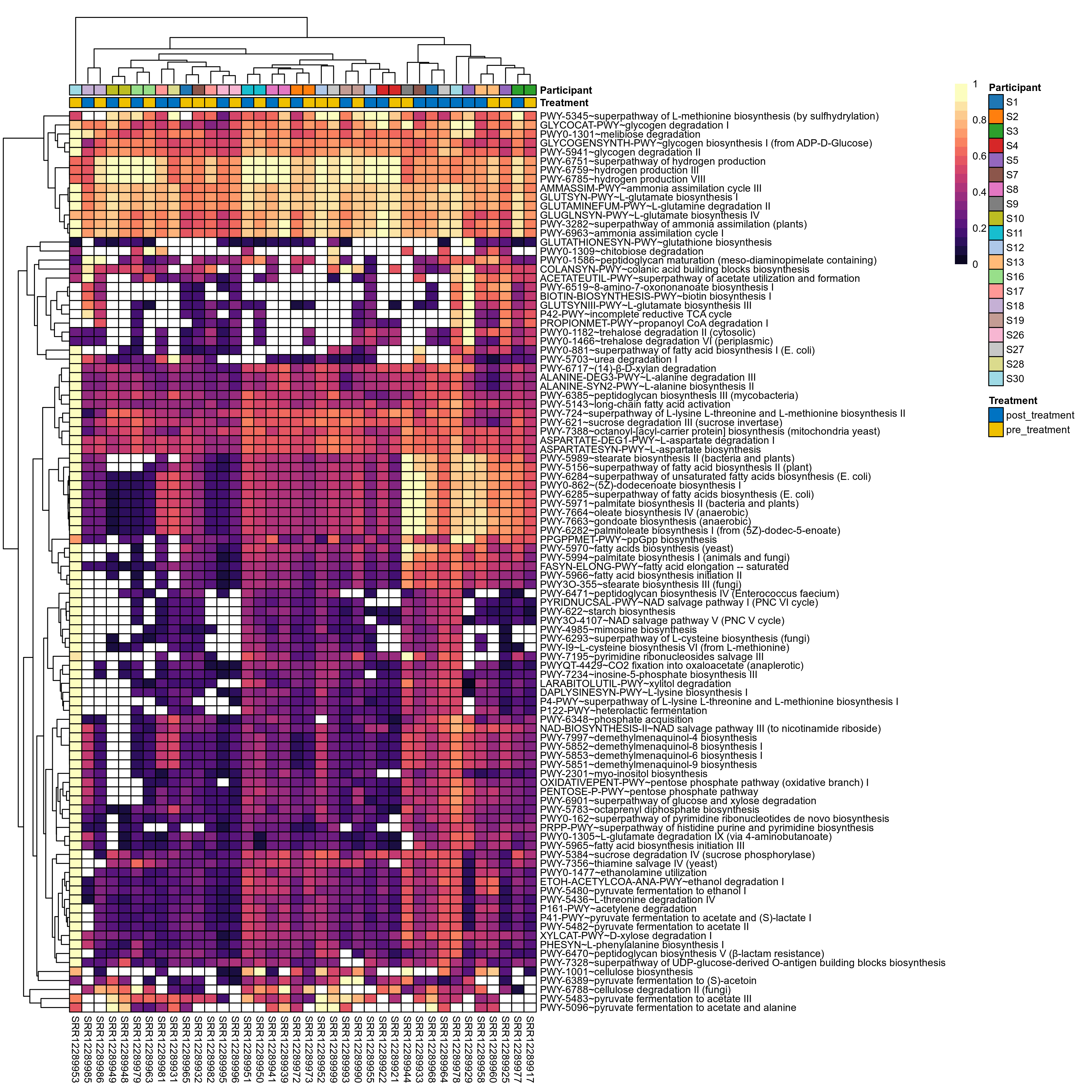
Quantitative visualisation of the top most abundant microbial functions identified in the analysed samples.
Click here to open full-sized image in new window.
Click here to open full-sized image in new window.
Click here to open full-sized image in new window.
Click here to open full-sized image in new window.
Click here to open full-sized image in new window.
Click here to open full-sized image in new window.
Click here to open full-sized image in new window.
Click here to open full-sized image in new window.
Click here to open full-sized image in new window.
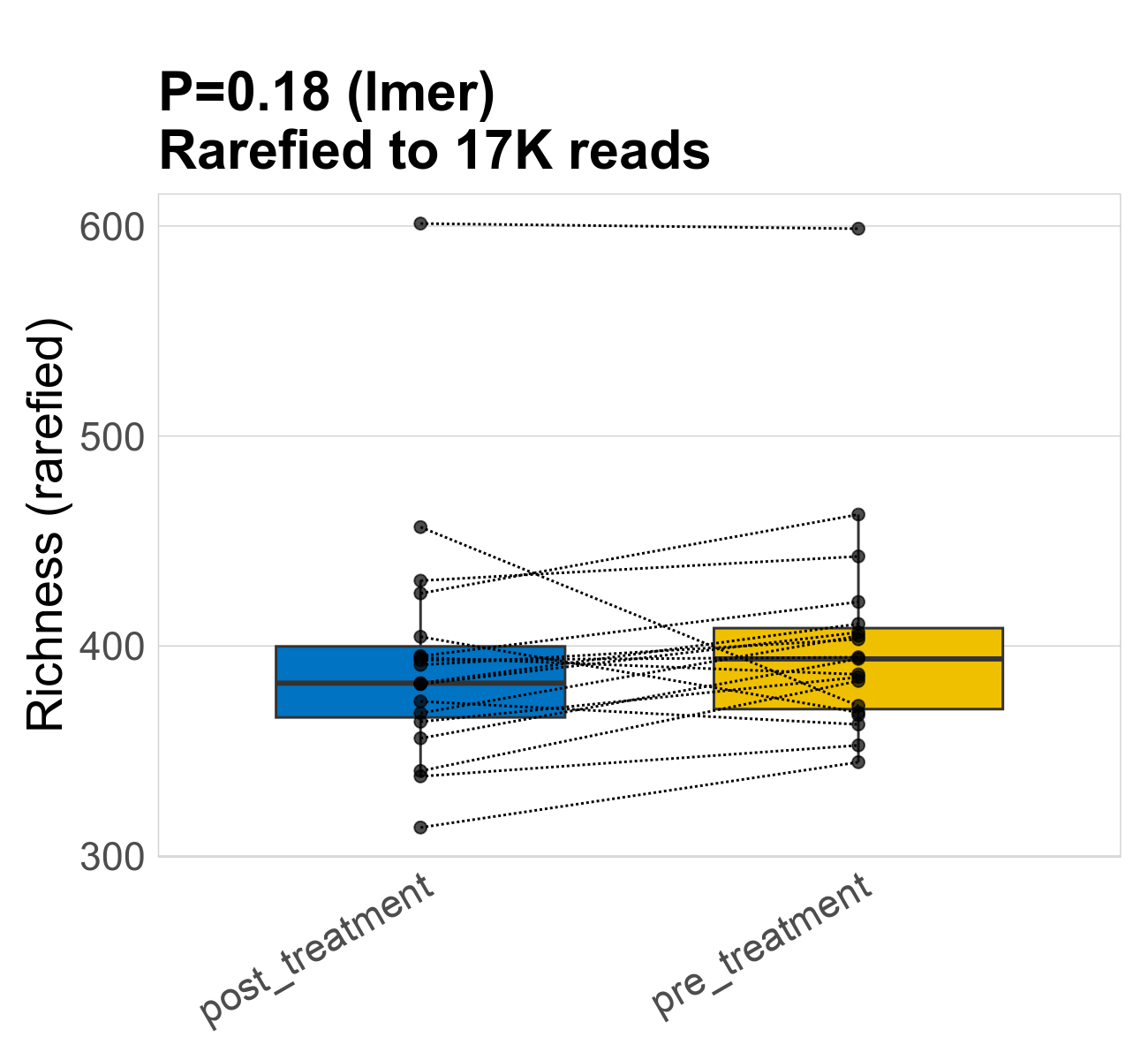
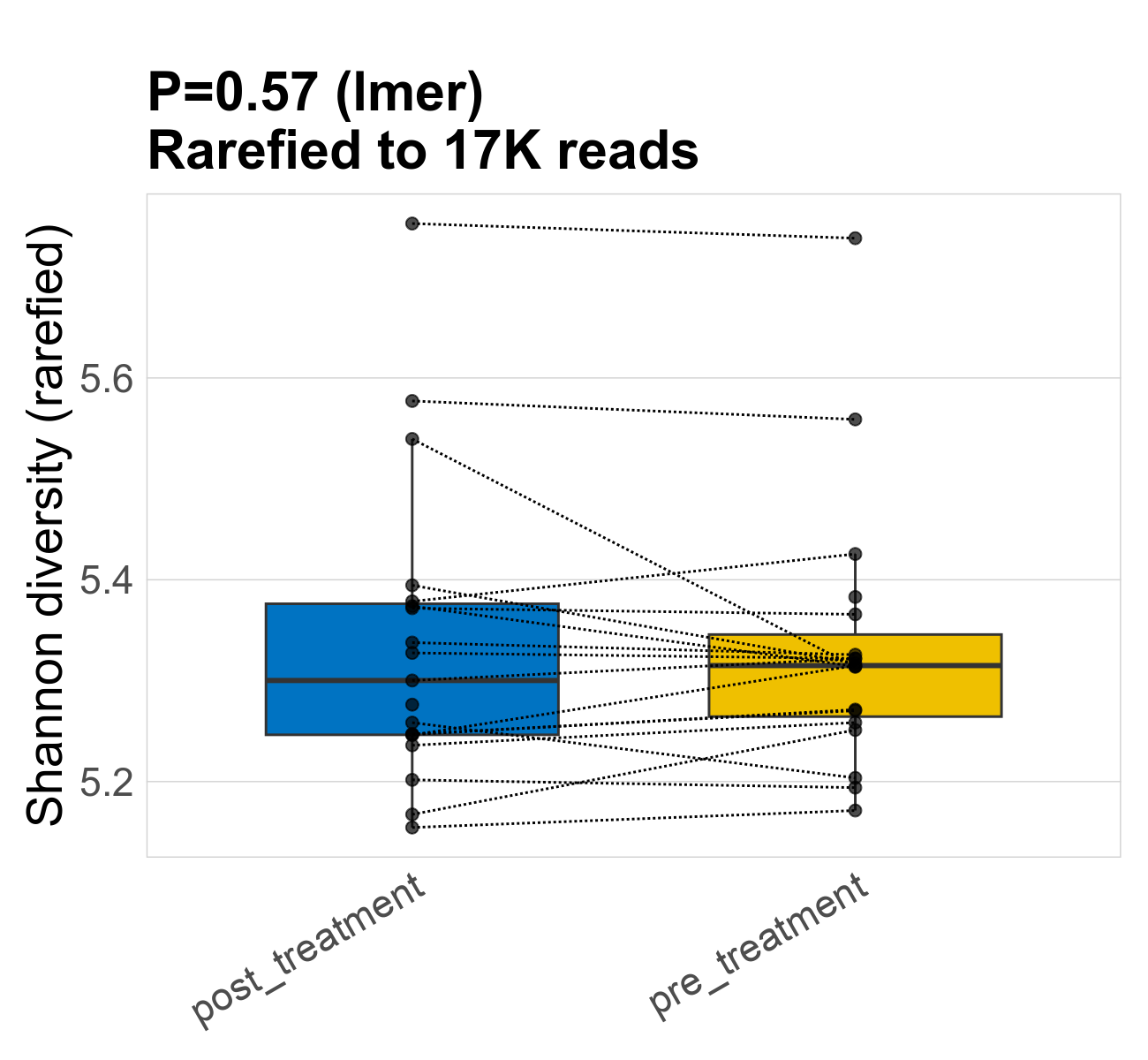
This page provides an overview of the functional alpha diversity of the analysed sample. Alpha diversity was measured using the Shannon index and functional richnes. Richness measures the total number of gene functions present in each sample. Shannon index combines richness and evenness.
| Index | rarefiedTo | P lmer condition effect | Mean Pos | Mean Abundance | Median Abundance | Mean Treatmentpost_treatment | Median Treatmentpost_treatment | SD Treatmentpost_treatment | Mean Treatmentpre_treatment | Median Treatmentpre_treatment | SD Treatmentpre_treatment | Fold Change Log2(Treatmentpre_treatment/Treatmentpost_treatment) | Positive samples | Positive Treatmentpost_treatment | Positive Treatmentpre_treatment | Positive_Treatmentpost_treatment_percent | Positive_Treatmentpre_treatment_percent |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Shannon | 16649 | 0.57 | 5.3 | 5.3 | 5.3 | 5.3 | 5.3 | 0.15 | 5.3 | 5.3 | 0.13 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| Richness | 16649 | 0.18 | 400 | 400 | 390 | 390 | 380 | 60 | 400 | 390 | 56 | 0.037 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
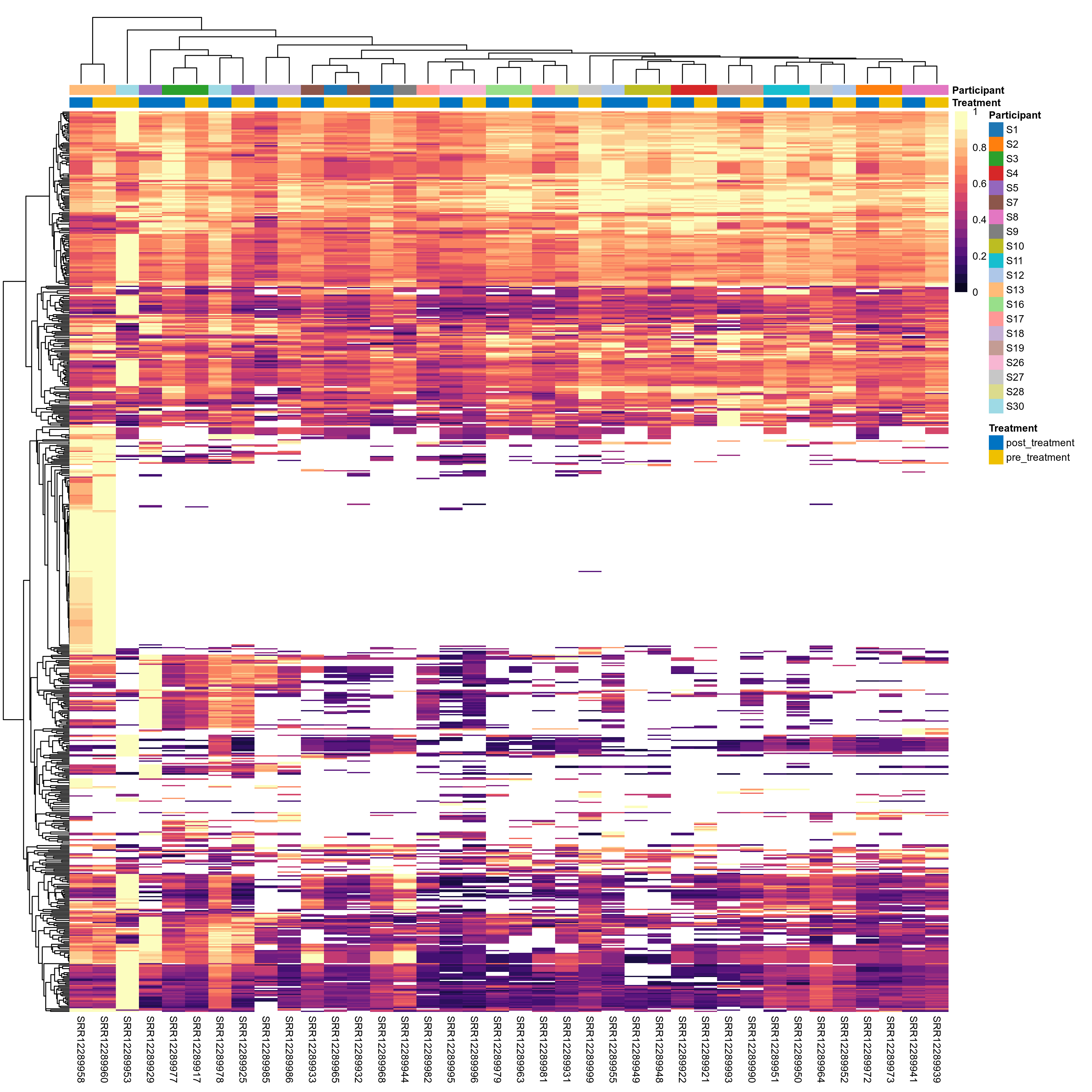
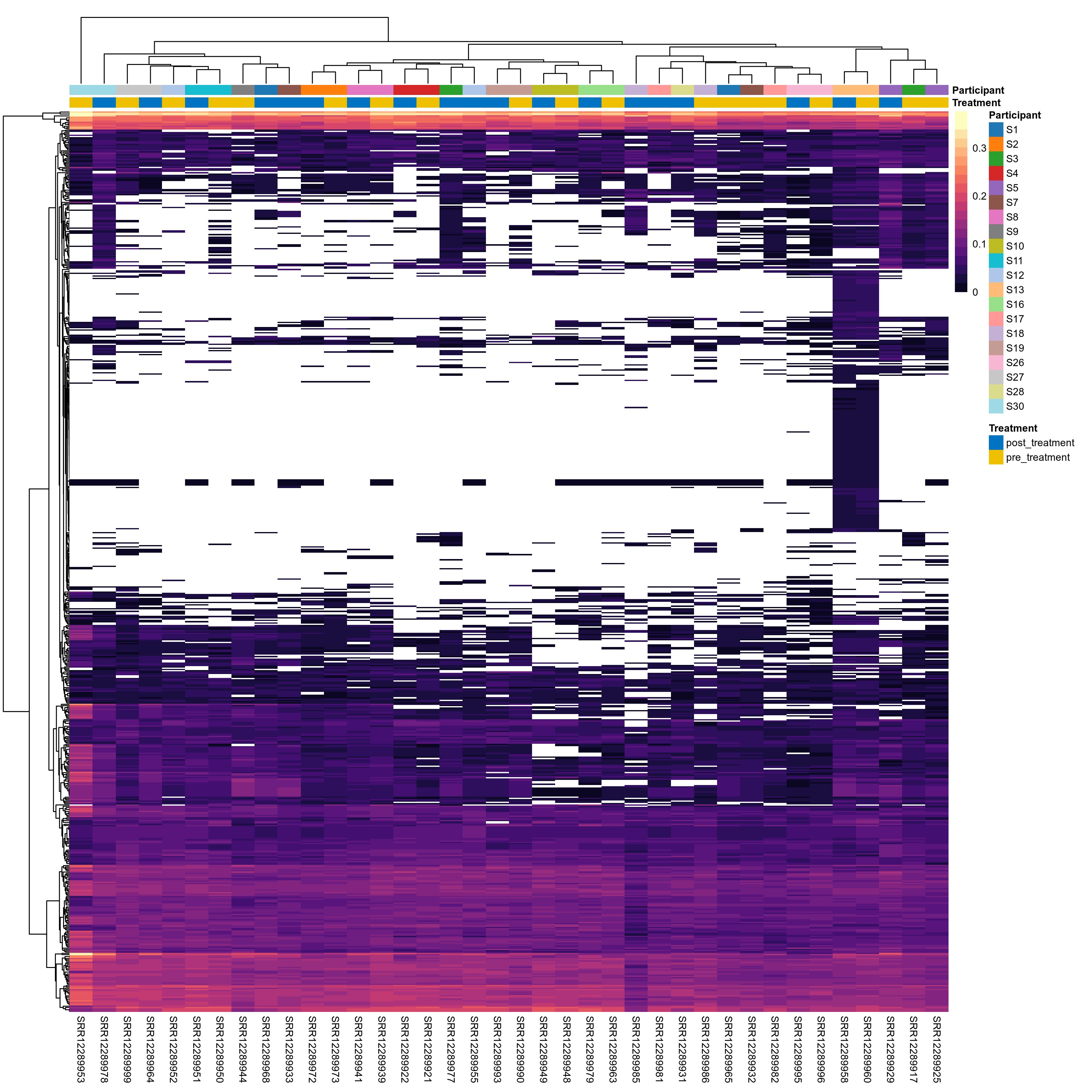
Microbial functions were analyzed using supervised and unsupervised multivariate techniques. Functional profiles were ordinated using the unsupervised methods Principal Coordinates Analysis (PCoA), Non-Metric Multidimensional Scaling (NMDS) and Principal Component Analysis (PCA). The supervised methods Adonis and Redundancy analysis (RDA) were used to assess if variance in the functional profiles was significantly associated with the study condition. A guide explaining these methods can be found here. Sparse Partial Least Square Discriminant Analysis (sPLS-DA) from the MixMc package was additionaly used to extract features associated with the condition of interest.
Differentially abundant microbial functions were identified using the univariate methods ANOVA or LMER (linear mixed effect regression) on clr transformed relative abundance data, Fisher's exact test, and Aldex2 (ANOVA-like Differential Expression). Aldex2 was run on read count data. Fisher's exact test was used to test for diffrerences in the presence and absence (detection rate) of microbial functions across study groups.
LMER is used for repeated measures data, using random effects to control for correlation between samples from the same subject. Fixed effects are included for treatment groups, time, and treatment over time, where appropriate. The LMER P values correspond to a nested model test of the significance of including the corresponding fixed effect.
ALDEx2 uses subsampling (Bayesian sampling) to estimate the underlying technical variation. For each subsample instance, center log-ratio transformed data is statistically compared across study groups and computed P values are corrected for multiple testing using the Benjamini–Hochberg procedure. The expected P value (mean P value) is reported, which are those that would likely have been observed if the same samples had been run multiple times. The expected values are reported for both the distribution of P values and for the distribution of Benjamini–Hochberg corrected values.
| Function | P lmer condition effect (sqrt) | FDR lmer condition effect (sqrt) | Pbonf lmer condition effect (sqrt) | P lmer condition effect (clr) | FDR lmer condition effect (clr) | Pbonf lmer condition effect (clr) | Mean Pos | Mean Abundance | Median Abundance | Mean Treatmentpost_treatment | Median Treatmentpost_treatment | SD Treatmentpost_treatment | Mean Treatmentpre_treatment | Median Treatmentpre_treatment | SD Treatmentpre_treatment | Fold Change Log2(Treatmentpre_treatment/Treatmentpost_treatment) | Positive samples | Positive Treatmentpost_treatment | Positive Treatmentpre_treatment | Positive_Treatmentpost_treatment_percent | Positive_Treatmentpre_treatment_percent |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PWY-5481~pyruvate fermentation to (S)-lactate | 0.37 | 0.97 | 1 | 0.75 | 0.98 | 1 | 0.013 | 0.013 | 0.014 | 0.012 | 0.014 | 0.0035 | 0.014 | 0.014 | 0.0052 | 0.22 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| P108-PWY~pyruvate fermentation to propanoate I | 0.89 | 0.98 | 1 | 0.48 | 0.91 | 1 | 0.002 | 0.00075 | 0 | 0.00089 | 0 | 0.0023 | 0.00061 | 0 | 0.0011 | -0.54 | 14 / 38 (37%) | 6 / 19 (32%) | 8 / 19 (42%) | 0.316 | 0.421 |
| CITRULLINE-DEG-PWY~L-citrulline degradation | 0.45 | 0.97 | 1 | 0.32 | 0.91 | 1 | 0.019 | 0.019 | 0.019 | 0.02 | 0.02 | 0.0041 | 0.019 | 0.019 | 0.0032 | -0.074 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5084~2-oxoglutarate decarboxylation to succinyl-CoA | 0.29 | 0.97 | 1 | 0.47 | 0.91 | 1 | 0.0016 | 0.00017 | 0 | 0.00014 | 0 | 6e-04 | 0.00019 | 0 | 0.00064 | 0.44 | 4 / 38 (11%) | 2 / 19 (11%) | 2 / 19 (11%) | 0.105 | 0.105 |
| PWY-7183~pyrimidine nucleobases salvage I | 0.29 | 0.97 | 1 | 0.65 | 0.96 | 1 | 0.011 | 0.011 | 0.011 | 0.011 | 0.01 | 0.0025 | 0.012 | 0.011 | 0.0035 | 0.13 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5026~indole-3-acetate biosynthesis V (bacteria and fungi) | 0.73 | 0.97 | 1 | 0.83 | 0.99 | 1 | 0.00054 | 0.00031 | 0.00012 | 3e-04 | 0.00012 | 0.00037 | 0.00032 | 0.00012 | 0.00058 | 0.093 | 22 / 38 (58%) | 10 / 19 (53%) | 12 / 19 (63%) | 0.526 | 0.632 |
| PWY-6385~peptidoglycan biosynthesis III (mycobacteria) | 0.86 | 0.97 | 1 | 0.69 | 0.96 | 1 | 0.0098 | 0.0098 | 0.008 | 0.0098 | 0.0096 | 0.0042 | 0.0098 | 0.0074 | 0.0083 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| COBALSYN-PWY~superpathway of adenosylcobalamin salvage from cobinamide I | 0.65 | 0.97 | 1 | 0.5 | 0.93 | 1 | 0.0048 | 0.0048 | 0.0045 | 0.0049 | 0.0048 | 0.0019 | 0.0046 | 0.0044 | 0.0014 | -0.091 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5653~NAD biosynthesis from 2-amino-3-carboxymuconate semialdehyde | 0.46 | 0.97 | 1 | 0.24 | 0.91 | 1 | 0.0093 | 0.0093 | 0.0094 | 0.0095 | 0.0094 | 0.0021 | 0.0091 | 0.0094 | 0.0026 | -0.062 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| ARABCAT-PWY~L-arabinose degradation I | 0.66 | 0.97 | 1 | 0.59 | 0.94 | 1 | 0.0041 | 0.0041 | 0.0029 | 0.0042 | 0.0032 | 0.0028 | 0.0041 | 0.0029 | 0.0045 | -0.035 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| SERSYN-PWY~L-serine biosynthesis I | 0.62 | 0.97 | 1 | 0.97 | 0.99 | 1 | 0.011 | 0.011 | 0.0095 | 0.01 | 0.0095 | 0.0033 | 0.011 | 0.0095 | 0.0067 | 0.14 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
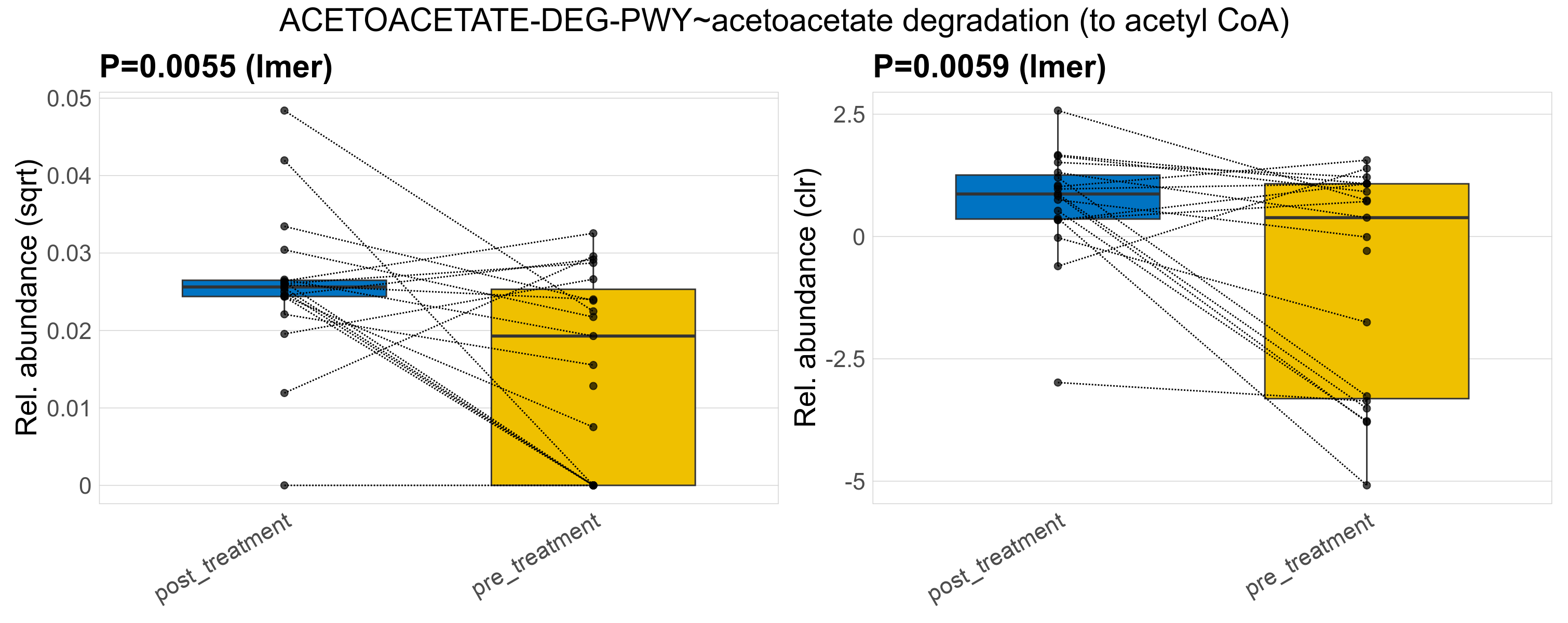
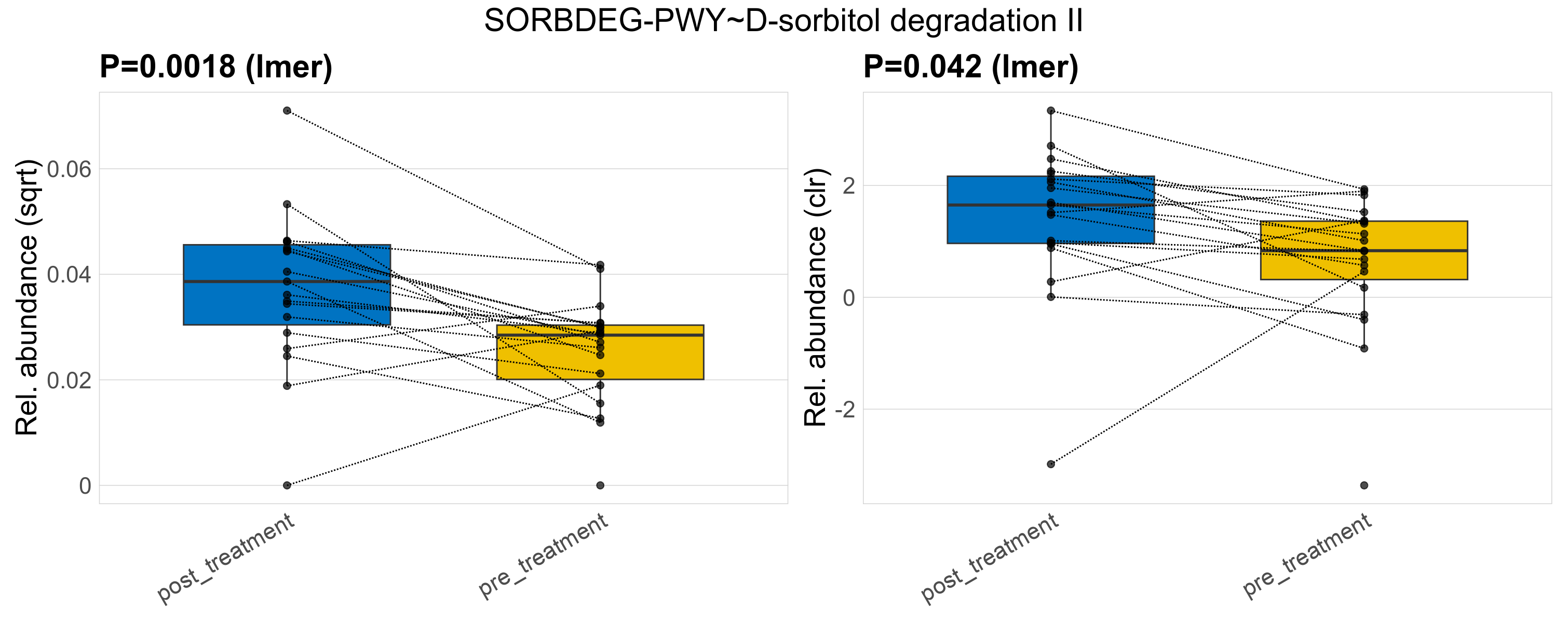
| SORBDEG-PWY~D-sorbitol degradation II | 0.0018 | 0.33 | 0.96 | 0.042 | 0.91 | 1 | 0.0012 | 0.0012 | 0.00092 | 0.0016 | 0.0015 | 0.0011 | 0.00074 | 0.00081 | 0.00047 | -1.1 | 36 / 38 (95%) | 18 / 19 (95%) | 18 / 19 (95%) | 0.947 | 0.947 |
| PWY-5198~factor 420 biosynthesis | 0.87 | 0.97 | 1 | 0.97 | 0.99 | 1 | 0.00022 | 2.3e-05 | 0 | 2.1e-05 | 0 | 7.3e-05 | 2.5e-05 | 0 | 7.6e-05 | 0.25 | 4 / 38 (11%) | 2 / 19 (11%) | 2 / 19 (11%) | 0.105 | 0.105 |
| NAD-BIOSYNTHESIS-III~NAD biosynthesis III (from nicotinamide) | 0.18 | 0.97 | 1 | 0.25 | 0.91 | 1 | 0.00019 | 1.5e-05 | 0 | 2.4e-06 | 0 | 1.1e-05 | 2.7e-05 | 0 | 8.3e-05 | 3.5 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| PWY-6620~guanine and guanosine salvage | 0.84 | 0.97 | 1 | 0.42 | 0.91 | 1 | 0.0092 | 0.0092 | 0.0095 | 0.0093 | 0.0097 | 0.0016 | 0.0092 | 0.0094 | 0.0023 | -0.016 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-40~putrescine biosynthesis I | 0.78 | 0.97 | 1 | 0.5 | 0.93 | 1 | 0.0058 | 0.0057 | 0.0046 | 0.0056 | 0.0045 | 0.0034 | 0.0057 | 0.0048 | 0.0038 | 0.026 | 37 / 38 (97%) | 18 / 19 (95%) | 19 / 19 (100%) | 0.947 | 1 |
| PWY-7969~5-hydroxybenzimidazolyl adenosylcobamide biosynthesis from adenosylcobinamide-GDP | 0.59 | 0.97 | 1 | 0.39 | 0.91 | 1 | 0.0049 | 0.0049 | 0.0047 | 0.005 | 0.0047 | 0.0018 | 0.0047 | 0.0046 | 0.0014 | -0.089 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-7226~guanosine deoxyribonucleotides de novo biosynthesis I | 0.53 | 0.97 | 1 | 0.25 | 0.91 | 1 | 0.0017 | 0.0014 | 0.00066 | 0.0014 | 0.00053 | 0.0022 | 0.0013 | 0.00067 | 0.0016 | -0.11 | 31 / 38 (82%) | 14 / 19 (74%) | 17 / 19 (89%) | 0.737 | 0.895 |
| PWY-6317~D-galactose degradation I (Leloir pathway) | 0.75 | 0.97 | 1 | 0.46 | 0.91 | 1 | 0.022 | 0.022 | 0.022 | 0.022 | 0.023 | 0.0054 | 0.022 | 0.022 | 0.0081 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6387~UDP-N-acetylmuramoyl-pentapeptide biosynthesis I (meso-diaminopimelate containing) | 0.84 | 0.97 | 1 | 0.71 | 0.97 | 1 | 0.017 | 0.017 | 0.017 | 0.017 | 0.017 | 0.0032 | 0.017 | 0.017 | 0.0054 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6964~ammonia assimilation cycle II | 0.48 | 0.97 | 1 | 0.28 | 0.91 | 1 | 0.044 | 0.044 | 0.045 | 0.045 | 0.045 | 0.0074 | 0.043 | 0.045 | 0.012 | -0.066 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6619~adenine and adenosine salvage VI | 0.52 | 0.97 | 1 | 0.36 | 0.91 | 1 | 0.00029 | 0.00011 | 0 | 1e-04 | 0 | 0.00021 | 0.00011 | 0 | 0.00016 | 0.14 | 14 / 38 (37%) | 7 / 19 (37%) | 7 / 19 (37%) | 0.368 | 0.368 |
| PWY-7663~gondoate biosynthesis (anaerobic) | 0.09 | 0.97 | 1 | 0.18 | 0.91 | 1 | 0.0049 | 0.0049 | 0.0035 | 0.0053 | 0.0042 | 0.0043 | 0.0044 | 0.003 | 0.0046 | -0.27 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6828~linezolid resistance | 0.58 | 0.97 | 1 | 0.64 | 0.96 | 1 | 0.00024 | 1e-04 | 0 | 8.8e-05 | 0 | 0.00017 | 0.00012 | 0 | 0.00024 | 0.45 | 16 / 38 (42%) | 7 / 19 (37%) | 9 / 19 (47%) | 0.368 | 0.474 |
| PWY0-1296~purine ribonucleosides degradation | 0.52 | 0.97 | 1 | 0.8 | 0.99 | 1 | 0.01 | 0.01 | 0.01 | 0.0099 | 0.01 | 0.003 | 0.01 | 0.0098 | 0.0031 | 0.014 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6578~8-amino-7-oxononanoate biosynthesis III | 0.21 | 0.97 | 1 | 0.21 | 0.91 | 1 | 0.00032 | 0.00016 | 1.4e-05 | 0.00013 | 0 | 0.00021 | 2e-04 | 0.00016 | 0.00023 | 0.62 | 19 / 38 (50%) | 8 / 19 (42%) | 11 / 19 (58%) | 0.421 | 0.579 |
| PWY0-1182~trehalose degradation II (cytosolic) | 0.86 | 0.97 | 1 | 0.79 | 0.99 | 1 | 0.002 | 0.0013 | 0.00056 | 0.0015 | 5e-04 | 0.0026 | 0.0011 | 6e-04 | 0.0013 | -0.45 | 25 / 38 (66%) | 12 / 19 (63%) | 13 / 19 (68%) | 0.632 | 0.684 |
| ASPARAGINE-DEG1-PWY-1~L-asparagine degradation III (mammalian) | 0.92 | 0.98 | 1 | 0.9 | 0.99 | 1 | 0.00038 | 0.00021 | 7.7e-05 | 2e-04 | 7.7e-05 | 0.00027 | 0.00022 | 7.6e-05 | 0.00036 | 0.14 | 21 / 38 (55%) | 11 / 19 (58%) | 10 / 19 (53%) | 0.579 | 0.526 |
| PWY-6282~palmitoleate biosynthesis I (from (5Z)-dodec-5-enoate) | 0.089 | 0.97 | 1 | 0.18 | 0.91 | 1 | 0.0048 | 0.0048 | 0.0035 | 0.0053 | 0.0042 | 0.0043 | 0.0044 | 0.003 | 0.0046 | -0.27 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-46~putrescine biosynthesis III | 0.84 | 0.97 | 1 | 0.85 | 0.99 | 1 | 0.0014 | 0.0012 | 0.00075 | 0.0013 | 0.00062 | 0.0014 | 0.0011 | 0.00087 | 0.0012 | -0.24 | 33 / 38 (87%) | 16 / 19 (84%) | 17 / 19 (89%) | 0.842 | 0.895 |
| PWY-2941~L-lysine biosynthesis II | 0.76 | 0.97 | 1 | 0.75 | 0.98 | 1 | 0.0017 | 0.0015 | 0.00083 | 0.0016 | 0.00084 | 0.0018 | 0.0014 | 0.00081 | 0.0013 | -0.19 | 33 / 38 (87%) | 17 / 19 (89%) | 16 / 19 (84%) | 0.895 | 0.842 |
| PWY-7509~cardiolipin and phosphatidylethanolamine biosynthesis (Xanthomonas) | 0.1 | 0.97 | 1 | 0.082 | 0.91 | 1 | 0.0029 | 0.0029 | 0.0029 | 0.0032 | 0.0033 | 0.0014 | 0.0026 | 0.0027 | 0.0012 | -0.3 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6759~hydrogen production III | 0.76 | 0.97 | 1 | 0.9 | 0.99 | 1 | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 | 0.009 | 0.03 | 0.031 | 0.008 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| GLYSYN-THR-PWY~glycine biosynthesis IV | 0.76 | 0.97 | 1 | 0.79 | 0.99 | 1 | 0.011 | 0.011 | 0.0098 | 0.011 | 0.0098 | 0.0035 | 0.012 | 0.0098 | 0.0055 | 0.13 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6839~2-aminoethylphosphonate biosynthesis | 0.9 | 0.98 | 1 | 0.84 | 0.99 | 1 | 0.00042 | 0.00013 | 0 | 0.00013 | 0 | 0.00025 | 0.00013 | 0 | 0.00031 | 0 | 12 / 38 (32%) | 6 / 19 (32%) | 6 / 19 (32%) | 0.316 | 0.316 |
| PWY-7965~2-methyladeninyl adenosylcobamide biosynthesis from adenosylcobinamide-GDP | 0.59 | 0.97 | 1 | 0.39 | 0.91 | 1 | 0.0049 | 0.0049 | 0.0047 | 0.005 | 0.0047 | 0.0018 | 0.0047 | 0.0046 | 0.0014 | -0.089 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY0-1535~D-serine degradation | 0.77 | 0.97 | 1 | 0.82 | 0.99 | 1 | 0.00018 | 0.00011 | 8.4e-05 | 0.00011 | 7.5e-05 | 0.00016 | 0.00011 | 8.7e-05 | 0.00016 | 0 | 23 / 38 (61%) | 11 / 19 (58%) | 12 / 19 (63%) | 0.579 | 0.632 |
| PWY-6543~4-aminobenzoate biosynthesis | 0.55 | 0.97 | 1 | 0.63 | 0.96 | 1 | 0.0023 | 0.0021 | 0.0014 | 0.0022 | 0.0015 | 0.0021 | 0.0021 | 0.0012 | 0.0033 | -0.067 | 36 / 38 (95%) | 18 / 19 (95%) | 18 / 19 (95%) | 0.947 | 0.947 |
| PWY0-1546~muropeptide degradation | 0.67 | 0.97 | 1 | 0.39 | 0.91 | 1 | 0.00045 | 9.5e-05 | 0 | 9.3e-05 | 0 | 0.00022 | 9.7e-05 | 0 | 0.00029 | 0.061 | 8 / 38 (21%) | 5 / 19 (26%) | 3 / 19 (16%) | 0.263 | 0.158 |
| PWY-5989~stearate biosynthesis II (bacteria and plants) | 0.062 | 0.97 | 1 | 0.041 | 0.91 | 1 | 0.005 | 0.0044 | 0.0029 | 0.0047 | 0.0029 | 0.0043 | 0.004 | 0.0029 | 0.0046 | -0.23 | 33 / 38 (87%) | 16 / 19 (84%) | 17 / 19 (89%) | 0.842 | 0.895 |
| PWY-6788~cellulose degradation II (fungi) | 0.63 | 0.97 | 1 | 0.084 | 0.91 | 1 | 0.0079 | 0.0056 | 0.0039 | 0.0054 | 0.0041 | 0.0055 | 0.0059 | 0.0018 | 0.007 | 0.13 | 27 / 38 (71%) | 15 / 19 (79%) | 12 / 19 (63%) | 0.789 | 0.632 |
| PWYQT-4429~CO2 fixation into oxaloacetate (anaplerotic) | 0.45 | 0.97 | 1 | 0.51 | 0.93 | 1 | 0.0035 | 0.0031 | 0.0018 | 0.0026 | 0.0019 | 0.0026 | 0.0036 | 0.0018 | 0.0062 | 0.47 | 34 / 38 (89%) | 16 / 19 (84%) | 18 / 19 (95%) | 0.842 | 0.947 |
| GALACT-GLUCUROCAT-PWY~superpathway of hexuronide and hexuronate degradation | 0.67 | 0.97 | 1 | 0.51 | 0.93 | 1 | 0.0029 | 0.0028 | 0.0026 | 0.0031 | 0.0027 | 0.0026 | 0.0024 | 0.0025 | 0.0018 | -0.37 | 36 / 38 (95%) | 17 / 19 (89%) | 19 / 19 (100%) | 0.895 | 1 |
| PWY-5122~geranyl diphosphate biosynthesis | 0.81 | 0.97 | 1 | 0.52 | 0.93 | 1 | 0.0012 | 0.0011 | 0.00077 | 0.0011 | 0.00084 | 0.0012 | 0.0011 | 0.00076 | 0.00091 | 0 | 33 / 38 (87%) | 16 / 19 (84%) | 17 / 19 (89%) | 0.842 | 0.895 |
| PWY-5891~menaquinol-11 biosynthesis | 0.24 | 0.97 | 1 | 0.48 | 0.91 | 1 | 0.0015 | 0.00073 | 6.1e-05 | 0.00093 | 0.00012 | 0.0015 | 0.00052 | 0 | 0.00089 | -0.84 | 19 / 38 (50%) | 10 / 19 (53%) | 9 / 19 (47%) | 0.526 | 0.474 |
| PWY-5493~reductive monocarboxylic acid cycle | 0.36 | 0.97 | 1 | 0.19 | 0.91 | 1 | 0.034 | 0.034 | 0.034 | 0.035 | 0.034 | 0.0081 | 0.032 | 0.034 | 0.0083 | -0.13 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
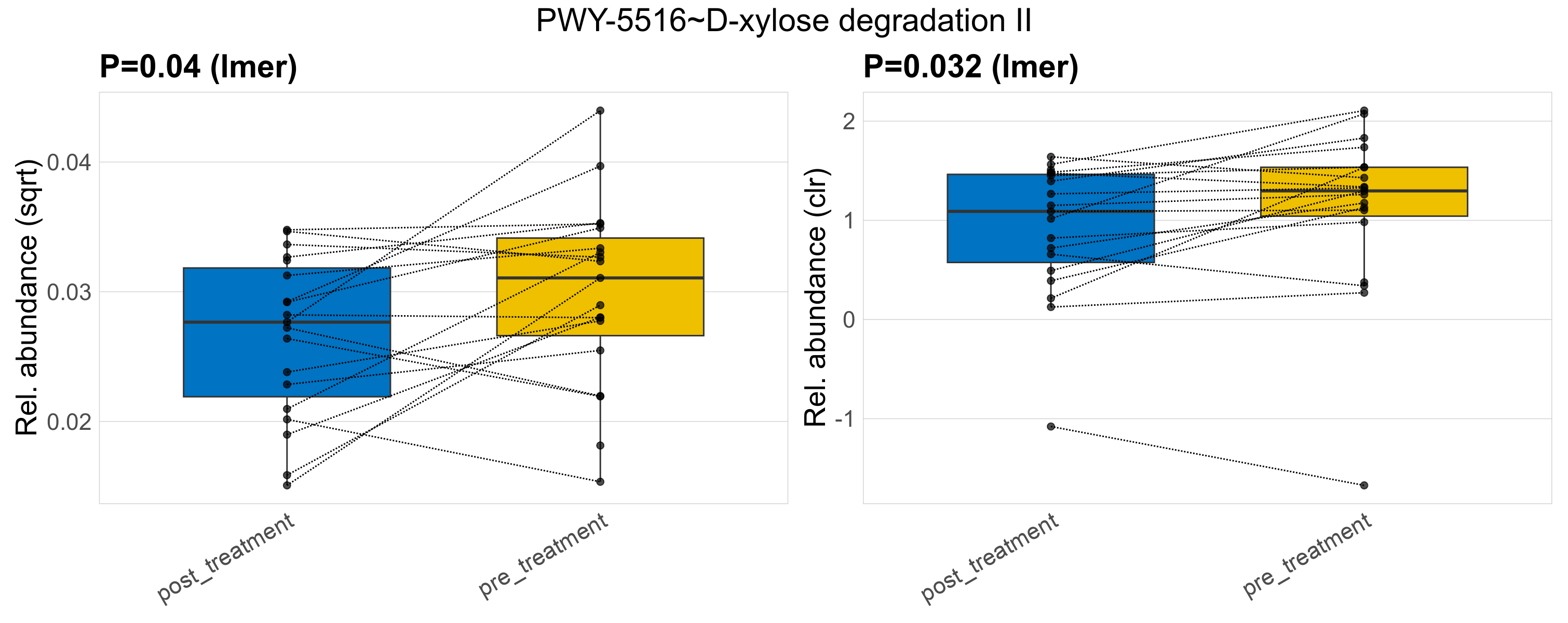
| XYLCAT-PWY~D-xylose degradation I | 0.81 | 0.97 | 1 | 0.8 | 0.99 | 1 | 0.0053 | 0.0053 | 0.0039 | 0.005 | 0.004 | 0.0038 | 0.0055 | 0.0034 | 0.0067 | 0.14 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY0-1295~pyrimidine ribonucleosides degradation | 0.2 | 0.97 | 1 | 0.089 | 0.91 | 1 | 0.0065 | 0.0065 | 0.0062 | 0.0068 | 0.0064 | 0.0021 | 0.0062 | 0.006 | 0.0021 | -0.13 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| GLNSYN-PWY~L-glutamine biosynthesis I | 0.38 | 0.97 | 1 | 0.26 | 0.91 | 1 | 0.047 | 0.047 | 0.048 | 0.048 | 0.05 | 0.0066 | 0.046 | 0.046 | 0.011 | -0.061 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-7219~adenosine ribonucleotides de novo biosynthesis | 0.87 | 0.97 | 1 | 0.71 | 0.97 | 1 | 0.016 | 0.016 | 0.016 | 0.016 | 0.017 | 0.0035 | 0.016 | 0.015 | 0.0042 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5382~hydrogen oxidation II (aerobic NAD) | 0.42 | 0.97 | 1 | 0.52 | 0.93 | 1 | 6e-04 | 0.00019 | 0 | 0.00024 | 0 | 0.00057 | 0.00013 | 0 | 3e-04 | -0.88 | 12 / 38 (32%) | 6 / 19 (32%) | 6 / 19 (32%) | 0.316 | 0.316 |
| PWY-5970~fatty acids biosynthesis (yeast) | 0.21 | 0.97 | 1 | 0.081 | 0.91 | 1 | 0.0041 | 0.0035 | 0.0028 | 0.0036 | 0.0031 | 0.0032 | 0.0034 | 0.0022 | 0.0044 | -0.082 | 32 / 38 (84%) | 16 / 19 (84%) | 16 / 19 (84%) | 0.842 | 0.842 |
| PWY-7971~adenosylcobinamide-GDP salvage from cobinamide I | 0.51 | 0.97 | 1 | 0.48 | 0.91 | 1 | 0.0026 | 0.0026 | 0.0022 | 0.0027 | 0.0023 | 0.0014 | 0.0025 | 0.0021 | 0.0012 | -0.11 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6758~hydrogen production II | 0.42 | 0.97 | 1 | 0.52 | 0.93 | 1 | 6e-04 | 0.00019 | 0 | 0.00024 | 0 | 0.00057 | 0.00013 | 0 | 3e-04 | -0.88 | 12 / 38 (32%) | 6 / 19 (32%) | 6 / 19 (32%) | 0.316 | 0.316 |
| PWY-6197~chlorinated phenols degradation | 0.26 | 0.97 | 1 | 0.17 | 0.91 | 1 | 0.0014 | 0.0014 | 0.0012 | 0.0015 | 0.0012 | 0.001 | 0.0013 | 0.0011 | 0.00086 | -0.21 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-7909~formaldehyde oxidation VII (THF pathway) | 0.33 | 0.97 | 1 | 0.48 | 0.91 | 1 | 0.00087 | 0.00039 | 0 | 5e-04 | 0 | 0.001 | 0.00027 | 0 | 0.00045 | -0.89 | 17 / 38 (45%) | 9 / 19 (47%) | 8 / 19 (42%) | 0.474 | 0.421 |
| PWY-4861~UDP-α-D-galacturonate biosynthesis I (from UDP-D-glucuronate) | 0.18 | 0.97 | 1 | 0.23 | 0.91 | 1 | 0.00015 | 1.6e-05 | 0 | 1.8e-05 | 0 | 5.7e-05 | 1.4e-05 | 0 | 4.6e-05 | -0.36 | 4 / 38 (11%) | 2 / 19 (11%) | 2 / 19 (11%) | 0.105 | 0.105 |
| PWY-7195~pyrimidine ribonucleosides salvage III | 0.92 | 0.98 | 1 | 0.93 | 0.99 | 1 | 0.0021 | 0.0014 | 0.00079 | 0.0012 | 0.00076 | 0.0013 | 0.0017 | 0.00081 | 0.0026 | 0.5 | 26 / 38 (68%) | 12 / 19 (63%) | 14 / 19 (74%) | 0.632 | 0.737 |
| OANTIGEN-PWY~O-antigen building blocks biosynthesis (E. coli) | 0.5 | 0.97 | 1 | 0.87 | 0.99 | 1 | 0.0036 | 0.0035 | 0.0031 | 0.0036 | 0.0032 | 0.0024 | 0.0034 | 0.0027 | 0.0028 | -0.082 | 37 / 38 (97%) | 18 / 19 (95%) | 19 / 19 (100%) | 0.947 | 1 |
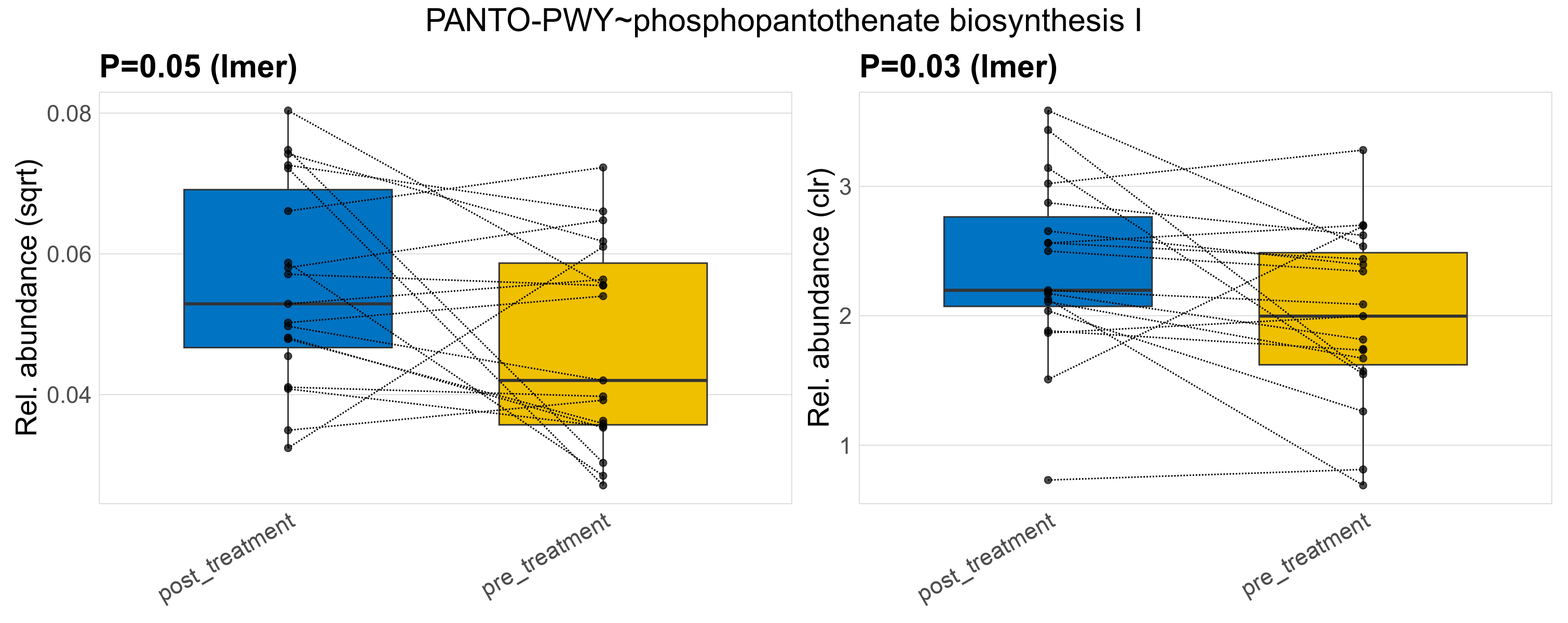
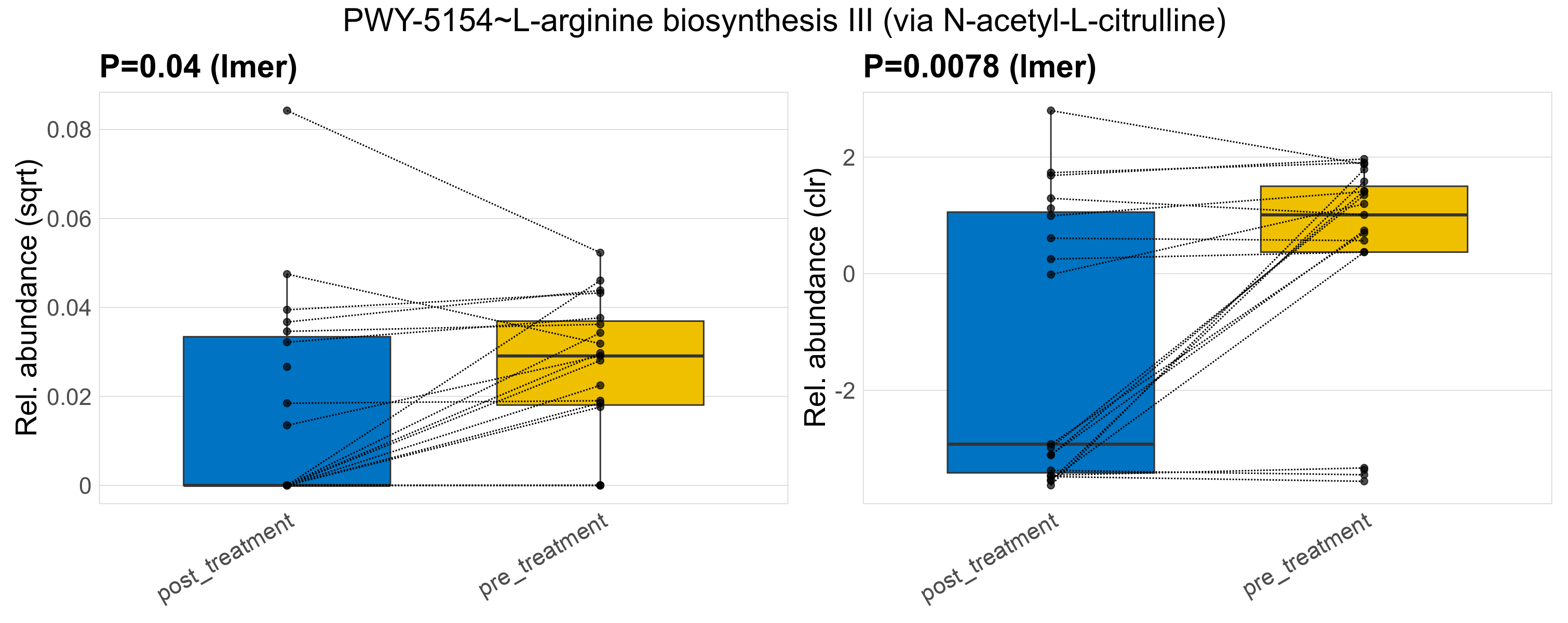
| PWY-5154~L-arginine biosynthesis III (via N-acetyl-L-citrulline) | 0.04 | 0.97 | 1 | 0.0078 | 0.62 | 1 | 0.0014 | 0.00088 | 0.00043 | 0.00083 | 0 | 0.0017 | 0.00093 | 0.00085 | 0.00081 | 0.16 | 24 / 38 (63%) | 9 / 19 (47%) | 15 / 19 (79%) | 0.474 | 0.789 |
| PWY-6122~5-aminoimidazole ribonucleotide biosynthesis II | 0.82 | 0.97 | 1 | 0.44 | 0.91 | 1 | 0.021 | 0.021 | 0.02 | 0.021 | 0.021 | 0.0046 | 0.021 | 0.02 | 0.0083 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| GLUTSYN-PWY~L-glutamate biosynthesis I | 0.8 | 0.97 | 1 | 0.48 | 0.91 | 1 | 0.091 | 0.091 | 0.093 | 0.091 | 0.092 | 0.014 | 0.09 | 0.094 | 0.021 | -0.016 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-2301~myo-inositol biosynthesis | 0.32 | 0.97 | 1 | 0.27 | 0.91 | 1 | 0.002 | 0.0018 | 0.001 | 0.0014 | 0.0011 | 0.0017 | 0.0022 | 0.00097 | 0.004 | 0.65 | 34 / 38 (89%) | 17 / 19 (89%) | 17 / 19 (89%) | 0.895 | 0.895 |
| PWY-6898~thiamine salvage III | 0.33 | 0.97 | 1 | 0.2 | 0.91 | 1 | 0.0079 | 0.0079 | 0.0076 | 0.008 | 0.0077 | 0.002 | 0.0077 | 0.0073 | 0.0031 | -0.055 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY0-1241~ADP-L-glycero-β-D-manno-heptose biosynthesis | 0.31 | 0.97 | 1 | 0.47 | 0.91 | 1 | 0.00067 | 0.00023 | 0 | 0.00016 | 0 | 0.00036 | 3e-04 | 0 | 0.00058 | 0.91 | 13 / 38 (34%) | 6 / 19 (32%) | 7 / 19 (37%) | 0.316 | 0.368 |
| PWY-7964~adeninyl adenosylcobamide biosynthesis from adenosylcobinamide-GDP | 0.59 | 0.97 | 1 | 0.39 | 0.91 | 1 | 0.0049 | 0.0049 | 0.0047 | 0.005 | 0.0047 | 0.0018 | 0.0047 | 0.0046 | 0.0014 | -0.089 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| NONMEVIPP-PWY~methylerythritol phosphate pathway I | 0.92 | 0.98 | 1 | 0.65 | 0.96 | 1 | 0.014 | 0.014 | 0.014 | 0.014 | 0.014 | 0.0034 | 0.014 | 0.013 | 0.0046 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| OXIDATIVEPENT-PWY~pentose phosphate pathway (oxidative branch) I | 0.7 | 0.97 | 1 | 0.56 | 0.93 | 1 | 0.0033 | 0.0029 | 0.0017 | 0.0029 | 0.0024 | 0.0027 | 0.003 | 0.0016 | 0.0051 | 0.049 | 34 / 38 (89%) | 16 / 19 (84%) | 18 / 19 (95%) | 0.842 | 0.947 |
| PWY-5384~sucrose degradation IV (sucrose phosphorylase) | 0.9 | 0.98 | 1 | 0.68 | 0.96 | 1 | 0.0063 | 0.0061 | 0.0058 | 0.0058 | 0.0072 | 0.0036 | 0.0064 | 0.0057 | 0.0066 | 0.14 | 37 / 38 (97%) | 18 / 19 (95%) | 19 / 19 (100%) | 0.947 | 1 |
| PWY0-1586~peptidoglycan maturation (meso-diaminopimelate containing) | 0.095 | 0.97 | 1 | 0.051 | 0.91 | 1 | 0.0033 | 0.0026 | 0.0014 | 0.0028 | 0.0016 | 0.0029 | 0.0024 | 0.00082 | 0.0034 | -0.22 | 30 / 38 (79%) | 17 / 19 (89%) | 13 / 19 (68%) | 0.895 | 0.684 |
| PWY-5785~di-transpoly-cis-undecaprenyl phosphate biosynthesis | 0.74 | 0.97 | 1 | 0.63 | 0.96 | 1 | 0.0017 | 0.0017 | 0.001 | 0.0015 | 0.0012 | 0.0014 | 0.0019 | 0.00097 | 0.0032 | 0.34 | 37 / 38 (97%) | 18 / 19 (95%) | 19 / 19 (100%) | 0.947 | 1 |
| TRESYN-PWY~trehalose biosynthesis I | 0.51 | 0.97 | 1 | 0.91 | 0.99 | 1 | 0.00097 | 0.00049 | 1.6e-05 | 0.00064 | 0 | 0.0016 | 0.00034 | 3.1e-05 | 0.00067 | -0.91 | 19 / 38 (50%) | 9 / 19 (47%) | 10 / 19 (53%) | 0.474 | 0.526 |
| PWY0-881~superpathway of fatty acid biosynthesis I (E. coli) | 0.91 | 0.98 | 1 | 0.96 | 0.99 | 1 | 0.0026 | 0.0015 | 0.00025 | 0.0015 | 0.00021 | 0.0026 | 0.0015 | 0.00029 | 0.0032 | 0 | 21 / 38 (55%) | 10 / 19 (53%) | 11 / 19 (58%) | 0.526 | 0.579 |
| PWY-6587~pyruvate fermentation to ethanol III | 0.19 | 0.97 | 1 | 0.55 | 0.93 | 1 | 0.0016 | 0.0011 | 0.00076 | 9e-04 | 0.00049 | 0.0011 | 0.0013 | 0.001 | 0.0015 | 0.53 | 26 / 38 (68%) | 13 / 19 (68%) | 13 / 19 (68%) | 0.684 | 0.684 |
| PWY-2622~trehalose biosynthesis IV | 0.059 | 0.97 | 1 | 0.1 | 0.91 | 1 | 0.0038 | 0.0038 | 0.0031 | 0.0035 | 0.0031 | 0.0019 | 0.004 | 0.0029 | 0.0022 | 0.19 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| COA-PWY-1~superpathway of coenzyme A biosynthesis III (mammals) | 0.7 | 0.97 | 1 | 0.45 | 0.91 | 1 | 0.0083 | 0.0083 | 0.0072 | 0.0084 | 0.0082 | 0.0029 | 0.0082 | 0.0071 | 0.0043 | -0.035 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| MET-SAM-PWY~superpathway of S-adenosyl-L-methionine biosynthesis | 0.82 | 0.97 | 1 | 0.92 | 0.99 | 1 | 0.00087 | 0.00037 | 0 | 0.00035 | 0 | 0.00073 | 0.00038 | 0 | 0.00071 | 0.12 | 16 / 38 (42%) | 8 / 19 (42%) | 8 / 19 (42%) | 0.421 | 0.421 |
| PWY-6124~inosine-5-phosphate biosynthesis II | 0.85 | 0.97 | 1 | 0.59 | 0.94 | 1 | 0.012 | 0.012 | 0.01 | 0.012 | 0.012 | 0.0035 | 0.012 | 0.01 | 0.0048 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY0-1325~superpathway of L-asparagine biosynthesis | 0.84 | 0.97 | 1 | 0.77 | 0.99 | 1 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.0027 | 0.01 | 0.011 | 0.0028 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| P21-PWY~pentose phosphate pathway (partial) | 0.63 | 0.97 | 1 | 0.36 | 0.91 | 1 | 0.021 | 0.021 | 0.021 | 0.021 | 0.021 | 0.0057 | 0.021 | 0.018 | 0.0062 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| FASYN-ELONG-PWY~fatty acid elongation -- saturated | 0.39 | 0.97 | 1 | 0.18 | 0.91 | 1 | 0.0059 | 0.0059 | 0.0044 | 0.0059 | 0.0049 | 0.0046 | 0.006 | 0.0041 | 0.008 | 0.024 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6608~guanosine nucleotides degradation III | 0.19 | 0.97 | 1 | 0.096 | 0.91 | 1 | 0.0039 | 0.0039 | 0.0039 | 0.0037 | 0.0039 | 0.0024 | 0.0041 | 0.0039 | 0.0015 | 0.15 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| GLUTSYNIII-PWY~L-glutamate biosynthesis III | 0.63 | 0.97 | 1 | 0.91 | 0.99 | 1 | 0.002 | 0.0013 | 0.00033 | 0.0016 | 3e-04 | 0.0032 | 0.001 | 0.00038 | 0.0016 | -0.68 | 25 / 38 (66%) | 12 / 19 (63%) | 13 / 19 (68%) | 0.632 | 0.684 |
| PWY-7211~superpathway of pyrimidine deoxyribonucleotides de novo biosynthesis | 0.69 | 0.97 | 1 | 0.78 | 0.99 | 1 | 0.0014 | 0.00085 | 0.00023 | 0.00094 | 0.00012 | 0.0016 | 0.00076 | 3e-04 | 0.0012 | -0.31 | 23 / 38 (61%) | 12 / 19 (63%) | 11 / 19 (58%) | 0.632 | 0.579 |
| PWY0-1319~CDP-diacylglycerol biosynthesis II | 0.91 | 0.98 | 1 | 0.54 | 0.93 | 1 | 0.00093 | 0.00083 | 0.00068 | 0.00084 | 0.00071 | 0.00069 | 0.00082 | 0.00055 | 0.00073 | -0.035 | 34 / 38 (89%) | 16 / 19 (84%) | 18 / 19 (95%) | 0.842 | 0.947 |
| PENTOSE-P-PWY~pentose phosphate pathway | 0.73 | 0.97 | 1 | 0.95 | 0.99 | 1 | 0.0031 | 0.0029 | 0.0023 | 0.0028 | 0.0024 | 0.0025 | 0.003 | 0.0018 | 0.0044 | 0.1 | 35 / 38 (92%) | 17 / 19 (89%) | 18 / 19 (95%) | 0.895 | 0.947 |
| PWY-5966~fatty acid biosynthesis initiation II | 0.61 | 0.97 | 1 | 0.19 | 0.91 | 1 | 0.0063 | 0.0054 | 0.0034 | 0.005 | 0.0037 | 0.0047 | 0.0059 | 0.0031 | 0.009 | 0.24 | 33 / 38 (87%) | 17 / 19 (89%) | 16 / 19 (84%) | 0.895 | 0.842 |
| PWY-5938~pyruvate fermentation to (R)-acetoin I | 0.93 | 0.98 | 1 | 0.99 | 1 | 1 | 0.00048 | 0.00013 | 0 | 0.00013 | 0 | 0.00034 | 0.00013 | 0 | 0.00026 | 0 | 10 / 38 (26%) | 5 / 19 (26%) | 5 / 19 (26%) | 0.263 | 0.263 |
| PWY-6125~superpathway of guanosine nucleotides de novo biosynthesis II | 0.62 | 0.97 | 1 | 0.3 | 0.91 | 1 | 0.0015 | 0.001 | 4e-04 | 0.001 | 0.00023 | 0.0018 | 0.00097 | 4e-04 | 0.0014 | -0.044 | 25 / 38 (66%) | 11 / 19 (58%) | 14 / 19 (74%) | 0.579 | 0.737 |
| POLYISOPRENSYN-PWY~polyisoprenoid biosynthesis (E. coli) | 0.71 | 0.97 | 1 | 0.1 | 0.91 | 1 | 0.0012 | 0.00087 | 0.00052 | 0.00091 | 0.00067 | 0.0012 | 0.00083 | 0.00048 | 0.00083 | -0.13 | 27 / 38 (71%) | 11 / 19 (58%) | 16 / 19 (84%) | 0.579 | 0.842 |
| PWY-6549~L-glutamine biosynthesis III | 0.51 | 0.97 | 1 | 0.91 | 0.99 | 1 | 0.0011 | 0.00042 | 0 | 0.00027 | 0 | 0.00043 | 0.00057 | 0 | 0.0013 | 1.1 | 14 / 38 (37%) | 7 / 19 (37%) | 7 / 19 (37%) | 0.368 | 0.368 |
| ASPARAGINE-DEG1-PWY~L-asparagine degradation I | 0.73 | 0.97 | 1 | 0.67 | 0.96 | 1 | 0.0086 | 0.0086 | 0.0078 | 0.0087 | 0.0079 | 0.0039 | 0.0085 | 0.0072 | 0.0048 | -0.034 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5807~heptaprenyl diphosphate biosynthesis | 0.62 | 0.97 | 1 | 0.83 | 0.99 | 1 | 0.00093 | 0.00088 | 0.00071 | 8e-04 | 0.00074 | 0.00064 | 0.00096 | 0.00052 | 9e-04 | 0.26 | 36 / 38 (95%) | 18 / 19 (95%) | 18 / 19 (95%) | 0.947 | 0.947 |
| PWY0-501~lipoate biosynthesis and incorporation I | 0.72 | 0.97 | 1 | 0.96 | 0.99 | 1 | 0.0011 | 0.00053 | 3.5e-05 | 6e-04 | 0 | 0.0011 | 0.00045 | 7.1e-05 | 0.00074 | -0.42 | 19 / 38 (50%) | 9 / 19 (47%) | 10 / 19 (53%) | 0.474 | 0.526 |
| PWY-4341~L-glutamate biosynthesis V | 0.75 | 0.97 | 1 | 0.46 | 0.91 | 1 | 0.045 | 0.045 | 0.046 | 0.046 | 0.047 | 0.0085 | 0.045 | 0.044 | 0.012 | -0.032 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| HOMOSER-THRESYN-PWY~L-threonine biosynthesis | 0.98 | 0.99 | 1 | 0.53 | 0.93 | 1 | 0.0081 | 0.0081 | 0.008 | 0.008 | 0.0079 | 0.0025 | 0.0083 | 0.008 | 0.0046 | 0.053 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5030~L-histidine degradation III | 0.56 | 0.97 | 1 | 0.91 | 0.99 | 1 | 0.0014 | 0.00075 | 0.00014 | 0.00093 | 0.00013 | 0.0018 | 0.00057 | 0.00015 | 0.001 | -0.71 | 20 / 38 (53%) | 10 / 19 (53%) | 10 / 19 (53%) | 0.526 | 0.526 |
| PWY-5097~L-lysine biosynthesis VI | 0.63 | 0.97 | 1 | 0.33 | 0.91 | 1 | 0.018 | 0.018 | 0.018 | 0.018 | 0.018 | 0.003 | 0.017 | 0.017 | 0.0044 | -0.082 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| ANAEROFRUCAT-PWY~homolactic fermentation | 0.75 | 0.97 | 1 | 0.36 | 0.91 | 1 | 0.014 | 0.014 | 0.014 | 0.014 | 0.014 | 0.0033 | 0.014 | 0.014 | 0.0048 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-7494~choline degradation IV | 0.65 | 0.97 | 1 | 0.21 | 0.91 | 1 | 0.00077 | 0.00051 | 0.00027 | 0.00055 | 0.00015 | 8e-04 | 0.00046 | 0.00029 | 5e-04 | -0.26 | 25 / 38 (66%) | 10 / 19 (53%) | 15 / 19 (79%) | 0.526 | 0.789 |
| PWY-5347~superpathway of L-methionine biosynthesis (transsulfuration) | 0.74 | 0.97 | 1 | 0.74 | 0.98 | 1 | 0.00098 | 0.00039 | 0 | 0.00038 | 0 | 0.00084 | 4e-04 | 0 | 0.00078 | 0.074 | 15 / 38 (39%) | 7 / 19 (37%) | 8 / 19 (42%) | 0.368 | 0.421 |
| PWY-7459~kojibiose degradation | 0.47 | 0.97 | 1 | 0.22 | 0.91 | 1 | 8e-04 | 0.00078 | 0.00057 | 8e-04 | 5e-04 | 0.00091 | 0.00076 | 0.00065 | 0.00053 | -0.074 | 37 / 38 (97%) | 18 / 19 (95%) | 19 / 19 (100%) | 0.947 | 1 |
| ASPARTATE-DEG1-PWY~L-aspartate degradation I | 0.57 | 0.97 | 1 | 0.27 | 0.91 | 1 | 0.02 | 0.02 | 0.019 | 0.02 | 0.02 | 0.0063 | 0.02 | 0.017 | 0.012 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6700~queuosine biosynthesis | 0.1 | 0.97 | 1 | 0.13 | 0.91 | 1 | 0.0026 | 0.0026 | 0.0023 | 0.0031 | 0.0025 | 0.0019 | 0.0022 | 0.002 | 0.0012 | -0.49 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6922~L-Nδ-acetylornithine biosynthesis | 0.67 | 0.97 | 1 | 0.96 | 0.99 | 1 | 0.0012 | 0.00035 | 0 | 0.00044 | 0 | 0.001 | 0.00026 | 0 | 0.00056 | -0.76 | 11 / 38 (29%) | 5 / 19 (26%) | 6 / 19 (32%) | 0.263 | 0.316 |
| GOLPDLCAT-PWY~superpathway of glycerol degradation to 13-propanediol | 0.98 | 0.99 | 1 | 0.83 | 0.99 | 1 | 0.00048 | 0.00011 | 0 | 0.00011 | 0 | 0.00023 | 0.00012 | 0 | 0.00028 | 0.13 | 9 / 38 (24%) | 5 / 19 (26%) | 4 / 19 (21%) | 0.263 | 0.211 |
| GLUTDEG-PWY~L-glutamate degradation II | 0.37 | 0.97 | 1 | 0.38 | 0.91 | 1 | 0.0087 | 0.0087 | 0.0082 | 0.009 | 0.0086 | 0.0043 | 0.0083 | 0.008 | 0.003 | -0.12 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6284~superpathway of unsaturated fatty acids biosynthesis (E. coli) | 0.086 | 0.97 | 1 | 0.14 | 0.91 | 1 | 0.0047 | 0.0047 | 0.0034 | 0.0052 | 0.0042 | 0.0042 | 0.0043 | 0.003 | 0.0045 | -0.27 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| P142-PWY~pyruvate fermentation to acetate I | 0.23 | 0.97 | 1 | 0.16 | 0.91 | 1 | 0.016 | 0.016 | 0.017 | 0.017 | 0.018 | 0.0043 | 0.016 | 0.016 | 0.0038 | -0.087 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| FASYN-INITIAL-PWY~superpathway of fatty acid biosynthesis initiation (E. coli) | 0.57 | 0.97 | 1 | 0.2 | 0.91 | 1 | 0.0013 | 0.00063 | 0 | 0.00068 | 0 | 0.0015 | 0.00058 | 0.00022 | 0.00091 | -0.23 | 18 / 38 (47%) | 7 / 19 (37%) | 11 / 19 (58%) | 0.368 | 0.579 |
| PWY0-461~L-lysine degradation I | 0.24 | 0.97 | 1 | 0.26 | 0.91 | 1 | 0.005 | 0.0049 | 0.0049 | 0.0048 | 0.0049 | 0.0024 | 0.0051 | 0.0051 | 0.0027 | 0.087 | 37 / 38 (97%) | 18 / 19 (95%) | 19 / 19 (100%) | 0.947 | 1 |
| PWY-5890~menaquinol-10 biosynthesis | 0.24 | 0.97 | 1 | 0.48 | 0.91 | 1 | 0.0015 | 0.00073 | 6.1e-05 | 0.00093 | 0.00012 | 0.0015 | 0.00052 | 0 | 0.00089 | -0.84 | 19 / 38 (50%) | 10 / 19 (53%) | 9 / 19 (47%) | 0.526 | 0.474 |
| PWY-6744~hydrogen production I | 0.76 | 0.97 | 1 | 0.97 | 0.99 | 1 | 0.00011 | 1.8e-05 | 0 | 1.3e-05 | 0 | 3.3e-05 | 2.2e-05 | 0 | 6.2e-05 | 0.76 | 6 / 38 (16%) | 3 / 19 (16%) | 3 / 19 (16%) | 0.158 | 0.158 |
| COA-PWY~coenzyme A biosynthesis I (prokaryotic) | 0.76 | 0.97 | 1 | 0.51 | 0.93 | 1 | 0.0081 | 0.0081 | 0.0075 | 0.0082 | 0.0077 | 0.0029 | 0.0081 | 0.0068 | 0.0041 | -0.018 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY0-1312~acetate and ATP formation from acetyl-CoA I | 0.99 | 0.99 | 1 | 0.62 | 0.96 | 1 | 0.013 | 0.013 | 0.013 | 0.013 | 0.013 | 0.0031 | 0.013 | 0.013 | 0.0051 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| METSYN-PWY~superpathway of L-homoserine and L-methionine biosynthesis | 0.86 | 0.97 | 1 | 0.93 | 0.99 | 1 | 0.00087 | 0.00037 | 0 | 0.00036 | 0 | 0.00075 | 0.00038 | 0 | 0.00071 | 0.078 | 16 / 38 (42%) | 8 / 19 (42%) | 8 / 19 (42%) | 0.421 | 0.421 |
| PWY4FS-8~phosphatidylglycerol biosynthesis II (non-plastidic) | 0.92 | 0.98 | 1 | 0.52 | 0.93 | 1 | 0.00092 | 0.00078 | 0.00073 | 0.00081 | 0.00091 | 0.00069 | 0.00074 | 0.00054 | 0.00066 | -0.13 | 32 / 38 (84%) | 15 / 19 (79%) | 17 / 19 (89%) | 0.789 | 0.895 |
| PWY0-1314~fructose degradation | 0.21 | 0.97 | 1 | 0.22 | 0.91 | 1 | 0.00042 | 0.00027 | 0.00013 | 2e-04 | 0.00011 | 0.00026 | 0.00034 | 0.00014 | 0.00052 | 0.77 | 25 / 38 (66%) | 11 / 19 (58%) | 14 / 19 (74%) | 0.579 | 0.737 |
| METHFORM-PWY~methyl-coenzyme M reduction to methane | 0.69 | 0.97 | 1 | 0.96 | 0.99 | 1 | 0.00068 | 7.2e-05 | 0 | 5.2e-05 | 0 | 0.00015 | 9.2e-05 | 0 | 0.00032 | 0.82 | 4 / 38 (11%) | 2 / 19 (11%) | 2 / 19 (11%) | 0.105 | 0.105 |
| GLUTAMATE-SYN2-PWY~L-glutamate biosynthesis II | 0.29 | 0.97 | 1 | 0.98 | 0.99 | 1 | 0.00091 | 0.00048 | 5.2e-05 | 0.00068 | 0 | 0.0014 | 0.00028 | 8e-05 | 0.00046 | -1.3 | 20 / 38 (53%) | 9 / 19 (47%) | 11 / 19 (58%) | 0.474 | 0.579 |
| PWY-7198~pyrimidine deoxyribonucleotides de novo biosynthesis IV | 0.32 | 0.97 | 1 | 0.29 | 0.91 | 1 | 0.00093 | 0.00034 | 0 | 0.00028 | 0 | 0.00081 | 4e-04 | 0 | 0.001 | 0.51 | 14 / 38 (37%) | 5 / 19 (26%) | 9 / 19 (47%) | 0.263 | 0.474 |
| PWY-5951~(RR)-butanediol biosynthesis | 0.62 | 0.97 | 1 | 0.79 | 0.99 | 1 | 0.00014 | 2.9e-05 | 0 | 2.1e-05 | 0 | 5.4e-05 | 3.7e-05 | 0 | 9.4e-05 | 0.82 | 8 / 38 (21%) | 4 / 19 (21%) | 4 / 19 (21%) | 0.211 | 0.211 |
| ORN-AMINOPENTANOATE-CAT-PWY~L-ornithine degradation I (L-proline biosynthesis) | 0.98 | 0.99 | 1 | 0.66 | 0.96 | 1 | 0.0011 | 0.00084 | 0.00057 | 0.00078 | 0.00057 | 0.00073 | 0.00091 | 0.00048 | 0.001 | 0.22 | 29 / 38 (76%) | 15 / 19 (79%) | 14 / 19 (74%) | 0.789 | 0.737 |
| PWY-6293~superpathway of L-cysteine biosynthesis (fungi) | 0.87 | 0.97 | 1 | 0.3 | 0.91 | 1 | 0.0038 | 0.0026 | 0.00097 | 0.0022 | 0.0011 | 0.0029 | 0.0031 | 0.00085 | 0.0072 | 0.49 | 26 / 38 (68%) | 14 / 19 (74%) | 12 / 19 (63%) | 0.737 | 0.632 |
| PWY-5418~phenol degradation I (aerobic) | 0.26 | 0.97 | 1 | 0.17 | 0.91 | 1 | 0.0014 | 0.0014 | 0.0012 | 0.0015 | 0.0012 | 0.001 | 0.0013 | 0.0011 | 0.00086 | -0.21 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5483~pyruvate fermentation to acetate III | 0.68 | 0.97 | 1 | 0.92 | 0.99 | 1 | 0.0036 | 0.0027 | 0.0025 | 0.0027 | 0.0022 | 0.0024 | 0.0028 | 0.0025 | 0.0026 | 0.052 | 29 / 38 (76%) | 15 / 19 (79%) | 14 / 19 (74%) | 0.789 | 0.737 |
| ARGSYN-PWY~L-arginine biosynthesis I (via L-ornithine) | 0.62 | 0.97 | 1 | 0.57 | 0.93 | 1 | 0.019 | 0.019 | 0.019 | 0.019 | 0.02 | 0.0056 | 0.019 | 0.018 | 0.0062 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-7858~(5Z)-dodecenoate biosynthesis II | 0.26 | 0.97 | 1 | 0.37 | 0.91 | 1 | 0.00036 | 7.5e-05 | 0 | 3e-05 | 0 | 8e-05 | 0.00012 | 0 | 0.00033 | 2 | 8 / 38 (21%) | 3 / 19 (16%) | 5 / 19 (26%) | 0.158 | 0.263 |
| COMPLETE-ARO-PWY~superpathway of aromatic amino acid biosynthesis | 0.44 | 0.97 | 1 | 0.35 | 0.91 | 1 | 0.0018 | 0.001 | 0.00055 | 0.00084 | 0 | 0.001 | 0.0012 | 0.00063 | 0.0015 | 0.51 | 21 / 38 (55%) | 9 / 19 (47%) | 12 / 19 (63%) | 0.474 | 0.632 |
| PWY-4985~mimosine biosynthesis | 0.53 | 0.97 | 1 | 0.44 | 0.91 | 1 | 0.0017 | 0.0011 | 0.00027 | 0.00078 | 0.00045 | 0.0011 | 0.0013 | 0.00026 | 0.0032 | 0.74 | 24 / 38 (63%) | 11 / 19 (58%) | 13 / 19 (68%) | 0.579 | 0.684 |
| PWY0-1305~L-glutamate degradation IX (via 4-aminobutanoate) | 0.88 | 0.98 | 1 | 0.78 | 0.99 | 1 | 0.0027 | 0.0026 | 0.0017 | 0.0025 | 0.0018 | 0.0024 | 0.0027 | 0.0017 | 0.005 | 0.11 | 36 / 38 (95%) | 18 / 19 (95%) | 18 / 19 (95%) | 0.947 | 0.947 |
| GLUTAMINDEG-PWY~L-glutamine degradation I | 0.08 | 0.97 | 1 | 0.065 | 0.91 | 1 | 0.014 | 0.014 | 0.014 | 0.015 | 0.015 | 0.0034 | 0.014 | 0.013 | 0.0034 | -0.1 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| BRANCHED-CHAIN-AA-SYN-PWY~superpathway of branched chain amino acid biosynthesis | 0.79 | 0.97 | 1 | 0.82 | 0.99 | 1 | 0.022 | 0.022 | 0.021 | 0.022 | 0.022 | 0.0051 | 0.022 | 0.021 | 0.0067 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-7309~acrylonitrile degradation II | 0.72 | 0.97 | 1 | 0.94 | 0.99 | 1 | 0.00034 | 0.00023 | 0.00012 | 0.00021 | 0.00014 | 0.00025 | 0.00025 | 0.00011 | 0.00039 | 0.25 | 26 / 38 (68%) | 13 / 19 (68%) | 13 / 19 (68%) | 0.684 | 0.684 |
| PWY-6510~methanol oxidation to formaldehyde II | 0.15 | 0.97 | 1 | 0.22 | 0.91 | 1 | 0.00094 | 0.00089 | 0.00087 | 0.00074 | 0.00067 | 0.00051 | 0.001 | 0.00097 | 0.00075 | 0.43 | 36 / 38 (95%) | 18 / 19 (95%) | 18 / 19 (95%) | 0.947 | 0.947 |
| PWY-6163~chorismate biosynthesis from 3-dehydroquinate | 0.79 | 0.97 | 1 | 0.59 | 0.94 | 1 | 0.011 | 0.011 | 0.011 | 0.011 | 0.012 | 0.003 | 0.011 | 0.0097 | 0.004 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5939~pyruvate fermentation to (R)-acetoin II | 0.95 | 0.99 | 1 | 0.68 | 0.96 | 1 | 0.00092 | 0.00039 | 0 | 0.00035 | 0 | 0.00065 | 0.00042 | 0 | 0.00089 | 0.26 | 16 / 38 (42%) | 9 / 19 (47%) | 7 / 19 (37%) | 0.474 | 0.368 |
| PWY-7400~L-arginine biosynthesis IV (archaebacteria) | 0.69 | 0.97 | 1 | 0.59 | 0.95 | 1 | 0.019 | 0.019 | 0.018 | 0.019 | 0.02 | 0.0054 | 0.018 | 0.018 | 0.0063 | -0.078 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY0-1315~L-lactaldehyde degradation (anaerobic) | 0.79 | 0.97 | 1 | 0.48 | 0.91 | 1 | 0.014 | 0.014 | 0.013 | 0.015 | 0.014 | 0.0039 | 0.014 | 0.013 | 0.0048 | -0.1 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PRPP-PWY~superpathway of histidine purine and pyrimidine biosynthesis | 0.79 | 0.97 | 1 | 0.92 | 0.99 | 1 | 0.003 | 0.0024 | 0.0011 | 0.002 | 0.00085 | 0.0022 | 0.0027 | 0.0013 | 0.0053 | 0.43 | 30 / 38 (79%) | 15 / 19 (79%) | 15 / 19 (79%) | 0.789 | 0.789 |
| ILEUSYN-PWY~L-isoleucine biosynthesis I (from threonine) | 0.79 | 0.97 | 1 | 0.82 | 0.99 | 1 | 0.023 | 0.023 | 0.023 | 0.023 | 0.023 | 0.0053 | 0.023 | 0.023 | 0.0069 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-66~GDP-L-fucose biosynthesis I (from GDP-D-mannose) | 0.15 | 0.97 | 1 | 0.12 | 0.91 | 1 | 0.0032 | 0.0032 | 0.0029 | 0.0035 | 0.0038 | 0.002 | 0.0028 | 0.0026 | 0.0016 | -0.32 | 37 / 38 (97%) | 19 / 19 (100%) | 18 / 19 (95%) | 1 | 0.947 |
| PWY-922~mevalonate pathway I | 0.79 | 0.97 | 1 | 1 | 1 | 1 | 0.00034 | 8.9e-05 | 0 | 6.6e-05 | 0 | 0.00013 | 0.00011 | 0 | 0.00031 | 0.74 | 10 / 38 (26%) | 5 / 19 (26%) | 5 / 19 (26%) | 0.263 | 0.263 |
| PWY-6630~superpathway of L-tyrosine biosynthesis | 0.23 | 0.97 | 1 | 0.1 | 0.91 | 1 | 0.0034 | 0.0034 | 0.0026 | 0.0033 | 0.0027 | 0.0021 | 0.0034 | 0.0025 | 0.0047 | 0.043 | 37 / 38 (97%) | 19 / 19 (100%) | 18 / 19 (95%) | 1 | 0.947 |
| PWY-5108~L-isoleucine biosynthesis V | 0.68 | 0.97 | 1 | 0.67 | 0.96 | 1 | 0.0012 | 0.00058 | 6.2e-05 | 0.00075 | 0 | 0.0014 | 0.00042 | 0.00019 | 0.00067 | -0.84 | 19 / 38 (50%) | 8 / 19 (42%) | 11 / 19 (58%) | 0.421 | 0.579 |
| THREONINE-DEG2-PWY~L-threonine degradation II | 0.27 | 0.97 | 1 | 0.17 | 0.91 | 1 | 0.0022 | 0.0022 | 0.0015 | 0.002 | 0.0012 | 0.0021 | 0.0023 | 0.0015 | 0.0016 | 0.2 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY0-1597~prenylated FMNH2 biosynthesis | 0.36 | 0.97 | 1 | 0.37 | 0.91 | 1 | 0.0013 | 0.0013 | 0.0012 | 0.0014 | 0.0014 | 0.00095 | 0.0012 | 0.0012 | 0.00053 | -0.22 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-7392~taxadiene biosynthesis (engineered) | 0.66 | 0.97 | 1 | 0.092 | 0.91 | 1 | 0.0012 | 0.00094 | 0.00056 | 0.00099 | 0.00052 | 0.0014 | 0.00088 | 0.00064 | 0.00083 | -0.17 | 29 / 38 (76%) | 12 / 19 (63%) | 17 / 19 (89%) | 0.632 | 0.895 |
| PWY-6753~S-methyl-5-thioadenosine degradation III | 0.38 | 0.97 | 1 | 0.55 | 0.93 | 1 | 0.00016 | 2.5e-05 | 0 | 1.5e-05 | 0 | 3.8e-05 | 3.5e-05 | 0 | 9.5e-05 | 1.2 | 6 / 38 (16%) | 3 / 19 (16%) | 3 / 19 (16%) | 0.158 | 0.158 |
| PWY-5783~octaprenyl diphosphate biosynthesis | 0.84 | 0.97 | 1 | 0.42 | 0.91 | 1 | 0.0033 | 0.0033 | 0.0021 | 0.0032 | 0.0026 | 0.0026 | 0.0035 | 0.0021 | 0.0053 | 0.13 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
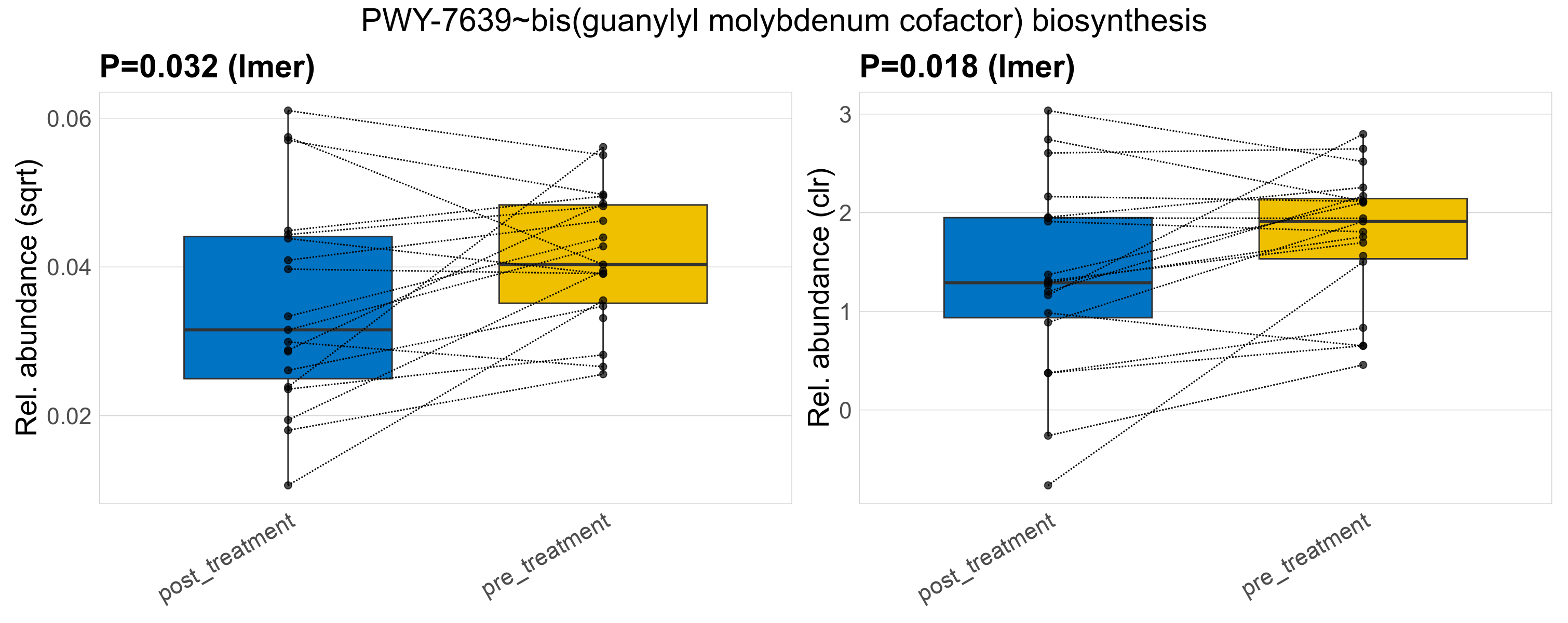
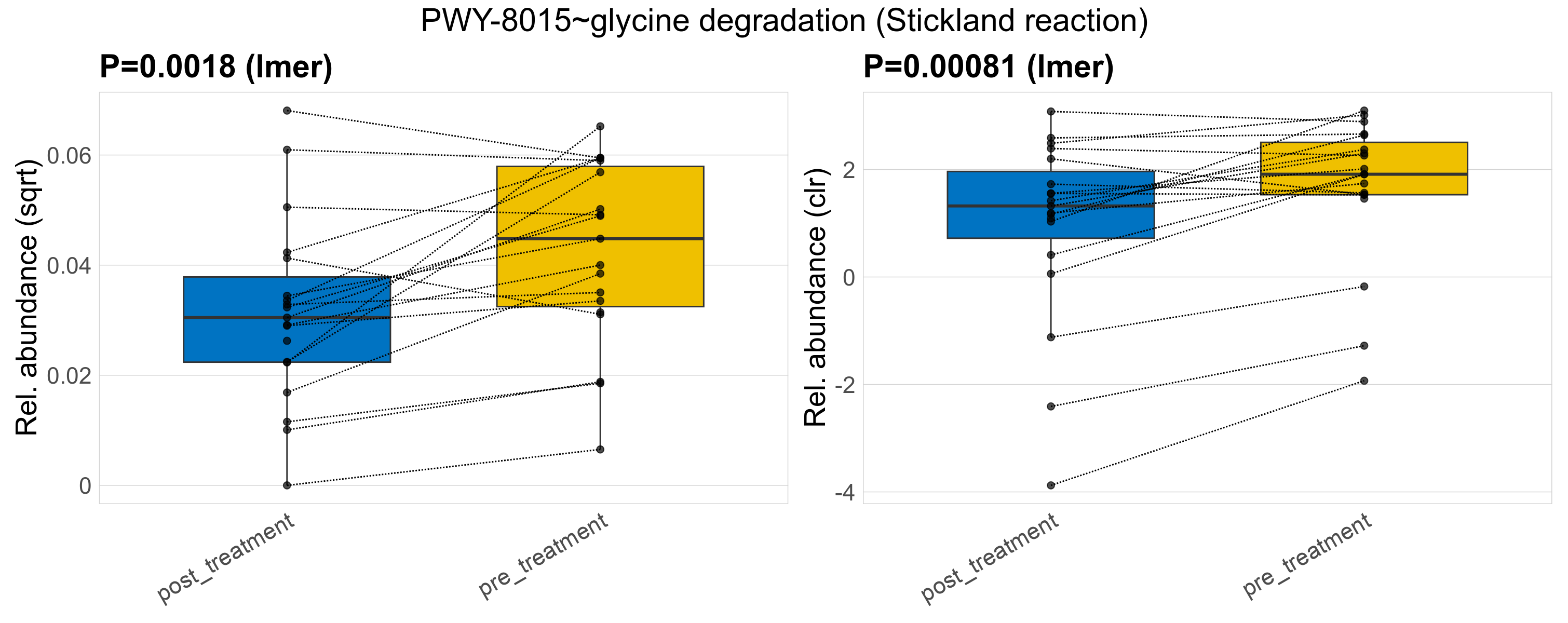
| PWY-8015~glycine degradation (Stickland reaction) | 0.0018 | 0.33 | 0.99 | 0.00081 | 0.45 | 0.45 | 0.0017 | 0.0016 | 0.0012 | 0.0012 | 0.00093 | 0.0012 | 0.0021 | 0.002 | 0.0013 | 0.81 | 37 / 38 (97%) | 18 / 19 (95%) | 19 / 19 (100%) | 0.947 | 1 |
| PWY-7806~glyphosate degradation II | 0.41 | 0.97 | 1 | 0.92 | 0.99 | 1 | 0.00081 | 0.00051 | 0.00019 | 4e-04 | 0.00014 | 0.00054 | 0.00062 | 0.00042 | 0.00075 | 0.63 | 24 / 38 (63%) | 12 / 19 (63%) | 12 / 19 (63%) | 0.632 | 0.632 |
| PWY-6173~histamine biosynthesis | 0.91 | 0.98 | 1 | 0.48 | 0.91 | 1 | 0.00038 | 0.00013 | 0 | 0.00016 | 0 | 0.00044 | 9.5e-05 | 0 | 0.00017 | -0.75 | 13 / 38 (34%) | 5 / 19 (26%) | 8 / 19 (42%) | 0.263 | 0.421 |
| PWY-6970~acetyl-CoA biosynthesis II (NADP-dependent pyruvate dehydrogenase) | 0.82 | 0.97 | 1 | 0.68 | 0.96 | 1 | 0.0017 | 0.00065 | 0 | 0.00066 | 0 | 0.0014 | 0.00065 | 0 | 0.0012 | -0.022 | 15 / 38 (39%) | 7 / 19 (37%) | 8 / 19 (42%) | 0.368 | 0.421 |
| PWY-5123~trans trans-farnesyl diphosphate biosynthesis | 0.52 | 0.97 | 1 | 0.73 | 0.97 | 1 | 0.00085 | 0.00036 | 0 | 0.00045 | 0 | 0.00095 | 0.00027 | 0 | 0.00048 | -0.74 | 16 / 38 (42%) | 8 / 19 (42%) | 8 / 19 (42%) | 0.421 | 0.421 |
| PWY-7790~UMP biosynthesis II | 0.079 | 0.97 | 1 | 0.074 | 0.91 | 1 | 0.024 | 0.024 | 0.025 | 0.025 | 0.025 | 0.0044 | 0.023 | 0.021 | 0.0071 | -0.12 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5005~biotin biosynthesis II | 0.37 | 0.97 | 1 | 0.39 | 0.91 | 1 | 0.00038 | 0.00016 | 0 | 0.00013 | 0 | 0.00022 | 0.00018 | 0 | 0.00023 | 0.47 | 16 / 38 (42%) | 7 / 19 (37%) | 9 / 19 (47%) | 0.368 | 0.474 |
| PWY-6901~superpathway of glucose and xylose degradation | 0.67 | 0.97 | 1 | 0.95 | 0.99 | 1 | 0.0031 | 0.0028 | 0.0022 | 0.0028 | 0.0024 | 0.0026 | 0.0028 | 0.002 | 0.0044 | 0 | 35 / 38 (92%) | 17 / 19 (89%) | 18 / 19 (95%) | 0.895 | 0.947 |
| METHANOGENESIS-PWY~methanogenesis from H2 and CO2 | 0.16 | 0.97 | 1 | 0.26 | 0.91 | 1 | 0.00037 | 2.9e-05 | 0 | 8.8e-06 | 0 | 3.8e-05 | 5e-05 | 0 | 0.00015 | 2.5 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| PWY0-781~aspartate superpathway | 0.64 | 0.97 | 1 | 0.91 | 0.99 | 1 | 0.0021 | 0.0018 | 0.001 | 0.0015 | 0.00091 | 0.0013 | 0.002 | 0.0012 | 0.0023 | 0.42 | 32 / 38 (84%) | 16 / 19 (84%) | 16 / 19 (84%) | 0.842 | 0.842 |
| GLUCUROCAT-PWY~superpathway of β-D-glucuronosides degradation | 0.37 | 0.97 | 1 | 0.9 | 0.99 | 1 | 0.0038 | 0.0037 | 0.0038 | 0.0043 | 0.0045 | 0.003 | 0.0032 | 0.0032 | 0.0023 | -0.43 | 37 / 38 (97%) | 18 / 19 (95%) | 19 / 19 (100%) | 0.947 | 1 |
| PWY-5676~acetyl-CoA fermentation to butanoate II | 0.099 | 0.97 | 1 | 0.12 | 0.91 | 1 | 0.0015 | 0.0014 | 0.001 | 0.0016 | 0.0013 | 0.0012 | 0.0012 | 9e-04 | 0.00095 | -0.42 | 37 / 38 (97%) | 19 / 19 (100%) | 18 / 19 (95%) | 1 | 0.947 |
| PWY-3821~D-galactose detoxification | 0.79 | 0.97 | 1 | 0.86 | 0.99 | 1 | 0.0013 | 0.00081 | 0.00055 | 0.00078 | 0.00062 | 0.00088 | 0.00085 | 0.00048 | 0.0012 | 0.12 | 24 / 38 (63%) | 11 / 19 (58%) | 13 / 19 (68%) | 0.579 | 0.684 |
| ARGSYNBSUB-PWY~L-arginine biosynthesis II (acetyl cycle) | 0.86 | 0.97 | 1 | 0.59 | 0.94 | 1 | 0.021 | 0.021 | 0.021 | 0.021 | 0.022 | 0.0048 | 0.021 | 0.02 | 0.0057 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-4121~glutathionylspermidine biosynthesis | 0.97 | 0.99 | 1 | 0.55 | 0.93 | 1 | 0.00081 | 0.00028 | 0 | 0.00033 | 0 | 0.00071 | 0.00022 | 0 | 0.00043 | -0.58 | 13 / 38 (34%) | 5 / 19 (26%) | 8 / 19 (42%) | 0.263 | 0.421 |
| BSUBPOLYAMSYN-PWY~spermidine biosynthesis I | 0.6 | 0.97 | 1 | 0.76 | 0.98 | 1 | 0.00069 | 0.00058 | 0.00053 | 0.00055 | 0.00048 | 0.00051 | 0.00062 | 0.00054 | 0.00045 | 0.17 | 32 / 38 (84%) | 16 / 19 (84%) | 16 / 19 (84%) | 0.842 | 0.842 |
| PWY-5686~UMP biosynthesis I | 0.088 | 0.97 | 1 | 0.088 | 0.91 | 1 | 0.024 | 0.024 | 0.025 | 0.025 | 0.026 | 0.0043 | 0.023 | 0.021 | 0.0071 | -0.12 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| P124-PWY~Bifidobacterium shunt | 0.95 | 0.99 | 1 | 0.48 | 0.91 | 1 | 0.011 | 0.011 | 0.011 | 0.01 | 0.011 | 0.0025 | 0.011 | 0.011 | 0.0054 | 0.14 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-1001~cellulose biosynthesis | 0.21 | 0.97 | 1 | 0.73 | 0.97 | 1 | 0.0021 | 0.0013 | 0.00012 | 0.00088 | 0.00011 | 0.0013 | 0.0017 | 0.00012 | 0.0023 | 0.95 | 23 / 38 (61%) | 11 / 19 (58%) | 12 / 19 (63%) | 0.579 | 0.632 |
| PWY-7968~5-methylbenzimidazolyl adenosylcobamide biosynthesis from adenosylcobinamide-GDP | 0.59 | 0.97 | 1 | 0.39 | 0.91 | 1 | 0.0049 | 0.0049 | 0.0047 | 0.005 | 0.0047 | 0.0018 | 0.0047 | 0.0046 | 0.0014 | -0.089 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-7197~pyrimidine deoxyribonucleotide phosphorylation | 0.26 | 0.97 | 1 | 0.29 | 0.91 | 1 | 0.00044 | 3e-04 | 2e-04 | 0.00024 | 9.4e-05 | 0.00028 | 0.00036 | 0.00026 | 0.00038 | 0.58 | 26 / 38 (68%) | 12 / 19 (63%) | 14 / 19 (74%) | 0.632 | 0.737 |
| DENOVOPURINE2-PWY~superpathway of purine nucleotides de novo biosynthesis II | 0.81 | 0.97 | 1 | 0.97 | 0.99 | 1 | 0.0015 | 0.001 | 0.00048 | 0.0011 | 0.00045 | 0.0019 | 0.00087 | 5e-04 | 0.0012 | -0.34 | 26 / 38 (68%) | 13 / 19 (68%) | 13 / 19 (68%) | 0.684 | 0.684 |
| PWY0-823~L-arginine degradation III (arginine decarboxylase/agmatinase pathway) | 0.78 | 0.97 | 1 | 0.5 | 0.93 | 1 | 0.0058 | 0.0057 | 0.0046 | 0.0056 | 0.0045 | 0.0034 | 0.0057 | 0.0048 | 0.0038 | 0.026 | 37 / 38 (97%) | 18 / 19 (95%) | 19 / 19 (100%) | 0.947 | 1 |
| PWY-7111~pyruvate fermentation to isobutanol (engineered) | 0.67 | 0.97 | 1 | 0.93 | 0.99 | 1 | 0.024 | 0.024 | 0.024 | 0.023 | 0.025 | 0.0067 | 0.024 | 0.024 | 0.0083 | 0.061 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY0-1298~superpathway of pyrimidine deoxyribonucleosides degradation | 0.051 | 0.97 | 1 | 0.12 | 0.91 | 1 | 0.0028 | 0.0027 | 0.0025 | 0.0031 | 0.0028 | 0.0015 | 0.0023 | 0.002 | 0.0013 | -0.43 | 37 / 38 (97%) | 19 / 19 (100%) | 18 / 19 (95%) | 1 | 0.947 |
| HISTSYN-PWY~L-histidine biosynthesis | 0.084 | 0.97 | 1 | 0.039 | 0.91 | 1 | 0.0095 | 0.0095 | 0.0092 | 0.01 | 0.0099 | 0.0024 | 0.009 | 0.0089 | 0.0033 | -0.15 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| ALANINE-DEG3-PWY~L-alanine degradation III | 0.67 | 0.97 | 1 | 0.19 | 0.91 | 1 | 0.0072 | 0.0072 | 0.0064 | 0.007 | 0.0069 | 0.0034 | 0.0075 | 0.005 | 0.008 | 0.1 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| MALTOSECAT-PWY~maltose degradation | 0.72 | 0.97 | 1 | 0.93 | 0.99 | 1 | 0.00048 | 0.00017 | 0 | 0.00019 | 0 | 0.00031 | 0.00014 | 0 | 0.00026 | -0.44 | 13 / 38 (34%) | 6 / 19 (32%) | 7 / 19 (37%) | 0.316 | 0.368 |
| PWY-4261~glycerol degradation I | 0.26 | 0.97 | 1 | 0.45 | 0.91 | 1 | 0.0014 | 0.0011 | 0.00064 | 0.00087 | 0.00044 | 0.0011 | 0.0013 | 0.00098 | 0.0013 | 0.58 | 29 / 38 (76%) | 14 / 19 (74%) | 15 / 19 (79%) | 0.737 | 0.789 |
| PWY-6470~peptidoglycan biosynthesis V (β-lactam resistance) | 0.45 | 0.97 | 1 | 0.26 | 0.91 | 1 | 0.006 | 0.0057 | 0.0036 | 0.0052 | 0.0035 | 0.0049 | 0.0062 | 0.0036 | 0.0097 | 0.25 | 36 / 38 (95%) | 17 / 19 (89%) | 19 / 19 (100%) | 0.895 | 1 |
| PWY-6376~desferrioxamine B biosynthesis | 0.24 | 0.97 | 1 | 0.26 | 0.91 | 1 | 0.005 | 0.0049 | 0.0049 | 0.0048 | 0.0049 | 0.0024 | 0.0051 | 0.0051 | 0.0027 | 0.087 | 37 / 38 (97%) | 18 / 19 (95%) | 19 / 19 (100%) | 0.947 | 1 |
| PWY-7388~octanoyl-[acyl-carrier protein] biosynthesis (mitochondria yeast) | 0.61 | 0.97 | 1 | 0.71 | 0.97 | 1 | 0.013 | 0.013 | 0.012 | 0.012 | 0.014 | 0.005 | 0.013 | 0.011 | 0.0084 | 0.12 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-7221~guanosine ribonucleotides de novo biosynthesis | 0.15 | 0.97 | 1 | 0.092 | 0.91 | 1 | 0.0012 | 0.00096 | 0.00075 | 0.00089 | 0.00063 | 0.0011 | 0.001 | 0.00085 | 0.00093 | 0.17 | 31 / 38 (82%) | 14 / 19 (74%) | 17 / 19 (89%) | 0.737 | 0.895 |
| PWY-7003~glycerol degradation to butanol | 0.49 | 0.97 | 1 | 0.63 | 0.96 | 1 | 0.0014 | 0.00056 | 0 | 0.00068 | 0 | 0.001 | 0.00044 | 0 | 0.00068 | -0.63 | 15 / 38 (39%) | 8 / 19 (42%) | 7 / 19 (37%) | 0.421 | 0.368 |
| ASPARAGINESYN-PWY~L-asparagine biosynthesis II | 0.84 | 0.97 | 1 | 0.77 | 0.99 | 1 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.0027 | 0.01 | 0.011 | 0.0028 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6527~stachyose degradation | 0.62 | 0.97 | 1 | 0.9 | 0.99 | 1 | 0.0019 | 0.0012 | 0.00068 | 0.0011 | 0.00066 | 0.0013 | 0.0012 | 0.00083 | 0.0018 | 0.13 | 23 / 38 (61%) | 11 / 19 (58%) | 12 / 19 (63%) | 0.579 | 0.632 |
| PWY-7176~UTP and CTP de novo biosynthesis | 0.15 | 0.97 | 1 | 0.06 | 0.91 | 1 | 0.00098 | 0.00082 | 0.00064 | 0.00078 | 0.00048 | 0.00098 | 0.00087 | 0.00065 | 0.00073 | 0.16 | 32 / 38 (84%) | 14 / 19 (74%) | 18 / 19 (95%) | 0.737 | 0.947 |
| PWY-6012-1~acyl carrier protein activation | 0.92 | 0.98 | 1 | 0.95 | 0.99 | 1 | 0.003 | 0.003 | 0.0026 | 0.0029 | 0.0027 | 0.0015 | 0.0031 | 0.0022 | 0.0025 | 0.096 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-7356~thiamine salvage IV (yeast) | 0.99 | 0.99 | 1 | 0.45 | 0.91 | 1 | 0.0044 | 0.0041 | 0.0036 | 0.0038 | 0.004 | 0.0025 | 0.0044 | 0.0031 | 0.0046 | 0.21 | 35 / 38 (92%) | 18 / 19 (95%) | 17 / 19 (89%) | 0.947 | 0.895 |
| PWY-4821~UDP-α-D-xylose biosynthesis | 0.55 | 0.97 | 1 | 0.62 | 0.96 | 1 | 0.0015 | 0.00039 | 0 | 2e-04 | 0 | 6e-04 | 0.00058 | 0 | 0.0023 | 1.5 | 10 / 38 (26%) | 4 / 19 (21%) | 6 / 19 (32%) | 0.211 | 0.316 |
| LARABITOLUTIL-PWY~xylitol degradation | 0.75 | 0.97 | 1 | 0.48 | 0.91 | 1 | 0.0036 | 0.003 | 0.0015 | 0.0027 | 0.0015 | 0.0028 | 0.0034 | 0.001 | 0.0059 | 0.33 | 32 / 38 (84%) | 16 / 19 (84%) | 16 / 19 (84%) | 0.842 | 0.842 |
| GLUTATHIONESYN-PWY~glutathione biosynthesis | 0.65 | 0.97 | 1 | 0.67 | 0.96 | 1 | 0.0015 | 0.00093 | 0.00015 | 0.0014 | 0.00012 | 0.0052 | 0.00044 | 0.00019 | 0.00065 | -1.7 | 23 / 38 (61%) | 11 / 19 (58%) | 12 / 19 (63%) | 0.579 | 0.632 |
| RHAMCAT-PWY~L-rhamnose degradation I | 0.29 | 0.97 | 1 | 0.79 | 0.99 | 1 | 0.0012 | 0.00083 | 0.00032 | 0.0012 | 0.00066 | 0.0017 | 0.00048 | 0.00028 | 0.00051 | -1.3 | 27 / 38 (71%) | 12 / 19 (63%) | 15 / 19 (79%) | 0.632 | 0.789 |
| PWY3O-355~stearate biosynthesis III (fungi) | 0.48 | 0.97 | 1 | 0.19 | 0.91 | 1 | 0.0058 | 0.0049 | 0.0034 | 0.0046 | 0.0035 | 0.0042 | 0.0051 | 0.0027 | 0.0076 | 0.15 | 32 / 38 (84%) | 16 / 19 (84%) | 16 / 19 (84%) | 0.842 | 0.842 |
| P121-PWY~adenine and adenosine salvage I | 0.72 | 0.97 | 1 | 0.4 | 0.91 | 1 | 0.007 | 0.007 | 0.0071 | 0.0071 | 0.0071 | 0.0015 | 0.0069 | 0.007 | 0.0021 | -0.041 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| ASPARTATESYN-PWY~L-aspartate biosynthesis | 0.57 | 0.97 | 1 | 0.27 | 0.91 | 1 | 0.02 | 0.02 | 0.019 | 0.02 | 0.02 | 0.0063 | 0.02 | 0.017 | 0.012 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY66-409~superpathway of purine nucleotide salvage | 0.87 | 0.97 | 1 | 0.85 | 0.99 | 1 | 0.0012 | 0.00085 | 0.00043 | 0.00098 | 0.00046 | 0.0016 | 0.00072 | 4e-04 | 0.00074 | -0.44 | 26 / 38 (68%) | 13 / 19 (68%) | 13 / 19 (68%) | 0.684 | 0.684 |
| PWY-6628~superpathway of L-phenylalanine biosynthesis | 0.7 | 0.97 | 1 | 0.37 | 0.91 | 1 | 0.01 | 0.01 | 0.01 | 0.01 | 0.011 | 0.0038 | 0.011 | 0.0086 | 0.006 | 0.14 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-7208~superpathway of pyrimidine nucleobases salvage | 0.19 | 0.97 | 1 | 0.07 | 0.91 | 1 | 0.001 | 0.00079 | 0.00056 | 0.00074 | 0.00051 | 0.00093 | 0.00083 | 6e-04 | 0.00079 | 0.17 | 30 / 38 (79%) | 13 / 19 (68%) | 17 / 19 (89%) | 0.684 | 0.895 |
| PWY-6277~superpathway of 5-aminoimidazole ribonucleotide biosynthesis | 0.54 | 0.97 | 1 | 0.26 | 0.91 | 1 | 0.014 | 0.014 | 0.013 | 0.014 | 0.015 | 0.0027 | 0.014 | 0.013 | 0.0036 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| ETOH-ACETYLCOA-ANA-PWY~ethanol degradation I | 0.57 | 0.97 | 1 | 0.72 | 0.97 | 1 | 0.0034 | 0.0034 | 0.0021 | 0.003 | 0.0017 | 0.0025 | 0.0037 | 0.0024 | 0.0048 | 0.3 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PROSYN-PWY~L-proline biosynthesis I | 0.77 | 0.97 | 1 | 0.64 | 0.96 | 1 | 0.011 | 0.011 | 0.01 | 0.011 | 0.011 | 0.0028 | 0.011 | 0.0099 | 0.0035 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| GLUCONEO-PWY~gluconeogenesis I | 0.12 | 0.97 | 1 | 0.036 | 0.91 | 1 | 0.0074 | 0.0074 | 0.0074 | 0.0081 | 0.0085 | 0.0028 | 0.0068 | 0.0068 | 0.0025 | -0.25 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-7199~pyrimidine deoxyribonucleosides salvage | 0.81 | 0.97 | 1 | 0.43 | 0.91 | 1 | 0.0011 | 0.00093 | 6e-04 | 0.001 | 6e-04 | 0.0012 | 0.00086 | 0.00059 | 0.00082 | -0.22 | 31 / 38 (82%) | 14 / 19 (74%) | 17 / 19 (89%) | 0.737 | 0.895 |
| RIBOKIN-PWY~ribose phosphorylation | 0.52 | 0.97 | 1 | 0.48 | 0.91 | 1 | 0.00064 | 0.00055 | 0.00036 | 0.00055 | 0.00033 | 0.00075 | 0.00055 | 0.00037 | 0.00067 | 0 | 33 / 38 (87%) | 16 / 19 (84%) | 17 / 19 (89%) | 0.842 | 0.895 |
| GLUTAMATE-DEG1-PWY~L-glutamate degradation I | 0.2 | 0.97 | 1 | 0.39 | 0.91 | 1 | 0.0026 | 0.0026 | 0.0023 | 0.0031 | 0.0029 | 0.0022 | 0.0021 | 0.0017 | 0.0012 | -0.56 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| FOLSYN-PWY~superpathway of tetrahydrofolate biosynthesis and salvage | 0.28 | 0.97 | 1 | 0.27 | 0.91 | 1 | 0.00084 | 2e-04 | 0 | 0.00026 | 0 | 0.00065 | 0.00014 | 0 | 0.00032 | -0.89 | 9 / 38 (24%) | 5 / 19 (26%) | 4 / 19 (21%) | 0.263 | 0.211 |
| PWY-4101~D-sorbitol degradation I | 0.93 | 0.98 | 1 | 0.92 | 0.99 | 1 | 0.013 | 0.013 | 0.013 | 0.014 | 0.013 | 0.0052 | 0.013 | 0.012 | 0.0041 | -0.11 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5831~CDP-abequose biosynthesis | 0.73 | 0.97 | 1 | 0.93 | 0.99 | 1 | 0.00028 | 4.4e-05 | 0 | 6.8e-05 | 0 | 0.00028 | 2e-05 | 0 | 5.6e-05 | -1.8 | 6 / 38 (16%) | 3 / 19 (16%) | 3 / 19 (16%) | 0.158 | 0.158 |
| PWY-5344~L-homocysteine biosynthesis | 0.4 | 0.97 | 1 | 0.28 | 0.91 | 1 | 0.023 | 0.023 | 0.023 | 0.023 | 0.024 | 0.0051 | 0.022 | 0.022 | 0.0065 | -0.064 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-7247~β-D-glucuronide and D-glucuronate degradation | 0.22 | 0.97 | 1 | 0.34 | 0.91 | 1 | 0.0073 | 0.0073 | 0.0068 | 0.0081 | 0.0089 | 0.004 | 0.0065 | 0.006 | 0.0029 | -0.32 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY0-1477~ethanolamine utilization | 0.31 | 0.97 | 1 | 0.24 | 0.91 | 1 | 0.0032 | 0.0031 | 0.0022 | 0.0027 | 0.0024 | 0.0022 | 0.0036 | 0.002 | 0.0047 | 0.42 | 37 / 38 (97%) | 18 / 19 (95%) | 19 / 19 (100%) | 0.947 | 1 |
| PWY-5490~paraoxon degradation | 0.59 | 0.97 | 1 | 0.75 | 0.98 | 1 | 0.00045 | 8.3e-05 | 0 | 1e-04 | 0 | 4e-04 | 6.2e-05 | 0 | 0.00022 | -0.69 | 7 / 38 (18%) | 4 / 19 (21%) | 3 / 19 (16%) | 0.211 | 0.158 |
| PWY-7229~superpathway of adenosine nucleotides de novo biosynthesis I | 0.94 | 0.99 | 1 | 0.71 | 0.97 | 1 | 0.0074 | 0.0074 | 0.0063 | 0.0072 | 0.0066 | 0.0029 | 0.0077 | 0.006 | 0.0046 | 0.097 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| LACTOSECAT-PWY~lactose and galactose degradation I | 0.21 | 0.97 | 1 | 0.19 | 0.91 | 1 | 0.00086 | 5e-04 | 0.00028 | 0.00038 | 0.00018 | 0.00049 | 0.00061 | 0.00033 | 0.00099 | 0.68 | 22 / 38 (58%) | 10 / 19 (53%) | 12 / 19 (63%) | 0.526 | 0.632 |
| PWY-6827~gellan degradation | 0.35 | 0.97 | 1 | 0.44 | 0.91 | 1 | 0.00016 | 1.3e-05 | 0 | 4.1e-06 | 0 | 1.8e-05 | 2.2e-05 | 0 | 8.3e-05 | 2.4 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| PWY-6057~dimethyl sulfide degradation III (oxidation) | 0.5 | 0.97 | 1 | 0.52 | 0.93 | 1 | 0.00021 | 1.7e-05 | 0 | 1.3e-05 | 0 | 5.5e-05 | 2.1e-05 | 0 | 6.8e-05 | 0.69 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| PWY0-662~PRPP biosynthesis | 0.3 | 0.97 | 1 | 0.68 | 0.96 | 1 | 0.025 | 0.025 | 0.025 | 0.024 | 0.026 | 0.0058 | 0.026 | 0.025 | 0.0081 | 0.12 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY4LZ-257~superpathway of fermentation (Chlamydomonas reinhardtii) | 0.84 | 0.97 | 1 | 0.78 | 0.99 | 1 | 0.0017 | 0.00031 | 0 | 0.00027 | 0 | 0.00081 | 0.00035 | 0 | 0.0012 | 0.37 | 7 / 38 (18%) | 4 / 19 (21%) | 3 / 19 (16%) | 0.211 | 0.158 |
| PWY-7581~N-acetylneuraminate and N-acetylmannosamine degradation II | 0.39 | 0.97 | 1 | 0.78 | 0.99 | 1 | 0.00064 | 0.00015 | 0 | 0.00025 | 0 | 0.00067 | 5.6e-05 | 0 | 1e-04 | -2.2 | 9 / 38 (24%) | 4 / 19 (21%) | 5 / 19 (26%) | 0.211 | 0.263 |
| PWY-7686~L-malate degradation II | 0.92 | 0.98 | 1 | 0.65 | 0.96 | 1 | 0.0069 | 0.0069 | 0.0068 | 0.0069 | 0.0068 | 0.002 | 0.0068 | 0.007 | 0.0019 | -0.021 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-7179-1~purine deoxyribonucleosides degradation II | 0.12 | 0.97 | 1 | 0.069 | 0.91 | 1 | 0.0035 | 0.0035 | 0.0032 | 0.0033 | 0.0031 | 0.0017 | 0.0038 | 0.0032 | 0.0021 | 0.2 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY0-1309~chitobiose degradation | 0.57 | 0.97 | 1 | 0.77 | 0.99 | 1 | 0.0069 | 0.002 | 0 | 0.0015 | 0 | 0.003 | 0.0025 | 0 | 0.005 | 0.74 | 11 / 38 (29%) | 5 / 19 (26%) | 6 / 19 (32%) | 0.263 | 0.316 |
| PWY-5853~demethylmenaquinol-6 biosynthesis I | 0.46 | 0.97 | 1 | 0.4 | 0.91 | 1 | 0.0029 | 0.0027 | 0.0018 | 0.0028 | 0.0028 | 0.0022 | 0.0027 | 0.00097 | 0.0041 | -0.052 | 36 / 38 (95%) | 18 / 19 (95%) | 18 / 19 (95%) | 0.947 | 0.947 |
| DTDPRHAMSYN-PWY~dTDP-L-rhamnose biosynthesis | 0.085 | 0.97 | 1 | 0.054 | 0.91 | 1 | 0.01 | 0.01 | 0.01 | 0.011 | 0.011 | 0.003 | 0.0093 | 0.009 | 0.0028 | -0.24 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| ACETATEUTIL-PWY~superpathway of acetate utilization and formation | 0.43 | 0.97 | 1 | 0.9 | 0.99 | 1 | 0.0019 | 0.0013 | 0.00056 | 0.0016 | 0.00047 | 0.0023 | 0.00097 | 0.00064 | 0.0012 | -0.72 | 25 / 38 (66%) | 12 / 19 (63%) | 13 / 19 (68%) | 0.632 | 0.684 |
| PWY-7167~choline degradation III | 0.61 | 0.97 | 1 | 0.88 | 0.99 | 1 | 0.0011 | 0.00022 | 0 | 2e-04 | 0 | 0.00067 | 0.00025 | 0 | 0.00086 | 0.32 | 8 / 38 (21%) | 4 / 19 (21%) | 4 / 19 (21%) | 0.211 | 0.211 |
| N2FIX-PWY~nitrogen fixation I (ferredoxin) | 0.21 | 0.97 | 1 | 0.29 | 0.91 | 1 | 0.009 | 0.009 | 0.0088 | 0.0085 | 0.0089 | 0.0034 | 0.0094 | 0.0088 | 0.0029 | 0.15 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY0-541~cyclopropane fatty acid (CFA) biosynthesis | 0.4 | 0.97 | 1 | 0.45 | 0.91 | 1 | 0.0037 | 0.0037 | 0.0032 | 0.0034 | 0.0029 | 0.0019 | 0.0039 | 0.0035 | 0.002 | 0.2 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-7761~NAD salvage pathway II (PNC IV cycle) | 0.086 | 0.97 | 1 | 0.033 | 0.91 | 1 | 0.001 | 0.00049 | 0 | 0.00041 | 0 | 0.00078 | 0.00057 | 0.00041 | 0.00065 | 0.48 | 18 / 38 (47%) | 7 / 19 (37%) | 11 / 19 (58%) | 0.368 | 0.579 |
| PWY-1042~glycolysis IV (plant cytosol) | 0.17 | 0.97 | 1 | 0.044 | 0.91 | 1 | 0.015 | 0.015 | 0.014 | 0.015 | 0.016 | 0.0026 | 0.014 | 0.014 | 0.0038 | -0.1 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| GLYCOLYSIS-E-D~superpathway of glycolysis and the Entner-Doudoroff pathway | 0.38 | 0.97 | 1 | 0.94 | 0.99 | 1 | 0.0021 | 0.00062 | 0 | 0.00084 | 0 | 0.0019 | 0.00039 | 0 | 0.00078 | -1.1 | 11 / 38 (29%) | 5 / 19 (26%) | 6 / 19 (32%) | 0.263 | 0.316 |
| HSERMETANA-PWY~L-methionine biosynthesis III | 0.67 | 0.97 | 1 | 0.47 | 0.91 | 1 | 0.011 | 0.011 | 0.012 | 0.012 | 0.012 | 0.0044 | 0.011 | 0.011 | 0.005 | -0.13 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| GLYCOCAT-PWY~glycogen degradation I | 0.37 | 0.97 | 1 | 0.22 | 0.91 | 1 | 0.014 | 0.014 | 0.014 | 0.015 | 0.014 | 0.0072 | 0.014 | 0.014 | 0.0055 | -0.1 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PYRIDOXSYN-PWY~pyridoxal 5-phosphate biosynthesis I | 0.15 | 0.97 | 1 | 0.19 | 0.91 | 1 | 0.0026 | 0.00062 | 0 | 0.00086 | 0 | 0.002 | 0.00038 | 0 | 0.00078 | -1.2 | 9 / 38 (24%) | 5 / 19 (26%) | 4 / 19 (21%) | 0.263 | 0.211 |
| PHOSPHONOTASE-PWY~2-aminoethylphosphonate degradation I | 0.06 | 0.97 | 1 | 0.077 | 0.91 | 1 | 0.00034 | 5.4e-05 | 0 | 6.7e-05 | 0 | 0.00017 | 4.2e-05 | 0 | 0.00013 | -0.67 | 6 / 38 (16%) | 4 / 19 (21%) | 2 / 19 (11%) | 0.211 | 0.105 |
| PWY-5838~superpathway of menaquinol-8 biosynthesis I | 0.47 | 0.97 | 1 | 0.31 | 0.91 | 1 | 0.00075 | 5.9e-05 | 0 | 6.5e-05 | 0 | 0.00019 | 5.4e-05 | 0 | 0.00023 | -0.27 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
| PWY-5100~pyruvate fermentation to acetate and lactate II | 0.62 | 0.97 | 1 | 0.64 | 0.96 | 1 | 0.011 | 0.011 | 0.011 | 0.01 | 0.011 | 0.0035 | 0.011 | 0.011 | 0.0034 | 0.14 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6285~superpathway of fatty acids biosynthesis (E. coli) | 0.1 | 0.97 | 1 | 0.2 | 0.91 | 1 | 0.0047 | 0.0047 | 0.0035 | 0.0051 | 0.0042 | 0.0041 | 0.0043 | 0.003 | 0.0045 | -0.25 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6019~pseudouridine degradation | 0.31 | 0.97 | 1 | 0.63 | 0.96 | 1 | 0.0014 | 0.00018 | 0 | 0.00013 | 0 | 0.00055 | 0.00023 | 0 | 0.00088 | 0.82 | 5 / 38 (13%) | 2 / 19 (11%) | 3 / 19 (16%) | 0.105 | 0.158 |
| MALATE-ASPARTATE-SHUTTLE-PWY~L-aspartate degradation II | 0.51 | 0.97 | 1 | 0.86 | 0.99 | 1 | 0.0027 | 0.0027 | 0.002 | 0.0031 | 0.0019 | 0.0034 | 0.0024 | 0.0021 | 0.0015 | -0.37 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6807~xyloglucan degradation II (exoglucanase) | 0.15 | 0.97 | 1 | 0.15 | 0.91 | 1 | 0.008 | 0.00063 | 0 | 0.0011 | 0 | 0.004 | 0.00017 | 0 | 0.00072 | -2.7 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
| FUC-RHAMCAT-PWY~superpathway of fucose and rhamnose degradation | 0.41 | 0.97 | 1 | 0.41 | 0.91 | 1 | 0.001 | 0.00019 | 0 | 0.00022 | 0 | 0.00049 | 0.00016 | 0 | 0.00042 | -0.46 | 7 / 38 (18%) | 4 / 19 (21%) | 3 / 19 (16%) | 0.211 | 0.158 |
| PHOSLIPSYN-PWY~superpathway of phospholipid biosynthesis I (bacteria) | 0.37 | 0.97 | 1 | 0.27 | 0.91 | 1 | 9e-04 | 7.1e-05 | 0 | 8.4e-05 | 0 | 0.00026 | 5.8e-05 | 0 | 0.00025 | -0.53 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
| GALACTUROCAT-PWY~D-galacturonate degradation I | 0.6 | 0.97 | 1 | 0.79 | 0.99 | 1 | 0.0033 | 0.0033 | 0.0029 | 0.0036 | 0.0028 | 0.0025 | 0.0031 | 0.003 | 0.0019 | -0.22 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5659~GDP-mannose biosynthesis | 0.79 | 0.97 | 1 | 0.87 | 0.99 | 1 | 0.0028 | 0.0025 | 0.0019 | 0.0025 | 0.0015 | 0.0023 | 0.0024 | 0.0021 | 0.0019 | -0.059 | 33 / 38 (87%) | 17 / 19 (89%) | 16 / 19 (84%) | 0.895 | 0.842 |
| PWY-5194~siroheme biosynthesis | 0.17 | 0.97 | 1 | 0.38 | 0.91 | 1 | 0.00048 | 0.00043 | 3e-04 | 0.00035 | 3e-04 | 3e-04 | 5e-04 | 0.00036 | 0.00043 | 0.51 | 34 / 38 (89%) | 17 / 19 (89%) | 17 / 19 (89%) | 0.895 | 0.895 |
| PWY0-1313~acetate conversion to acetyl-CoA | 0.55 | 0.97 | 1 | 0.8 | 0.99 | 1 | 0.0034 | 0.0032 | 0.003 | 0.0034 | 0.0045 | 0.0023 | 0.003 | 0.0026 | 0.002 | -0.18 | 36 / 38 (95%) | 18 / 19 (95%) | 18 / 19 (95%) | 0.947 | 0.947 |
| PWY-5669~phosphatidylethanolamine biosynthesis I | 0.1 | 0.97 | 1 | 0.082 | 0.91 | 1 | 0.0029 | 0.0029 | 0.0029 | 0.0032 | 0.0033 | 0.0014 | 0.0026 | 0.0027 | 0.0012 | -0.3 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5156~superpathway of fatty acid biosynthesis II (plant) | 0.16 | 0.97 | 1 | 0.94 | 0.99 | 1 | 0.0048 | 0.0044 | 0.003 | 0.0047 | 0.0031 | 0.0041 | 0.0042 | 0.0029 | 0.0044 | -0.16 | 35 / 38 (92%) | 17 / 19 (89%) | 18 / 19 (95%) | 0.895 | 0.947 |
| PWY-7970~benzimidazolyl adenosylcobamide biosynthesis from adenosylcobinamide-GDP | 0.59 | 0.97 | 1 | 0.39 | 0.91 | 1 | 0.0049 | 0.0049 | 0.0047 | 0.005 | 0.0047 | 0.0018 | 0.0047 | 0.0046 | 0.0014 | -0.089 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-7205~CMP phosphorylation | 0.47 | 0.97 | 1 | 0.35 | 0.91 | 1 | 0.00064 | 0.00057 | 0.00044 | 0.00057 | 0.00046 | 0.00065 | 0.00058 | 0.00044 | 0.00046 | 0.025 | 34 / 38 (89%) | 16 / 19 (84%) | 18 / 19 (95%) | 0.842 | 0.947 |
| GLUGLNSYN-PWY~L-glutamate biosynthesis IV | 0.58 | 0.97 | 1 | 0.42 | 0.91 | 1 | 0.039 | 0.039 | 0.039 | 0.039 | 0.039 | 0.014 | 0.038 | 0.04 | 0.011 | -0.037 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6617~adenosine nucleotides degradation III | 0.94 | 0.99 | 1 | 0.67 | 0.96 | 1 | 0.001 | 0.00044 | 0 | 0.00049 | 0 | 0.00092 | 0.00038 | 0 | 0.00069 | -0.37 | 16 / 38 (42%) | 7 / 19 (37%) | 9 / 19 (47%) | 0.368 | 0.474 |
| PWY-5148~acyl-CoA hydrolysis | 0.42 | 0.97 | 1 | 0.43 | 0.91 | 1 | 0.00027 | 7.8e-05 | 0 | 9.5e-05 | 0 | 0.00015 | 6.1e-05 | 0 | 0.00013 | -0.64 | 11 / 38 (29%) | 6 / 19 (32%) | 5 / 19 (26%) | 0.316 | 0.263 |
| GDPRHAMSYN-PWY~GDP-D-rhamnose biosynthesis | 0.61 | 0.97 | 1 | 0.29 | 0.91 | 1 | 0.0011 | 0.00034 | 0 | 0.00037 | 0 | 0.00084 | 0.00031 | 0 | 0.00054 | -0.26 | 12 / 38 (32%) | 4 / 19 (21%) | 8 / 19 (42%) | 0.211 | 0.421 |
| PWY-6406~salicylate biosynthesis I | 0.15 | 0.97 | 1 | 0.3 | 0.91 | 1 | 8e-04 | 6.3e-05 | 0 | 3.9e-05 | 0 | 0.00017 | 8.7e-05 | 0 | 0.00035 | 1.2 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| PLPSAL-PWY~pyridoxal 5-phosphate salvage I | 0.089 | 0.97 | 1 | 0.29 | 0.91 | 1 | 0.0011 | 0.00075 | 0.00047 | 0.00089 | 0.00066 | 0.00098 | 0.00062 | 0.00045 | 0.00068 | -0.52 | 26 / 38 (68%) | 13 / 19 (68%) | 13 / 19 (68%) | 0.684 | 0.684 |
| PWY-7242~D-fructuronate degradation | 0.24 | 0.97 | 1 | 0.57 | 0.93 | 1 | 0.0024 | 0.002 | 0.0013 | 0.0024 | 0.0021 | 0.0022 | 0.0016 | 0.0012 | 0.0017 | -0.58 | 32 / 38 (84%) | 16 / 19 (84%) | 16 / 19 (84%) | 0.842 | 0.842 |
| PWY-6164~3-dehydroquinate biosynthesis I | 0.67 | 0.97 | 1 | 0.86 | 0.99 | 1 | 0.019 | 0.019 | 0.018 | 0.018 | 0.019 | 0.0052 | 0.019 | 0.018 | 0.0089 | 0.078 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-724~superpathway of L-lysine L-threonine and L-methionine biosynthesis II | 0.97 | 0.99 | 1 | 0.95 | 0.99 | 1 | 0.0094 | 0.0094 | 0.0097 | 0.0092 | 0.01 | 0.0037 | 0.0096 | 0.0088 | 0.0062 | 0.061 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-7346~UDP-α-D-glucuronate biosynthesis (from UDP-glucose) | 0.6 | 0.97 | 1 | 0.63 | 0.96 | 1 | 0.011 | 0.011 | 0.01 | 0.011 | 0.011 | 0.0043 | 0.01 | 0.0097 | 0.0038 | -0.14 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| DAPLYSINESYN-PWY~L-lysine biosynthesis I | 0.95 | 0.99 | 1 | 0.47 | 0.91 | 1 | 0.0033 | 0.0028 | 0.0016 | 0.0024 | 0.0017 | 0.0023 | 0.0033 | 0.0016 | 0.0061 | 0.46 | 33 / 38 (87%) | 17 / 19 (89%) | 16 / 19 (84%) | 0.895 | 0.842 |
| HISDEG-PWY~L-histidine degradation I | 0.15 | 0.97 | 1 | 0.1 | 0.91 | 1 | 0.00023 | 5.4e-05 | 0 | 7.4e-05 | 0 | 0.00015 | 3.3e-05 | 0 | 8.5e-05 | -1.2 | 9 / 38 (24%) | 6 / 19 (32%) | 3 / 19 (16%) | 0.316 | 0.158 |
| PWY-7518~atromentin biosynthesis | 0.28 | 0.97 | 1 | 0.26 | 0.91 | 1 | 0.00082 | 0.00065 | 0.00046 | 0.00053 | 0.00045 | 0.00056 | 0.00077 | 0.00053 | 0.00089 | 0.54 | 30 / 38 (79%) | 14 / 19 (74%) | 16 / 19 (84%) | 0.737 | 0.842 |
| PWY-2161~folate polyglutamylation | 0.74 | 0.97 | 1 | 0.81 | 0.99 | 1 | 0.013 | 0.013 | 0.012 | 0.013 | 0.013 | 0.0041 | 0.013 | 0.012 | 0.0061 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-7432~L-phenylalanine biosynthesis III (cytosolic plants) | 0.26 | 0.97 | 1 | 0.27 | 0.91 | 1 | 0.001 | 0.00076 | 0.00052 | 0.00061 | 0.00044 | 0.00063 | 0.00091 | 0.00057 | 0.0011 | 0.58 | 28 / 38 (74%) | 13 / 19 (68%) | 15 / 19 (79%) | 0.684 | 0.789 |
| PWY66-162~ethanol degradation IV | 0.16 | 0.97 | 1 | 0.36 | 0.91 | 1 | 0.0015 | 0.00012 | 0 | 1e-04 | 0 | 0.00045 | 0.00014 | 0 | 0.00058 | 0.49 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| PWY-622~starch biosynthesis | 0.97 | 0.99 | 1 | 0.56 | 0.93 | 1 | 0.0083 | 0.0064 | 0.0025 | 0.0053 | 0.0021 | 0.0065 | 0.0075 | 0.0038 | 0.017 | 0.5 | 29 / 38 (76%) | 15 / 19 (79%) | 14 / 19 (74%) | 0.789 | 0.737 |
| PWY0-1545~cardiolipin biosynthesis III | 0.26 | 0.97 | 1 | 0.28 | 0.91 | 1 | 0.00033 | 0.00012 | 0 | 0.00016 | 0 | 0.00032 | 8.4e-05 | 0 | 0.00016 | -0.93 | 14 / 38 (37%) | 8 / 19 (42%) | 6 / 19 (32%) | 0.421 | 0.316 |
| PWY-7685~L-malate degradation I | 0.15 | 0.97 | 1 | 0.19 | 0.91 | 1 | 0.00013 | 9.9e-06 | 0 | 1.5e-05 | 0 | 5.3e-05 | 5.1e-06 | 0 | 2.2e-05 | -1.6 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
| PWY-6612~superpathway of tetrahydrofolate biosynthesis | 0.18 | 0.97 | 1 | 0.15 | 0.91 | 1 | 0.00075 | 2e-04 | 0 | 0.00026 | 0 | 0.00062 | 0.00013 | 0 | 0.00031 | -1 | 10 / 38 (26%) | 6 / 19 (32%) | 4 / 19 (21%) | 0.316 | 0.211 |
| PWY-621~sucrose degradation III (sucrose invertase) | 0.97 | 0.99 | 1 | 0.53 | 0.93 | 1 | 0.015 | 0.015 | 0.015 | 0.014 | 0.015 | 0.0055 | 0.015 | 0.014 | 0.0095 | 0.1 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-2781~cis-zeatin biosynthesis | 0.41 | 0.97 | 1 | 0.86 | 0.99 | 1 | 0.012 | 0.012 | 0.012 | 0.012 | 0.012 | 0.0025 | 0.013 | 0.012 | 0.0036 | 0.12 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-283~benzoate degradation II (aerobic and anaerobic) | 0.21 | 0.97 | 1 | 0.42 | 0.91 | 1 | 0.0036 | 0.00047 | 0 | 0.00085 | 0 | 0.0023 | 8.8e-05 | 0 | 0.00026 | -3.3 | 5 / 38 (13%) | 3 / 19 (16%) | 2 / 19 (11%) | 0.158 | 0.105 |
| PWY-6910~hydroxymethylpyrimidine salvage | 0.48 | 0.97 | 1 | 0.96 | 0.99 | 1 | 0.011 | 0.011 | 0.011 | 0.011 | 0.01 | 0.0029 | 0.011 | 0.011 | 0.0036 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| GLYSYN-PWY~glycine biosynthesis I | 0.24 | 0.97 | 1 | 0.74 | 0.98 | 1 | 0.014 | 0.014 | 0.014 | 0.014 | 0.013 | 0.0027 | 0.014 | 0.014 | 0.004 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| GLUAMCAT-PWY~N-acetylglucosamine degradation I | 0.57 | 0.97 | 1 | 0.31 | 0.91 | 1 | 0.013 | 0.013 | 0.012 | 0.013 | 0.013 | 0.0038 | 0.012 | 0.011 | 0.0026 | -0.12 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5895~menaquinol-13 biosynthesis | 0.24 | 0.97 | 1 | 0.48 | 0.91 | 1 | 0.0015 | 0.00073 | 6.1e-05 | 0.00093 | 0.00012 | 0.0015 | 0.00052 | 0 | 0.00089 | -0.84 | 19 / 38 (50%) | 10 / 19 (53%) | 9 / 19 (47%) | 0.526 | 0.474 |
| PWY-6050~dimethyl sulfoxide degradation | 0.2 | 0.97 | 1 | 0.28 | 0.91 | 1 | 0.00081 | 4e-04 | 4.7e-05 | 0.00032 | 0 | 0.00075 | 0.00049 | 0.00012 | 0.00086 | 0.61 | 19 / 38 (50%) | 8 / 19 (42%) | 11 / 19 (58%) | 0.421 | 0.579 |
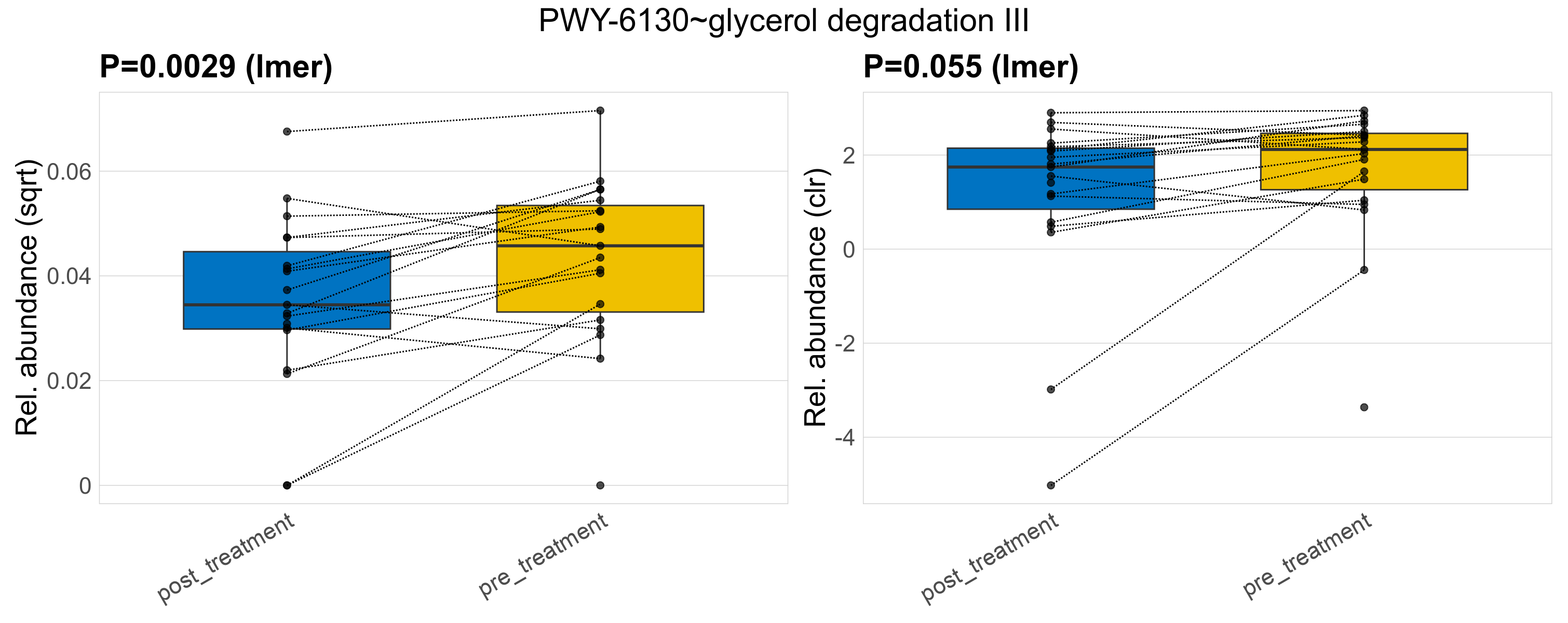
| PWY-6130~glycerol degradation III | 0.0029 | 0.41 | 1 | 0.055 | 0.91 | 1 | 0.0019 | 0.0018 | 0.0017 | 0.0015 | 0.0012 | 0.0011 | 0.0021 | 0.0021 | 0.0012 | 0.49 | 35 / 38 (92%) | 17 / 19 (89%) | 18 / 19 (95%) | 0.895 | 0.947 |
| PWY-6535~4-aminobutanoate degradation I | 0.8 | 0.97 | 1 | 0.38 | 0.91 | 1 | 0.00044 | 3.5e-05 | 0 | 3e-05 | 0 | 0.00012 | 4e-05 | 0 | 0.00018 | 0.42 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
| PWY18C3-24~methylsalicylate degradation | 0.14 | 0.97 | 1 | 0.14 | 0.91 | 1 | 0.0019 | 0.0018 | 0.0016 | 0.0019 | 0.0016 | 0.0013 | 0.0017 | 0.0017 | 0.0014 | -0.16 | 37 / 38 (97%) | 19 / 19 (100%) | 18 / 19 (95%) | 1 | 0.947 |
| GLYCLEAV-PWY~glycine cleavage | 0.69 | 0.97 | 1 | 0.45 | 0.91 | 1 | 0.0021 | 0.0019 | 0.00089 | 0.0021 | 0.00082 | 0.0028 | 0.0018 | 0.00097 | 0.0022 | -0.22 | 35 / 38 (92%) | 18 / 19 (95%) | 17 / 19 (89%) | 0.947 | 0.895 |
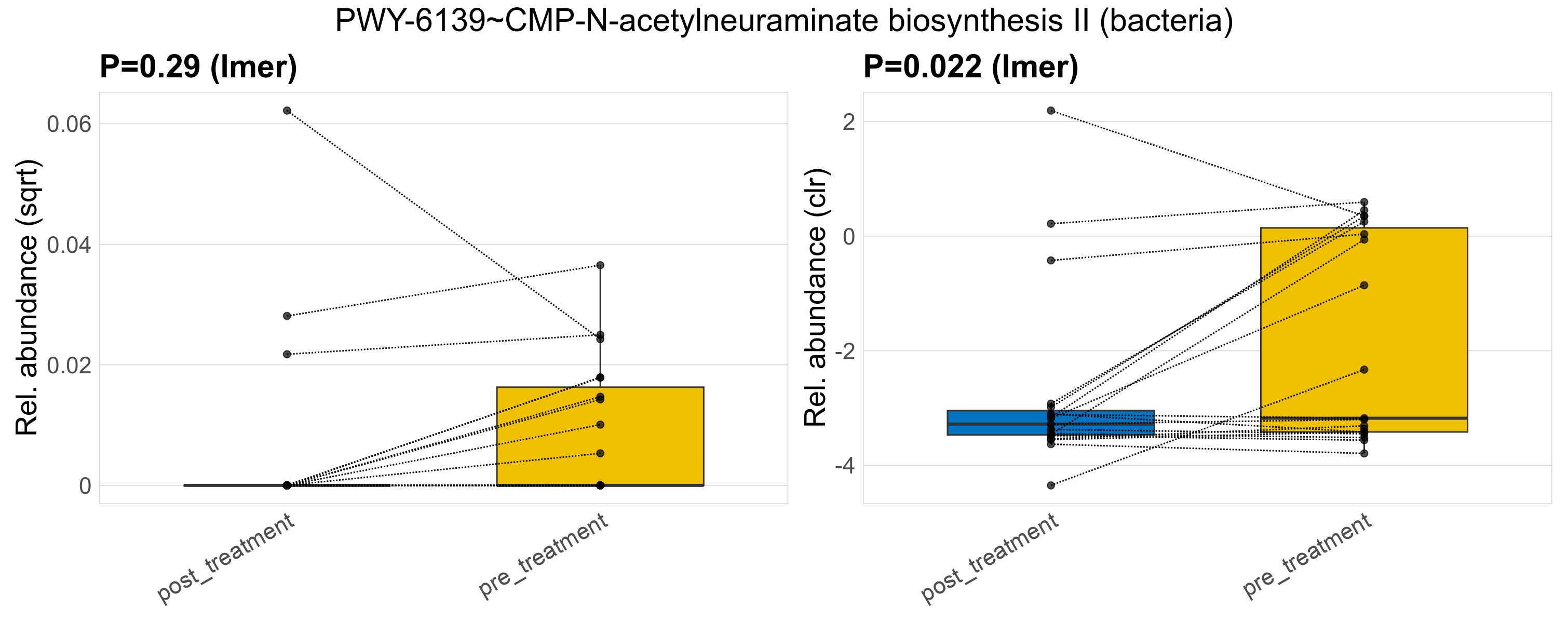
| PWY-6139~CMP-N-acetylneuraminate biosynthesis II (bacteria) | 0.29 | 0.97 | 1 | 0.022 | 0.91 | 1 | 0.00074 | 0.00023 | 0 | 0.00027 | 0 | 9e-04 | 2e-04 | 0 | 0.00034 | -0.43 | 12 / 38 (32%) | 3 / 19 (16%) | 9 / 19 (47%) | 0.158 | 0.474 |
| PWY-7184~pyrimidine deoxyribonucleotides de novo biosynthesis I | 0.56 | 0.97 | 1 | 0.71 | 0.97 | 1 | 0.00091 | 0.00043 | 0 | 0.00034 | 0 | 0.00075 | 0.00052 | 0 | 0.0011 | 0.61 | 18 / 38 (47%) | 9 / 19 (47%) | 9 / 19 (47%) | 0.474 | 0.474 |
| PWY-8004~Entner-Doudoroff pathway I | 0.16 | 0.97 | 1 | 0.19 | 0.91 | 1 | 0.0018 | 0.00024 | 0 | 4e-04 | 0 | 0.0011 | 7.4e-05 | 0 | 0.00025 | -2.4 | 5 / 38 (13%) | 3 / 19 (16%) | 2 / 19 (11%) | 0.158 | 0.105 |
| PWY-3801~sucrose degradation II (sucrose synthase) | 0.57 | 0.97 | 1 | 0.9 | 0.99 | 1 | 0.0051 | 0.0051 | 0.0038 | 0.0047 | 0.0043 | 0.0028 | 0.0054 | 0.0037 | 0.0056 | 0.2 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6519~8-amino-7-oxononanoate biosynthesis I | 0.25 | 0.97 | 1 | 0.38 | 0.91 | 1 | 0.0028 | 0.0011 | 0 | 0.0013 | 0 | 0.0027 | 0.00084 | 0 | 0.0018 | -0.63 | 15 / 38 (39%) | 8 / 19 (42%) | 7 / 19 (37%) | 0.421 | 0.368 |
| PWY4FS-7~phosphatidylglycerol biosynthesis I (plastidic) | 0.92 | 0.98 | 1 | 0.52 | 0.93 | 1 | 0.00092 | 0.00078 | 0.00073 | 0.00081 | 0.00091 | 0.00069 | 0.00074 | 0.00054 | 0.00066 | -0.13 | 32 / 38 (84%) | 15 / 19 (79%) | 17 / 19 (89%) | 0.789 | 0.895 |
| PWY0-162~superpathway of pyrimidine ribonucleotides de novo biosynthesis | 0.78 | 0.97 | 1 | 0.59 | 0.94 | 1 | 0.0041 | 0.0038 | 0.0026 | 0.0035 | 0.0024 | 0.0033 | 0.0042 | 0.003 | 0.0073 | 0.26 | 36 / 38 (95%) | 18 / 19 (95%) | 18 / 19 (95%) | 0.947 | 0.947 |
| PWY-5674~nitrate reduction IV (dissimilatory) | 0.37 | 0.97 | 1 | 0.55 | 0.93 | 1 | 0.00055 | 0.00016 | 0 | 0.00014 | 0 | 0.00043 | 0.00017 | 0 | 0.00045 | 0.28 | 11 / 38 (29%) | 5 / 19 (26%) | 6 / 19 (32%) | 0.263 | 0.316 |
| PWY-3282~superpathway of ammonia assimilation (plants) | 0.57 | 0.97 | 1 | 0.33 | 0.91 | 1 | 0.033 | 0.033 | 0.032 | 0.033 | 0.033 | 0.0096 | 0.032 | 0.032 | 0.01 | -0.044 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6545~pyrimidine deoxyribonucleotides de novo biosynthesis III | 0.17 | 0.97 | 1 | 0.14 | 0.91 | 1 | 0.0013 | 0.00027 | 0 | 2e-04 | 0 | 7e-04 | 0.00034 | 0 | 0.00088 | 0.77 | 8 / 38 (21%) | 2 / 19 (11%) | 6 / 19 (32%) | 0.105 | 0.316 |
| PWY-6610~adenine salvage | 0.062 | 0.97 | 1 | 0.081 | 0.91 | 1 | 0.0056 | 0.0056 | 0.0058 | 0.0053 | 0.0054 | 0.0019 | 0.0058 | 0.006 | 0.0015 | 0.13 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6590~superpathway of Clostridium acetobutylicum acidogenic fermentation | 0.61 | 0.97 | 1 | 0.52 | 0.93 | 1 | 6e-04 | 0.00035 | 0.00022 | 0.00036 | 0.00034 | 0.00039 | 0.00033 | 0.00014 | 0.00053 | -0.13 | 22 / 38 (58%) | 12 / 19 (63%) | 10 / 19 (53%) | 0.632 | 0.526 |
| PWY-6348~phosphate acquisition | 0.28 | 0.97 | 1 | 0.2 | 0.91 | 1 | 0.0026 | 0.0024 | 0.0017 | 0.0026 | 0.0014 | 0.0028 | 0.0022 | 0.0018 | 0.0037 | -0.24 | 35 / 38 (92%) | 18 / 19 (95%) | 17 / 19 (89%) | 0.947 | 0.895 |
| PWY-6389~pyruvate fermentation to (S)-acetoin | 0.31 | 0.97 | 1 | 0.22 | 0.91 | 1 | 0.0022 | 0.0018 | 0.001 | 0.0015 | 0.00081 | 0.0026 | 0.002 | 0.0011 | 0.0028 | 0.42 | 30 / 38 (79%) | 14 / 19 (74%) | 16 / 19 (84%) | 0.737 | 0.842 |
| P23-PWY~reductive TCA cycle I | 0.36 | 0.97 | 1 | 0.25 | 0.91 | 1 | 0.0013 | 0.00021 | 0 | 0.00026 | 0 | 0.00066 | 0.00017 | 0 | 0.00052 | -0.61 | 6 / 38 (16%) | 3 / 19 (16%) | 3 / 19 (16%) | 0.158 | 0.158 |
| PWY-5297~siroheme amide biosynthesis | 0.6 | 0.97 | 1 | 0.48 | 0.91 | 1 | 0.00026 | 4e-05 | 0 | 4e-05 | 0 | 0.00015 | 4.1e-05 | 0 | 8.6e-05 | 0.036 | 6 / 38 (16%) | 2 / 19 (11%) | 4 / 19 (21%) | 0.105 | 0.211 |
| PWY3O-4106~NAD salvage pathway IV (from nicotinamide riboside) | 0.69 | 0.97 | 1 | 0.93 | 0.99 | 1 | 0.00052 | 0.00022 | 0 | 0.00017 | 0 | 0.00039 | 0.00026 | 0 | 0.00065 | 0.61 | 16 / 38 (42%) | 8 / 19 (42%) | 8 / 19 (42%) | 0.421 | 0.421 |
| PWY66-366~flavin biosynthesis IV (mammalian) | 0.6 | 0.97 | 1 | 0.34 | 0.91 | 1 | 0.011 | 0.011 | 0.011 | 0.011 | 0.011 | 0.0017 | 0.011 | 0.011 | 0.0019 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5981~CDP-diacylglycerol biosynthesis III | 0.87 | 0.97 | 1 | 0.74 | 0.98 | 1 | 0.0097 | 0.0097 | 0.0094 | 0.0098 | 0.011 | 0.0027 | 0.0096 | 0.009 | 0.0021 | -0.03 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY0-1587~N6-L-threonylcarbamoyladenosine37-modified tRNA biosynthesis | 0.31 | 0.97 | 1 | 0.048 | 0.91 | 1 | 0.0098 | 0.0098 | 0.01 | 0.01 | 0.0098 | 0.0019 | 0.0095 | 0.01 | 0.0024 | -0.074 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| MANNCAT-PWY~D-mannose degradation | 0.52 | 0.97 | 1 | 0.61 | 0.96 | 1 | 0.007 | 0.007 | 0.0065 | 0.0074 | 0.0066 | 0.0034 | 0.0066 | 0.0064 | 0.0027 | -0.17 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-7335~UDP-N-acetyl-α-D-mannosaminouronate biosynthesis | 0.52 | 0.97 | 1 | 0.5 | 0.93 | 1 | 0.0015 | 0.00044 | 0 | 0.00054 | 0 | 0.0011 | 0.00035 | 0 | 0.00081 | -0.63 | 11 / 38 (29%) | 6 / 19 (32%) | 5 / 19 (26%) | 0.316 | 0.263 |
| UDPNAGSYN-PWY~UDP-N-acetyl-D-glucosamine biosynthesis I | 0.69 | 0.97 | 1 | 0.86 | 0.99 | 1 | 0.0071 | 0.0071 | 0.0059 | 0.0067 | 0.0065 | 0.0035 | 0.0074 | 0.0051 | 0.0056 | 0.14 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-7204~pyridoxal 5-phosphate salvage II (plants) | 0.6 | 0.97 | 1 | 0.98 | 0.99 | 1 | 0.00083 | 0.00044 | 0.00019 | 0.00043 | 0 | 0.00058 | 0.00044 | 0.00026 | 0.00071 | 0.033 | 20 / 38 (53%) | 9 / 19 (47%) | 11 / 19 (58%) | 0.474 | 0.579 |
| GLYCOGENSYNTH-PWY~glycogen biosynthesis I (from ADP-D-Glucose) | 0.55 | 0.97 | 1 | 0.92 | 0.99 | 1 | 0.036 | 0.036 | 0.037 | 0.035 | 0.039 | 0.01 | 0.037 | 0.037 | 0.015 | 0.08 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY0-1391~S-methyl-5-thioadenosine degradation IV | 0.4 | 0.97 | 1 | 0.34 | 0.91 | 1 | 0.00024 | 5.7e-05 | 0 | 5.5e-05 | 0 | 0.00015 | 5.9e-05 | 0 | 0.00014 | 0.1 | 9 / 38 (24%) | 3 / 19 (16%) | 6 / 19 (32%) | 0.158 | 0.316 |
| PWY-6381~bisucaberin biosynthesis | 0.24 | 0.97 | 1 | 0.26 | 0.91 | 1 | 0.005 | 0.0049 | 0.0049 | 0.0048 | 0.0049 | 0.0024 | 0.0051 | 0.0051 | 0.0027 | 0.087 | 37 / 38 (97%) | 18 / 19 (95%) | 19 / 19 (100%) | 0.947 | 1 |
| PWY-5910~superpathway of geranylgeranyldiphosphate biosynthesis I (via mevalonate) | 0.64 | 0.97 | 1 | 0.54 | 0.93 | 1 | 0.00025 | 3.3e-05 | 0 | 3.9e-05 | 0 | 0.00011 | 2.7e-05 | 0 | 9.6e-05 | -0.53 | 5 / 38 (13%) | 3 / 19 (16%) | 2 / 19 (11%) | 0.158 | 0.105 |
| PWY-4381~fatty acid biosynthesis initiation I | 0.52 | 0.97 | 1 | 0.48 | 0.91 | 1 | 0.009 | 0.009 | 0.0084 | 0.0092 | 0.0088 | 0.0029 | 0.0087 | 0.0073 | 0.0026 | -0.081 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-7664~oleate biosynthesis IV (anaerobic) | 0.095 | 0.97 | 1 | 0.2 | 0.91 | 1 | 0.0048 | 0.0048 | 0.0034 | 0.0052 | 0.0042 | 0.0042 | 0.0043 | 0.003 | 0.0045 | -0.27 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY66-426~hydrogen sulfide biosynthesis II (mammalian) | 0.26 | 0.97 | 1 | 0.32 | 0.91 | 1 | 0.0023 | 0.00018 | 0 | 0.00013 | 0 | 0.00056 | 0.00024 | 0 | 0.00074 | 0.88 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| PWY-6595~superpathway of guanosine nucleotides degradation (plants) | 0.27 | 0.97 | 1 | 0.46 | 0.91 | 1 | 0.0014 | 0.00046 | 0 | 0.00035 | 0 | 0.00066 | 0.00058 | 0 | 0.00091 | 0.73 | 13 / 38 (34%) | 6 / 19 (32%) | 7 / 19 (37%) | 0.316 | 0.368 |
| SAM-PWY~S-adenosyl-L-methionine biosynthesis | 0.92 | 0.98 | 1 | 0.6 | 0.95 | 1 | 0.024 | 0.024 | 0.023 | 0.024 | 0.024 | 0.0052 | 0.023 | 0.023 | 0.0059 | -0.061 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-7179~purine deoxyribonucleosides degradation I | 0.13 | 0.97 | 1 | 0.069 | 0.91 | 1 | 0.0036 | 0.0036 | 0.0032 | 0.0034 | 0.0031 | 0.0018 | 0.0039 | 0.0032 | 0.0022 | 0.2 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY0-1275~lipoate biosynthesis and incorporation II | 0.33 | 0.97 | 1 | 0.33 | 0.91 | 1 | 0.00039 | 8.1e-05 | 0 | 4.3e-05 | 0 | 0.00012 | 0.00012 | 0 | 0.00036 | 1.5 | 8 / 38 (21%) | 3 / 19 (16%) | 5 / 19 (26%) | 0.158 | 0.263 |
| PANTO-PWY~phosphopantothenate biosynthesis I | 0.05 | 0.97 | 1 | 0.03 | 0.91 | 1 | 0.0029 | 0.0029 | 0.0027 | 0.0033 | 0.0028 | 0.0016 | 0.0024 | 0.0018 | 0.0014 | -0.46 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-7996~menaquinol-4 biosynthesis I | 0.83 | 0.97 | 1 | 0.96 | 0.99 | 1 | 0.0057 | 0.0057 | 0.0056 | 0.0056 | 0.0056 | 0.0021 | 0.0057 | 0.0055 | 0.0018 | 0.026 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| GLYCOLYSIS~glycolysis I (from glucose 6-phosphate) | 0.077 | 0.97 | 1 | 0.011 | 0.75 | 1 | 0.011 | 0.011 | 0.012 | 0.012 | 0.013 | 0.0031 | 0.01 | 0.01 | 0.003 | -0.26 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-3001~superpathway of L-isoleucine biosynthesis I | 0.49 | 0.97 | 1 | 0.2 | 0.91 | 1 | 0.017 | 0.017 | 0.017 | 0.018 | 0.018 | 0.0048 | 0.017 | 0.016 | 0.0071 | -0.082 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5695~inosine 5-phosphate degradation | 0.97 | 0.99 | 1 | 0.82 | 0.99 | 1 | 0.0064 | 0.0064 | 0.0061 | 0.0064 | 0.0057 | 0.0026 | 0.0064 | 0.0064 | 0.0023 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| GLCMANNANAUT-PWY~superpathway of N-acetylglucosamine N-acetylmannosamine and N-acetylneuraminate degradation | 0.96 | 0.99 | 1 | 0.83 | 0.99 | 1 | 0.0045 | 0.0045 | 0.0045 | 0.0045 | 0.0037 | 0.0022 | 0.0044 | 0.0046 | 0.0018 | -0.032 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5849~menaquinol-6 biosynthesis | 0.83 | 0.97 | 1 | 0.96 | 0.99 | 1 | 0.0057 | 0.0057 | 0.0056 | 0.0056 | 0.0056 | 0.0021 | 0.0057 | 0.0055 | 0.0018 | 0.026 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5516~D-xylose degradation II | 0.04 | 0.97 | 1 | 0.032 | 0.91 | 1 | 0.00084 | 0.00084 | 0.00082 | 0.00074 | 0.00076 | 0.00032 | 0.00094 | 0.00097 | 0.00042 | 0.35 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY0-1317~L-lactaldehyde degradation (aerobic) | 0.81 | 0.97 | 1 | 0.56 | 0.93 | 1 | 0.0012 | 0.00042 | 0 | 0.00046 | 0 | 0.0011 | 0.00039 | 0 | 0.00071 | -0.24 | 14 / 38 (37%) | 6 / 19 (32%) | 8 / 19 (42%) | 0.316 | 0.421 |
| BIOTIN-BIOSYNTHESIS-PWY~biotin biosynthesis I | 0.27 | 0.97 | 1 | 0.58 | 0.94 | 1 | 0.0026 | 0.00096 | 0 | 0.0012 | 0 | 0.0024 | 7e-04 | 0 | 0.0015 | -0.78 | 14 / 38 (37%) | 7 / 19 (37%) | 7 / 19 (37%) | 0.368 | 0.368 |
| P41-PWY~pyruvate fermentation to acetate and (S)-lactate I | 0.32 | 0.97 | 1 | 0.3 | 0.91 | 1 | 0.0028 | 0.0028 | 0.0017 | 0.0023 | 0.0016 | 0.0021 | 0.0033 | 0.002 | 0.0047 | 0.52 | 37 / 38 (97%) | 18 / 19 (95%) | 19 / 19 (100%) | 0.947 | 1 |
| PWY-5482~pyruvate fermentation to acetate II | 0.33 | 0.97 | 1 | 0.32 | 0.91 | 1 | 0.0029 | 0.0029 | 0.0018 | 0.0024 | 0.0015 | 0.0021 | 0.0034 | 0.0022 | 0.0048 | 0.5 | 37 / 38 (97%) | 18 / 19 (95%) | 19 / 19 (100%) | 0.947 | 1 |
| PWY-5886~4-hydroxyphenylpyruvate biosynthesis | 0.28 | 0.97 | 1 | 0.26 | 0.91 | 1 | 0.00082 | 0.00065 | 0.00046 | 0.00053 | 0.00045 | 0.00056 | 0.00077 | 0.00053 | 0.00089 | 0.54 | 30 / 38 (79%) | 14 / 19 (74%) | 16 / 19 (84%) | 0.737 | 0.842 |
| PWY-723~alkylnitronates degradation | 0.08 | 0.97 | 1 | 0.59 | 0.94 | 1 | 0.00026 | 4.1e-05 | 0 | 3.6e-05 | 0 | 0.00011 | 4.6e-05 | 0 | 0.00014 | 0.35 | 6 / 38 (16%) | 3 / 19 (16%) | 3 / 19 (16%) | 0.158 | 0.158 |
| PWY-5851~demethylmenaquinol-9 biosynthesis | 0.46 | 0.97 | 1 | 0.4 | 0.91 | 1 | 0.0029 | 0.0027 | 0.0018 | 0.0028 | 0.0028 | 0.0022 | 0.0027 | 0.00097 | 0.0041 | -0.052 | 36 / 38 (95%) | 18 / 19 (95%) | 18 / 19 (95%) | 0.947 | 0.947 |
| PWY-6028~acetoin degradation | 0.95 | 0.99 | 1 | 0.8 | 0.99 | 1 | 0.00036 | 8.5e-05 | 0 | 7.8e-05 | 0 | 0.00023 | 9.1e-05 | 0 | 0.00023 | 0.22 | 9 / 38 (24%) | 5 / 19 (26%) | 4 / 19 (21%) | 0.263 | 0.211 |
| PWY-6536~4-aminobutanoate degradation III | 0.76 | 0.97 | 1 | 0.35 | 0.91 | 1 | 0.00092 | 7.3e-05 | 0 | 6.5e-05 | 0 | 0.00027 | 8.1e-05 | 0 | 0.00035 | 0.32 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
| PWY0-845~superpathway of pyridoxal 5-phosphate biosynthesis and salvage | 0.13 | 0.97 | 1 | 0.14 | 0.91 | 1 | 0.0022 | 0.00047 | 0 | 7e-04 | 0 | 0.0016 | 0.00024 | 0 | 0.00058 | -1.5 | 8 / 38 (21%) | 5 / 19 (26%) | 3 / 19 (16%) | 0.263 | 0.158 |
| PWY-6353~purine nucleotides degradation II (aerobic) | 0.66 | 0.97 | 1 | 0.9 | 0.99 | 1 | 0.0058 | 0.0058 | 0.0057 | 0.0057 | 0.0052 | 0.0022 | 0.0059 | 0.0065 | 0.002 | 0.05 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| P4-PWY~superpathway of L-lysine L-threonine and L-methionine biosynthesis I | 0.9 | 0.98 | 1 | 0.73 | 0.97 | 1 | 0.0038 | 0.0028 | 0.0014 | 0.0024 | 0.0012 | 0.0027 | 0.0032 | 0.0016 | 0.0066 | 0.42 | 28 / 38 (74%) | 14 / 19 (74%) | 14 / 19 (74%) | 0.737 | 0.737 |
| ARGDEG-III-PWY~L-arginine degradation IV (arginine decarboxylase/agmatine deiminase pathway) | 0.85 | 0.97 | 1 | 0.68 | 0.96 | 1 | 0.0067 | 0.0067 | 0.0062 | 0.0067 | 0.0059 | 0.0026 | 0.0068 | 0.0066 | 0.0026 | 0.021 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-701~L-methionine degradation II | 0.77 | 0.97 | 1 | 0.82 | 0.99 | 1 | 0.00018 | 0.00011 | 8.4e-05 | 0.00011 | 7.5e-05 | 0.00016 | 0.00011 | 8.7e-05 | 0.00016 | 0 | 23 / 38 (61%) | 11 / 19 (58%) | 12 / 19 (63%) | 0.579 | 0.632 |
| ALACAT2-PWY~L-alanine degradation II (to D-lactate) | 0.77 | 0.97 | 1 | 0.61 | 0.96 | 1 | 0.0012 | 0.00043 | 0 | 0.00055 | 0 | 0.0012 | 0.00031 | 0 | 0.00057 | -0.83 | 14 / 38 (37%) | 6 / 19 (32%) | 8 / 19 (42%) | 0.316 | 0.421 |
| PWY0-1466~trehalose degradation VI (periplasmic) | 0.88 | 0.98 | 1 | 0.76 | 0.99 | 1 | 0.0028 | 0.0018 | 0.00094 | 0.0022 | 0.00086 | 0.0043 | 0.0015 | 0.001 | 0.0019 | -0.55 | 25 / 38 (66%) | 12 / 19 (63%) | 13 / 19 (68%) | 0.632 | 0.684 |
| PWY-6897~thiamine salvage II | 0.84 | 0.97 | 1 | 0.36 | 0.91 | 1 | 0.01 | 0.01 | 0.01 | 0.01 | 0.011 | 0.0028 | 0.01 | 0.0094 | 0.0045 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5964~guanylyl molybdenum cofactor biosynthesis | 0.032 | 0.97 | 1 | 0.018 | 0.91 | 1 | 0.0016 | 0.0016 | 0.0015 | 0.0014 | 0.001 | 0.0011 | 0.0018 | 0.0016 | 0.00074 | 0.36 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5345~superpathway of L-methionine biosynthesis (by sulfhydrylation) | 0.76 | 0.97 | 1 | 0.47 | 0.91 | 1 | 0.0053 | 0.0049 | 0.0043 | 0.0051 | 0.0043 | 0.0031 | 0.0048 | 0.0043 | 0.0025 | -0.087 | 35 / 38 (92%) | 18 / 19 (95%) | 17 / 19 (89%) | 0.947 | 0.895 |
| PWY-6892~thiazole biosynthesis I (facultative anaerobic bacteria) | 0.0096 | 0.76 | 1 | 0.0063 | 0.62 | 1 | 0.00058 | 0.00034 | 0.00019 | 0.00025 | 0 | 0.00043 | 0.00043 | 0.00045 | 0.00037 | 0.78 | 22 / 38 (58%) | 8 / 19 (42%) | 14 / 19 (74%) | 0.421 | 0.737 |
| PWY-5489~methyl parathion degradation | 0.59 | 0.97 | 1 | 0.75 | 0.98 | 1 | 0.00045 | 8.3e-05 | 0 | 1e-04 | 0 | 4e-04 | 6.2e-05 | 0 | 0.00022 | -0.69 | 7 / 38 (18%) | 4 / 19 (21%) | 3 / 19 (16%) | 0.211 | 0.158 |
| PWY-6333~acetaldehyde biosynthesis I | 1 | 1 | 1 | 0.72 | 0.97 | 1 | 0.027 | 0.027 | 0.026 | 0.027 | 0.027 | 0.0081 | 0.027 | 0.026 | 0.0078 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-7622~UDP-α-D-galactofuranose biosynthesis | 0.74 | 0.97 | 1 | 0.9 | 0.99 | 1 | 0.022 | 0.022 | 0.02 | 0.022 | 0.02 | 0.0072 | 0.023 | 0.021 | 0.0089 | 0.064 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-1881~formate oxidation to CO2 | 0.19 | 0.97 | 1 | 0.57 | 0.93 | 1 | 0.00079 | 0.00058 | 2e-04 | 0.00044 | 0.00019 | 0.00069 | 0.00072 | 0.00024 | 0.0011 | 0.71 | 28 / 38 (74%) | 14 / 19 (74%) | 14 / 19 (74%) | 0.737 | 0.737 |
| PWY-6893~thiamine diphosphate biosynthesis II (Bacillus) | 0.072 | 0.97 | 1 | 0.19 | 0.91 | 1 | 0.0015 | 0.0012 | 0.00073 | 0.0015 | 0.001 | 0.0017 | 0.00083 | 0.00048 | 0.001 | -0.85 | 30 / 38 (79%) | 16 / 19 (84%) | 14 / 19 (74%) | 0.842 | 0.737 |
| RIBOSYN2-PWY~flavin biosynthesis I (bacteria and plants) | 0.15 | 0.97 | 1 | 0.11 | 0.91 | 1 | 0.0054 | 0.0054 | 0.0055 | 0.0058 | 0.0059 | 0.002 | 0.005 | 0.005 | 0.0017 | -0.21 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| ARGDEGRAD-PWY~L-arginine degradation V (arginine deiminase pathway) | 0.036 | 0.97 | 1 | 0.15 | 0.91 | 1 | 0.0015 | 0.0013 | 0.0011 | 0.00094 | 0.00082 | 0.00076 | 0.0017 | 0.0019 | 0.0013 | 0.85 | 33 / 38 (87%) | 16 / 19 (84%) | 17 / 19 (89%) | 0.842 | 0.895 |
| PWY-6126~superpathway of adenosine nucleotides de novo biosynthesis II | 0.87 | 0.97 | 1 | 0.68 | 0.96 | 1 | 0.0016 | 0.0011 | 0.00061 | 0.0012 | 4e-04 | 0.0019 | 0.0011 | 0.00062 | 0.0013 | -0.13 | 27 / 38 (71%) | 13 / 19 (68%) | 14 / 19 (74%) | 0.684 | 0.737 |
| P185-PWY~formaldehyde assimilation III (dihydroxyacetone cycle) | 0.95 | 0.99 | 1 | 0.84 | 0.99 | 1 | 0.0073 | 0.0073 | 0.0075 | 0.0073 | 0.0073 | 0.0026 | 0.0073 | 0.0077 | 0.0027 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5852~demethylmenaquinol-8 biosynthesis I | 0.46 | 0.97 | 1 | 0.4 | 0.91 | 1 | 0.0029 | 0.0027 | 0.0018 | 0.0028 | 0.0028 | 0.0022 | 0.0027 | 0.00097 | 0.0041 | -0.052 | 36 / 38 (95%) | 18 / 19 (95%) | 18 / 19 (95%) | 0.947 | 0.947 |
| PWY-7796~pentose phosphate pathway (oxidative branch) II | 0.034 | 0.97 | 1 | 0.034 | 0.91 | 1 | 0.00051 | 0.00043 | 0.00035 | 0.00057 | 0.00048 | 0.00054 | 0.00029 | 0.00027 | 0.00027 | -0.97 | 32 / 38 (84%) | 18 / 19 (95%) | 14 / 19 (74%) | 0.947 | 0.737 |
| HOMOSERSYN-PWY~L-homoserine biosynthesis | 0.32 | 0.97 | 1 | 0.24 | 0.91 | 1 | 0.02 | 0.02 | 0.02 | 0.021 | 0.02 | 0.0036 | 0.02 | 0.019 | 0.0051 | -0.07 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6894~thiamine diphosphate biosynthesis I (E. coli) | 0.072 | 0.97 | 1 | 0.19 | 0.91 | 1 | 0.0015 | 0.0012 | 0.00073 | 0.0015 | 0.001 | 0.0017 | 0.00083 | 0.00048 | 0.001 | -0.85 | 30 / 38 (79%) | 16 / 19 (84%) | 14 / 19 (74%) | 0.842 | 0.737 |
| P441-PWY~superpathway of N-acetylneuraminate degradation | 0.51 | 0.97 | 1 | 0.64 | 0.96 | 1 | 0.0058 | 0.0058 | 0.0056 | 0.0056 | 0.0045 | 0.0035 | 0.006 | 0.0066 | 0.003 | 0.1 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-7181~pyrimidine deoxyribonucleosides degradation | 0.03 | 0.97 | 1 | 0.11 | 0.91 | 1 | 0.0029 | 0.0029 | 0.0028 | 0.0034 | 0.0028 | 0.0016 | 0.0024 | 0.0021 | 0.0012 | -0.5 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-7210~pyrimidine deoxyribonucleotides biosynthesis from CTP | 0.12 | 0.97 | 1 | 0.093 | 0.91 | 1 | 0.00067 | 5.3e-05 | 0 | 0 | 0 | 0 | 0.00011 | 0 | 0.00038 | Inf | 3 / 38 (7.9%) | 0 / 19 (0%) | 3 / 19 (16%) | 0 | 0.158 |
| PWY-6349~CDP-archaeol biosynthesis | 0.17 | 0.97 | 1 | 0.26 | 0.91 | 1 | 0.00019 | 1.5e-05 | 0 | 3.9e-06 | 0 | 1.7e-05 | 2.6e-05 | 0 | 7.7e-05 | 2.7 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| SER-GLYSYN-PWY~superpathway of L-serine and glycine biosynthesis I | 0.55 | 0.97 | 1 | 0.97 | 0.99 | 1 | 0.0087 | 0.0087 | 0.008 | 0.0082 | 0.0082 | 0.0031 | 0.0092 | 0.0078 | 0.0059 | 0.17 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| GLYCINE-SYN2-PWY~glycine biosynthesis II | 0.69 | 0.97 | 1 | 0.45 | 0.91 | 1 | 0.0021 | 0.0019 | 0.00089 | 0.0021 | 0.00082 | 0.0028 | 0.0018 | 0.00097 | 0.0022 | -0.22 | 35 / 38 (92%) | 18 / 19 (95%) | 17 / 19 (89%) | 0.947 | 0.895 |
| PWY-7997~demethylmenaquinol-4 biosynthesis | 0.46 | 0.97 | 1 | 0.4 | 0.91 | 1 | 0.0029 | 0.0027 | 0.0018 | 0.0028 | 0.0028 | 0.0022 | 0.0027 | 0.00097 | 0.0041 | -0.052 | 36 / 38 (95%) | 18 / 19 (95%) | 18 / 19 (95%) | 0.947 | 0.947 |
| ASPASN-PWY~superpathway of L-aspartate and L-asparagine biosynthesis | 0.57 | 0.97 | 1 | 0.62 | 0.96 | 1 | 0.0029 | 0.0028 | 0.0024 | 0.0031 | 0.0023 | 0.0024 | 0.0026 | 0.0026 | 0.0016 | -0.25 | 37 / 38 (97%) | 19 / 19 (100%) | 18 / 19 (95%) | 1 | 0.947 |
| PWY-6053~dimethylsulfoniopropanoate biosynthesis III (algae) | 0.41 | 0.97 | 1 | 0.32 | 0.91 | 1 | 0.00044 | 9.3e-05 | 0 | 9e-05 | 0 | 0.00026 | 9.7e-05 | 0 | 3e-04 | 0.11 | 8 / 38 (21%) | 3 / 19 (16%) | 5 / 19 (26%) | 0.158 | 0.263 |
| PWY-6507~4-deoxy-L-threo-hex-4-enopyranuronate degradation | 0.85 | 0.97 | 1 | 0.55 | 0.93 | 1 | 0.00096 | 2e-04 | 0 | 0.00018 | 0 | 0.00041 | 0.00022 | 0 | 0.00055 | 0.29 | 8 / 38 (21%) | 5 / 19 (26%) | 3 / 19 (16%) | 0.263 | 0.158 |
| PWY-7228~superpathway of guanosine nucleotides de novo biosynthesis I | 0.21 | 0.97 | 1 | 0.056 | 0.91 | 1 | 0.0014 | 0.0012 | 0.00084 | 0.0012 | 0.001 | 0.0015 | 0.0013 | 0.00072 | 0.0011 | 0.12 | 34 / 38 (89%) | 15 / 19 (79%) | 19 / 19 (100%) | 0.789 | 1 |
| PWY-7194~pyrimidine nucleobases salvage II | 0.64 | 0.97 | 1 | 0.52 | 0.93 | 1 | 0.0013 | 0.0011 | 0.00064 | 0.001 | 0.00066 | 0.0011 | 0.0012 | 0.00059 | 0.0015 | 0.26 | 33 / 38 (87%) | 16 / 19 (84%) | 17 / 19 (89%) | 0.842 | 0.895 |
| PWY-5731~atrazine degradation III | 0.67 | 0.97 | 1 | 0.85 | 0.99 | 1 | 0.00055 | 0.00025 | 0 | 0.00031 | 0 | 0.00051 | 0.00019 | 0 | 0.00026 | -0.71 | 17 / 38 (45%) | 8 / 19 (42%) | 9 / 19 (47%) | 0.421 | 0.474 |
| PWY-7791~UMP biosynthesis III | 0.056 | 0.97 | 1 | 0.063 | 0.91 | 1 | 0.024 | 0.024 | 0.024 | 0.025 | 0.025 | 0.0045 | 0.023 | 0.021 | 0.0071 | -0.12 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6131~glycerol degradation II | 0.018 | 0.97 | 1 | 0.023 | 0.91 | 1 | 0.0022 | 0.0022 | 0.0019 | 0.0026 | 0.0022 | 0.0017 | 0.0017 | 0.0016 | 0.00095 | -0.61 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| TYRSYN~L-tyrosine biosynthesis I | 0.43 | 0.97 | 1 | 0.48 | 0.91 | 1 | 0.00095 | 6e-04 | 0.00031 | 0.00049 | 0.00018 | 0.00057 | 7e-04 | 0.00036 | 0.00082 | 0.51 | 24 / 38 (63%) | 11 / 19 (58%) | 13 / 19 (68%) | 0.579 | 0.684 |
| PWY-6609~adenine and adenosine salvage III | 0.2 | 0.97 | 1 | 0.25 | 0.91 | 1 | 0.0031 | 0.0031 | 0.0027 | 0.0029 | 0.0028 | 0.0016 | 0.0032 | 0.0027 | 0.0019 | 0.14 | 37 / 38 (97%) | 18 / 19 (95%) | 19 / 19 (100%) | 0.947 | 1 |
| PWY-7227~adenosine deoxyribonucleotides de novo biosynthesis | 0.53 | 0.97 | 1 | 0.25 | 0.91 | 1 | 0.0017 | 0.0014 | 0.00066 | 0.0014 | 0.00053 | 0.0022 | 0.0013 | 0.00067 | 0.0016 | -0.11 | 31 / 38 (82%) | 14 / 19 (74%) | 17 / 19 (89%) | 0.737 | 0.895 |
| NAD-BIOSYNTHESIS-II~NAD salvage pathway III (to nicotinamide riboside) | 0.049 | 0.97 | 1 | 0.044 | 0.91 | 1 | 0.0043 | 0.0041 | 0.0032 | 0.0046 | 0.0045 | 0.0035 | 0.0036 | 0.0021 | 0.0049 | -0.35 | 36 / 38 (95%) | 18 / 19 (95%) | 18 / 19 (95%) | 0.947 | 0.947 |
| ARGININE-SYN4-PWY~L-ornithine biosynthesis II | 0.43 | 0.97 | 1 | 0.68 | 0.96 | 1 | 0.0011 | 0.00018 | 0 | 0.00026 | 0 | 0.00078 | 0.00011 | 0 | 0.00029 | -1.2 | 6 / 38 (16%) | 3 / 19 (16%) | 3 / 19 (16%) | 0.158 | 0.158 |
| PWY-7639~bis(guanylyl molybdenum cofactor) biosynthesis | 0.032 | 0.97 | 1 | 0.018 | 0.91 | 1 | 0.0016 | 0.0016 | 0.0015 | 0.0014 | 0.001 | 0.0011 | 0.0018 | 0.0016 | 0.00074 | 0.36 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5142~acyl-[acyl-carrier protein] thioesterase pathway | 0.98 | 0.99 | 1 | 0.56 | 0.93 | 1 | 0.0017 | 0.0016 | 0.0015 | 0.0017 | 0.0015 | 0.0012 | 0.0016 | 0.0016 | 0.0012 | -0.087 | 36 / 38 (95%) | 17 / 19 (89%) | 19 / 19 (100%) | 0.895 | 1 |
| PWY-1722~formate assimilation into 510-methylenetetrahydrofolate | 0.88 | 0.98 | 1 | 0.72 | 0.97 | 1 | 0.016 | 0.016 | 0.016 | 0.016 | 0.015 | 0.0025 | 0.016 | 0.016 | 0.0037 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY3O-4107~NAD salvage pathway V (PNC V cycle) | 0.84 | 0.97 | 1 | 0.71 | 0.97 | 1 | 0.0023 | 0.0018 | 0.00093 | 0.0014 | 0.00093 | 0.0017 | 0.0021 | 0.00093 | 0.005 | 0.58 | 29 / 38 (76%) | 15 / 19 (79%) | 14 / 19 (74%) | 0.789 | 0.737 |
| SALVADEHYPOX-PWY~adenosine nucleotides degradation II | 0.38 | 0.97 | 1 | 0.5 | 0.93 | 1 | 0.0078 | 0.0078 | 0.0076 | 0.0076 | 0.0061 | 0.0028 | 0.008 | 0.0084 | 0.0025 | 0.074 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5994~palmitate biosynthesis I (animals and fungi) | 0.21 | 0.97 | 1 | 0.091 | 0.91 | 1 | 0.0049 | 0.0043 | 0.0033 | 0.0043 | 0.0034 | 0.0038 | 0.0043 | 0.0025 | 0.0057 | 0 | 33 / 38 (87%) | 17 / 19 (89%) | 16 / 19 (84%) | 0.895 | 0.842 |
| CITRULBIO-PWY~L-citrulline biosynthesis | 0.67 | 0.97 | 1 | 0.96 | 0.99 | 1 | 0.0013 | 0.00038 | 0 | 0.00047 | 0 | 0.0011 | 0.00029 | 0 | 0.00061 | -0.7 | 11 / 38 (29%) | 5 / 19 (26%) | 6 / 19 (32%) | 0.263 | 0.316 |
| PWY0-1301~melibiose degradation | 0.57 | 0.97 | 1 | 0.44 | 0.91 | 1 | 0.057 | 0.057 | 0.054 | 0.058 | 0.055 | 0.02 | 0.056 | 0.054 | 0.021 | -0.051 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PROUT-PWY~L-proline degradation | 0.99 | 0.99 | 1 | 0.6 | 0.95 | 1 | 0.0019 | 0.00069 | 0 | 8e-04 | 0 | 0.0018 | 0.00058 | 0 | 0.001 | -0.46 | 14 / 38 (37%) | 6 / 19 (32%) | 8 / 19 (42%) | 0.316 | 0.421 |
| PWY-6614~tetrahydrofolate biosynthesis | 0.17 | 0.97 | 1 | 0.95 | 0.99 | 1 | 0.0017 | 0.0016 | 0.0013 | 0.0017 | 0.0014 | 0.0011 | 0.0015 | 0.0012 | 9e-04 | -0.18 | 37 / 38 (97%) | 18 / 19 (95%) | 19 / 19 (100%) | 0.947 | 1 |
| PWY-6646~fluoroacetate degradation | 0.86 | 0.97 | 1 | 0.81 | 0.99 | 1 | 0.0018 | 0.0016 | 0.0012 | 0.0016 | 0.0011 | 0.0017 | 0.0016 | 0.0014 | 0.0013 | 0 | 34 / 38 (89%) | 17 / 19 (89%) | 17 / 19 (89%) | 0.895 | 0.895 |
| PWY-6121~5-aminoimidazole ribonucleotide biosynthesis I | 0.83 | 0.97 | 1 | 0.66 | 0.96 | 1 | 0.023 | 0.023 | 0.023 | 0.023 | 0.023 | 0.0044 | 0.024 | 0.023 | 0.008 | 0.061 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PANTOSYN-PWY~superpathway of coenzyme A biosynthesis I (bacteria) | 0.16 | 0.97 | 1 | 0.11 | 0.91 | 1 | 0.004 | 0.004 | 0.0042 | 0.0043 | 0.0043 | 0.0015 | 0.0037 | 0.0039 | 0.0014 | -0.22 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY0-1582~glycerol-3-phosphate to fumarate electron transfer | 0.87 | 0.97 | 1 | 0.79 | 0.99 | 1 | 0.0012 | 0.00049 | 0 | 0.00049 | 0 | 0.00085 | 5e-04 | 0 | 0.00089 | 0.029 | 15 / 38 (39%) | 7 / 19 (37%) | 8 / 19 (42%) | 0.368 | 0.421 |
| GLUCONSUPER-PWY~D-gluconate degradation | 0.31 | 0.97 | 1 | 0.026 | 0.91 | 1 | 0.0014 | 0.0011 | 0.00039 | 0.0011 | 0.00049 | 0.0012 | 0.0012 | 0.00028 | 0.0031 | 0.13 | 32 / 38 (84%) | 17 / 19 (89%) | 15 / 19 (79%) | 0.895 | 0.789 |
| PWY-5892~menaquinol-12 biosynthesis | 0.24 | 0.97 | 1 | 0.48 | 0.91 | 1 | 0.0015 | 0.00073 | 6.1e-05 | 0.00093 | 0.00012 | 0.0015 | 0.00052 | 0 | 0.00089 | -0.84 | 19 / 38 (50%) | 10 / 19 (53%) | 9 / 19 (47%) | 0.526 | 0.474 |
| ARG%2bPOLYAMINE-SYN~superpathway of arginine and polyamine biosynthesis | 0.44 | 0.97 | 1 | 0.3 | 0.91 | 1 | 0.0014 | 0.00072 | 0.00014 | 0.00069 | 0 | 0.0013 | 0.00075 | 6e-04 | 0.00083 | 0.12 | 19 / 38 (50%) | 8 / 19 (42%) | 11 / 19 (58%) | 0.421 | 0.579 |
| PWY0-1264~biotin-carboxyl carrier protein assembly | 0.78 | 0.97 | 1 | 0.95 | 0.99 | 1 | 0.0085 | 0.0085 | 0.0075 | 0.0084 | 0.0075 | 0.0033 | 0.0085 | 0.0074 | 0.0028 | 0.017 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| GLYCEROLMETAB-PWY~glycerol degradation V | 0.059 | 0.97 | 1 | 0.1 | 0.91 | 1 | 0.0027 | 0.0027 | 0.0023 | 0.0032 | 0.0025 | 0.0023 | 0.0022 | 0.0019 | 0.0013 | -0.54 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY0-1280~ethylene glycol degradation | 0.86 | 0.97 | 1 | 0.43 | 0.91 | 1 | 0.001 | 0.00035 | 0 | 0.00041 | 0 | 0.00093 | 0.00029 | 0 | 0.00049 | -0.5 | 13 / 38 (34%) | 5 / 19 (26%) | 8 / 19 (42%) | 0.263 | 0.421 |
| PWY-3841~folate transformations II | 0.67 | 0.97 | 1 | 0.32 | 0.91 | 1 | 0.0096 | 0.0096 | 0.0094 | 0.0096 | 0.0099 | 0.0019 | 0.0095 | 0.0092 | 0.0041 | -0.015 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-4981~L-proline biosynthesis II (from arginine) | 0.66 | 0.97 | 1 | 0.29 | 0.91 | 1 | 0.00097 | 7.6e-05 | 0 | 6.9e-05 | 0 | 0.00028 | 8.4e-05 | 0 | 0.00036 | 0.28 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
| PWY0-1300~2-O-α-mannosyl-D-glycerate degradation | 0.25 | 0.97 | 1 | 0.29 | 0.91 | 1 | 0.00021 | 4.4e-05 | 0 | 2.6e-05 | 0 | 7.9e-05 | 6.2e-05 | 0 | 0.00013 | 1.3 | 8 / 38 (21%) | 3 / 19 (16%) | 5 / 19 (26%) | 0.158 | 0.263 |
| PWY-7200~superpathway of pyrimidine deoxyribonucleoside salvage | 0.1 | 0.97 | 1 | 0.1 | 0.91 | 1 | 0.00044 | 5.8e-05 | 0 | 3.1e-05 | 0 | 0.00014 | 8.5e-05 | 0 | 0.00022 | 1.5 | 5 / 38 (13%) | 1 / 19 (5.3%) | 4 / 19 (21%) | 0.0526 | 0.211 |
| ALANINE-SYN2-PWY~L-alanine biosynthesis II | 0.67 | 0.97 | 1 | 0.19 | 0.91 | 1 | 0.0072 | 0.0072 | 0.0064 | 0.007 | 0.0069 | 0.0034 | 0.0075 | 0.005 | 0.008 | 0.1 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-7625~phosphatidylinositol biosynthesis II (eukaryotes) | 0.75 | 0.97 | 1 | 0.3 | 0.91 | 1 | 0.0012 | 0.001 | 0.00058 | 0.00094 | 6e-04 | 0.00097 | 0.0011 | 0.00042 | 0.0026 | 0.23 | 32 / 38 (84%) | 16 / 19 (84%) | 16 / 19 (84%) | 0.842 | 0.842 |
| P122-PWY~heterolactic fermentation | 0.91 | 0.98 | 1 | 0.65 | 0.96 | 1 | 0.003 | 0.0022 | 0.0011 | 0.0019 | 0.0012 | 0.002 | 0.0025 | 0.00088 | 0.0049 | 0.4 | 28 / 38 (74%) | 14 / 19 (74%) | 14 / 19 (74%) | 0.737 | 0.737 |
| TREDEGLOW-PWY~trehalose degradation I (low osmolarity) | 0.5 | 0.97 | 1 | 0.57 | 0.93 | 1 | 0.0035 | 0.0035 | 0.0024 | 0.0034 | 0.0024 | 0.0025 | 0.0036 | 0.0023 | 0.0037 | 0.082 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5512~UDP-N-acetyl-D-galactosamine biosynthesis I | 0.75 | 0.97 | 1 | 0.85 | 0.99 | 1 | 0.00041 | 9.8e-05 | 0 | 0.00012 | 0 | 0.00031 | 7.7e-05 | 0 | 2e-04 | -0.64 | 9 / 38 (24%) | 4 / 19 (21%) | 5 / 19 (26%) | 0.211 | 0.263 |
| PWY-6151~S-adenosyl-L-methionine cycle I | 0.7 | 0.97 | 1 | 0.88 | 0.99 | 1 | 0.0087 | 0.0087 | 0.0085 | 0.0082 | 0.0085 | 0.0029 | 0.0091 | 0.008 | 0.0059 | 0.15 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5350~thiosulfate disproportionation IV (rhodanese) | 0.61 | 0.97 | 1 | 0.9 | 0.99 | 1 | 0.0095 | 0.0095 | 0.0088 | 0.0092 | 0.0085 | 0.0035 | 0.0097 | 0.0091 | 0.0039 | 0.076 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| VALSYN-PWY~L-valine biosynthesis | 0.74 | 0.97 | 1 | 0.87 | 0.99 | 1 | 0.023 | 0.023 | 0.023 | 0.023 | 0.023 | 0.0053 | 0.023 | 0.023 | 0.0069 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| THIOREDOX-PWY~thioredoxin pathway | 0.73 | 0.97 | 1 | 0.85 | 0.99 | 1 | 0.028 | 0.028 | 0.029 | 0.027 | 0.029 | 0.007 | 0.028 | 0.028 | 0.0086 | 0.052 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-7842~UDP-yelosamine biosynthesis | 0.021 | 0.97 | 1 | 0.0072 | 0.62 | 1 | 0.00057 | 7.5e-05 | 0 | 0.00015 | 0 | 0.00031 | 0 | 0 | 0 | -Inf | 5 / 38 (13%) | 5 / 19 (26%) | 0 / 19 (0%) | 0.263 | 0 |
| PWY-6698~oxalate degradation V | 0.49 | 0.97 | 1 | 0.36 | 0.91 | 1 | 0.00075 | 0.00075 | 0.00058 | 0.00078 | 0.00062 | 0.00059 | 0.00071 | 0.00052 | 0.00061 | -0.14 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| ARO-PWY~chorismate biosynthesis I | 0.75 | 0.97 | 1 | 0.85 | 0.99 | 1 | 0.011 | 0.011 | 0.011 | 0.011 | 0.011 | 0.0036 | 0.012 | 0.0098 | 0.0052 | 0.13 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6143~CMP-pseudaminate biosynthesis | 0.21 | 0.97 | 1 | 0.2 | 0.91 | 1 | 0.00083 | 0.00015 | 0 | 0.00019 | 0 | 0.00047 | 0.00011 | 0 | 0.00029 | -0.79 | 7 / 38 (18%) | 4 / 19 (21%) | 3 / 19 (16%) | 0.211 | 0.158 |
| ANAGLYCOLYSIS-PWY~glycolysis III (from glucose) | 0.43 | 0.97 | 1 | 0.22 | 0.91 | 1 | 0.015 | 0.015 | 0.015 | 0.016 | 0.015 | 0.003 | 0.015 | 0.015 | 0.0047 | -0.093 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY0-381~glycerol and glycerophosphodiester degradation | 0.5 | 0.97 | 1 | 0.31 | 0.91 | 1 | 0.00096 | 0.00038 | 0 | 4e-04 | 0 | 0.00076 | 0.00036 | 0 | 0.00073 | -0.15 | 15 / 38 (39%) | 9 / 19 (47%) | 6 / 19 (32%) | 0.474 | 0.316 |
| GLYSYN-ALA-PWY~glycine biosynthesis III | 0.16 | 0.97 | 1 | 0.31 | 0.91 | 1 | 0.00077 | 0.00042 | 0.00012 | 0.00027 | 5.4e-05 | 0.00043 | 0.00058 | 0.00035 | 0.00076 | 1.1 | 21 / 38 (55%) | 10 / 19 (53%) | 11 / 19 (58%) | 0.526 | 0.579 |
| PWY-6613~tetrahydrofolate salvage from 510-methenyltetrahydrofolate | 0.67 | 0.97 | 1 | 0.88 | 0.99 | 1 | 0.0093 | 0.0093 | 0.0094 | 0.0091 | 0.0092 | 0.0014 | 0.0094 | 0.0099 | 0.0021 | 0.047 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6785~hydrogen production VIII | 0.76 | 0.97 | 1 | 0.9 | 0.99 | 1 | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 | 0.009 | 0.03 | 0.031 | 0.008 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6588~pyruvate fermentation to acetone | 0.0015 | 0.33 | 0.84 | 0.0035 | 0.62 | 1 | 0.0014 | 0.0011 | 0.0011 | 0.0016 | 0.0015 | 0.0011 | 0.00068 | 5e-04 | 0.00072 | -1.2 | 31 / 38 (82%) | 18 / 19 (95%) | 13 / 19 (68%) | 0.947 | 0.684 |
| PWY-6512~hydrogen oxidation III (anaerobic NADP) | 0.63 | 0.97 | 1 | 0.28 | 0.91 | 1 | 0.0012 | 0.00062 | 1.6e-05 | 0.00066 | 0 | 0.0013 | 0.00057 | 0.00017 | 9e-04 | -0.21 | 19 / 38 (50%) | 8 / 19 (42%) | 11 / 19 (58%) | 0.421 | 0.579 |
| PWY-43~putrescine biosynthesis II | 0.85 | 0.97 | 1 | 0.68 | 0.96 | 1 | 0.0067 | 0.0067 | 0.0062 | 0.0067 | 0.0059 | 0.0026 | 0.0068 | 0.0066 | 0.0026 | 0.021 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5965~fatty acid biosynthesis initiation III | 0.68 | 0.97 | 1 | 0.47 | 0.91 | 1 | 0.0044 | 0.0041 | 0.0024 | 0.004 | 0.0024 | 0.0038 | 0.0043 | 0.0025 | 0.0087 | 0.1 | 36 / 38 (95%) | 18 / 19 (95%) | 18 / 19 (95%) | 0.947 | 0.947 |
| PROPIONMET-PWY~propanoyl CoA degradation I | 0.82 | 0.97 | 1 | 0.52 | 0.93 | 1 | 0.0022 | 0.0011 | 0 | 0.0012 | 0 | 0.0027 | 0.00094 | 0.00038 | 0.0016 | -0.35 | 18 / 38 (47%) | 8 / 19 (42%) | 10 / 19 (53%) | 0.421 | 0.526 |
| PWY-5121~superpathway of geranylgeranyl diphosphate biosynthesis II (via MEP) | 0.53 | 0.97 | 1 | 0.81 | 0.99 | 1 | 0.0077 | 0.0077 | 0.008 | 0.0081 | 0.0085 | 0.0035 | 0.0073 | 0.0076 | 0.0026 | -0.15 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5278~sulfite oxidation III | 0.67 | 0.97 | 1 | 0.87 | 0.99 | 1 | 0.012 | 0.012 | 0.011 | 0.012 | 0.011 | 0.0039 | 0.012 | 0.012 | 0.0052 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5436~L-threonine degradation IV | 0.56 | 0.97 | 1 | 0.77 | 0.99 | 1 | 0.0025 | 0.0025 | 0.0015 | 0.0021 | 0.0014 | 0.002 | 0.0029 | 0.0016 | 0.0047 | 0.47 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY66-428~L-threonine degradation V | 0.72 | 0.97 | 1 | 0.35 | 0.91 | 1 | 0.00089 | 0.00054 | 0.00013 | 0.00058 | 0 | 9e-04 | 0.00049 | 0.00019 | 0.00059 | -0.24 | 23 / 38 (61%) | 9 / 19 (47%) | 14 / 19 (74%) | 0.474 | 0.737 |
| PWY-5703~urea degradation I | 0.76 | 0.97 | 1 | 0.71 | 0.97 | 1 | 0.0034 | 0.0026 | 0.00057 | 0.0031 | 0.00057 | 0.0047 | 0.0021 | 0.00058 | 0.0045 | -0.56 | 29 / 38 (76%) | 14 / 19 (74%) | 15 / 19 (79%) | 0.737 | 0.789 |
| PWY-7345~superpathway of anaerobic sucrose degradation | 0.56 | 0.97 | 1 | 0.99 | 1 | 1 | 0.0014 | 0.00037 | 0 | 0.00024 | 0 | 0.00047 | 5e-04 | 0 | 0.001 | 1.1 | 10 / 38 (26%) | 5 / 19 (26%) | 5 / 19 (26%) | 0.263 | 0.263 |
| PWY0-1324~N-acetylneuraminate and N-acetylmannosamine degradation I | 0.25 | 0.97 | 1 | 0.23 | 0.91 | 1 | 0.00084 | 4e-04 | 0 | 0.00033 | 0 | 0.00057 | 0.00046 | 9.4e-05 | 0.00065 | 0.48 | 18 / 38 (47%) | 8 / 19 (42%) | 10 / 19 (53%) | 0.421 | 0.526 |
| PYRIDNUCSAL-PWY~NAD salvage pathway I (PNC VI cycle) | 0.78 | 0.97 | 1 | 0.95 | 0.99 | 1 | 0.0025 | 0.0017 | 9e-04 | 0.0014 | 0.00088 | 0.0018 | 0.002 | 0.00092 | 0.0049 | 0.51 | 26 / 38 (68%) | 13 / 19 (68%) | 13 / 19 (68%) | 0.684 | 0.684 |
| PWY-2722~trehalose degradation IV | 0.27 | 0.97 | 1 | 0.12 | 0.91 | 1 | 0.0011 | 0.00017 | 0 | 0.00019 | 0 | 0.00049 | 0.00016 | 0 | 0.00062 | -0.25 | 6 / 38 (16%) | 4 / 19 (21%) | 2 / 19 (11%) | 0.211 | 0.105 |
| GLUTORN-PWY~L-ornithine biosynthesis I | 0.55 | 0.97 | 1 | 0.43 | 0.91 | 1 | 0.015 | 0.015 | 0.015 | 0.015 | 0.017 | 0.0038 | 0.015 | 0.014 | 0.0043 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PEPTIDOGLYCANSYN-PWY~peptidoglycan biosynthesis I (meso-diaminopimelate containing) | 0.52 | 0.97 | 1 | 1 | 1 | 1 | 0.018 | 0.018 | 0.017 | 0.018 | 0.018 | 0.0034 | 0.018 | 0.017 | 0.0054 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY1-2~L-alanine degradation IV | 0.69 | 0.97 | 1 | 0.65 | 0.96 | 1 | 0.0014 | 0.0014 | 9e-04 | 0.0017 | 0.00074 | 0.0018 | 0.0011 | 9e-04 | 0.00078 | -0.63 | 37 / 38 (97%) | 18 / 19 (95%) | 19 / 19 (100%) | 0.947 | 1 |
| PWY-7560~methylerythritol phosphate pathway II | 0.49 | 0.97 | 1 | 0.22 | 0.91 | 1 | 0.0073 | 0.0073 | 0.0078 | 0.0075 | 0.0079 | 0.0027 | 0.0072 | 0.0072 | 0.0029 | -0.059 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5973~cis-vaccenate biosynthesis | 0.1 | 0.97 | 1 | 0.19 | 0.91 | 1 | 0.0044 | 0.0042 | 0.0039 | 0.0048 | 0.0048 | 0.0031 | 0.0036 | 0.0033 | 0.0022 | -0.42 | 37 / 38 (97%) | 19 / 19 (100%) | 18 / 19 (95%) | 1 | 0.947 |
| PWY-7234~inosine-5-phosphate biosynthesis III | 0.44 | 0.97 | 1 | 0.33 | 0.91 | 1 | 0.0021 | 0.0019 | 0.0011 | 0.0016 | 0.0011 | 0.0016 | 0.0023 | 0.0011 | 0.0042 | 0.52 | 34 / 38 (89%) | 16 / 19 (84%) | 18 / 19 (95%) | 0.842 | 0.947 |
| PWY-2942~L-lysine biosynthesis III | 0.41 | 0.97 | 1 | 0.37 | 0.91 | 1 | 0.0062 | 0.0062 | 0.0058 | 0.0066 | 0.0058 | 0.003 | 0.0058 | 0.0055 | 0.0025 | -0.19 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| P283-PWY~hydrogen oxidation I (aerobic) | 0.35 | 0.97 | 1 | 0.38 | 0.91 | 1 | 0.0013 | 0.00051 | 0 | 0.00044 | 0 | 0.0012 | 0.00058 | 0 | 0.0017 | 0.4 | 15 / 38 (39%) | 7 / 19 (37%) | 8 / 19 (42%) | 0.368 | 0.421 |
| PWY-7328~superpathway of UDP-glucose-derived O-antigen building blocks biosynthesis | 0.79 | 0.97 | 1 | 0.89 | 0.99 | 1 | 0.0052 | 0.005 | 0.0038 | 0.0045 | 0.0044 | 0.0029 | 0.0054 | 0.0034 | 0.0073 | 0.26 | 36 / 38 (95%) | 18 / 19 (95%) | 18 / 19 (95%) | 0.947 | 0.947 |
| PWY-5805~nonaprenyl diphosphate biosynthesis I | 0.45 | 0.97 | 1 | 0.56 | 0.93 | 1 | 0.00041 | 7.6e-05 | 0 | 5.2e-05 | 0 | 0.00013 | 1e-04 | 0 | 0.00028 | 0.94 | 7 / 38 (18%) | 3 / 19 (16%) | 4 / 19 (21%) | 0.158 | 0.211 |
| PWY-1269~CMP-3-deoxy-D-manno-octulosonate biosynthesis | 0.27 | 0.97 | 1 | 0.37 | 0.91 | 1 | 0.0013 | 0.00038 | 0 | 0.00049 | 0 | 0.00089 | 0.00026 | 0 | 0.00052 | -0.91 | 11 / 38 (29%) | 6 / 19 (32%) | 5 / 19 (26%) | 0.316 | 0.263 |
| PWY-5941~glycogen degradation II | 0.79 | 0.97 | 1 | 0.71 | 0.97 | 1 | 0.036 | 0.036 | 0.036 | 0.035 | 0.036 | 0.0092 | 0.037 | 0.036 | 0.013 | 0.08 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6765~hydrogen production IV | 0.63 | 0.97 | 1 | 0.28 | 0.91 | 1 | 0.0012 | 0.00062 | 1.6e-05 | 0.00066 | 0 | 0.0013 | 0.00057 | 0.00017 | 9e-04 | -0.21 | 19 / 38 (50%) | 8 / 19 (42%) | 11 / 19 (58%) | 0.421 | 0.579 |
| PWY-6606~guanosine nucleotides degradation II | 0.27 | 0.97 | 1 | 0.46 | 0.91 | 1 | 0.0016 | 0.00056 | 0 | 0.00042 | 0 | 0.00079 | 7e-04 | 0 | 0.0011 | 0.74 | 13 / 38 (34%) | 6 / 19 (32%) | 7 / 19 (37%) | 0.316 | 0.368 |
| PWY-5535~acetate and ATP formation from acetyl-CoA II | 0.2 | 0.97 | 1 | 0.3 | 0.91 | 1 | 0.00093 | 0.00071 | 0.00033 | 0.00061 | 0.00022 | 0.001 | 8e-04 | 0.00038 | 0.00095 | 0.39 | 29 / 38 (76%) | 15 / 19 (79%) | 14 / 19 (74%) | 0.789 | 0.737 |
| PWY-5704~urea degradation II | 0.7 | 0.97 | 1 | 0.62 | 0.96 | 1 | 0.0039 | 0.0039 | 0.0035 | 0.0038 | 0.0028 | 0.0022 | 0.0039 | 0.0037 | 0.0018 | 0.037 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| 1CMET2-PWY~N10-formyl-tetrahydrofolate biosynthesis | 0.52 | 0.97 | 1 | 0.45 | 0.91 | 1 | 0.0051 | 0.0051 | 0.0054 | 0.0053 | 0.0055 | 0.0019 | 0.0049 | 0.0049 | 0.0016 | -0.11 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6717~(14)-β-D-xylan degradation | 0.63 | 0.97 | 1 | 0.26 | 0.91 | 1 | 0.026 | 0.026 | 0.023 | 0.025 | 0.024 | 0.014 | 0.026 | 0.02 | 0.022 | 0.057 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6890~4-amino-2-methyl-5-diphosphomethylpyrimidine biosynthesis | 0.78 | 0.97 | 1 | 0.73 | 0.97 | 1 | 0.012 | 0.012 | 0.013 | 0.012 | 0.013 | 0.0031 | 0.013 | 0.013 | 0.0046 | 0.12 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| LEUSYN-PWY~L-leucine biosynthesis | 0.74 | 0.97 | 1 | 0.81 | 0.99 | 1 | 0.02 | 0.02 | 0.019 | 0.019 | 0.019 | 0.0044 | 0.02 | 0.018 | 0.0062 | 0.074 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY0-1299~arginine dependent acid resistance | 0.98 | 0.99 | 1 | 0.57 | 0.93 | 1 | 0.025 | 0.025 | 0.025 | 0.025 | 0.025 | 0.005 | 0.025 | 0.026 | 0.0068 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5364~sulfur reduction II (via polysulfide) | 0.47 | 0.97 | 1 | 0.35 | 0.91 | 1 | 0.0012 | 0.00078 | 0.00029 | 0.00063 | 0.00012 | 0.00093 | 0.00094 | 0.00041 | 0.0024 | 0.58 | 24 / 38 (63%) | 11 / 19 (58%) | 13 / 19 (68%) | 0.579 | 0.684 |
| TRYPDEG-PWY~L-tryptophan degradation II (via pyruvate) | 0.77 | 0.97 | 1 | 0.82 | 0.99 | 1 | 0.00018 | 0.00011 | 8.4e-05 | 0.00011 | 7.5e-05 | 0.00016 | 0.00011 | 8.7e-05 | 0.00016 | 0 | 23 / 38 (61%) | 11 / 19 (58%) | 12 / 19 (63%) | 0.579 | 0.632 |
| PWY-6466~pyridoxal 5-phosphate biosynthesis II | 0.73 | 0.97 | 1 | 0.61 | 0.96 | 1 | 0.01 | 0.01 | 0.01 | 0.01 | 0.011 | 0.0039 | 0.01 | 0.01 | 0.0058 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5332~sulfur reduction I | 0.47 | 0.97 | 1 | 0.35 | 0.91 | 1 | 0.0012 | 0.00078 | 0.00029 | 0.00063 | 0.00012 | 0.00093 | 0.00094 | 0.00041 | 0.0024 | 0.58 | 24 / 38 (63%) | 11 / 19 (58%) | 13 / 19 (68%) | 0.579 | 0.684 |
| SULFATE-CYS-PWY~superpathway of sulfate assimilation and cysteine biosynthesis | 0.084 | 0.97 | 1 | 0.085 | 0.91 | 1 | 0.00086 | 0.00011 | 0 | 9.6e-05 | 0 | 0.00042 | 0.00013 | 0 | 4e-04 | 0.44 | 5 / 38 (13%) | 1 / 19 (5.3%) | 4 / 19 (21%) | 0.0526 | 0.211 |
| PWY-7013~(S)-propane-12-diol degradation | 0.037 | 0.97 | 1 | 0.037 | 0.91 | 1 | 0.0017 | 0.0016 | 0.0013 | 0.0014 | 0.0011 | 0.0011 | 0.0018 | 0.002 | 8e-04 | 0.36 | 36 / 38 (95%) | 17 / 19 (89%) | 19 / 19 (100%) | 0.895 | 1 |
| NONOXIPENT-PWY~pentose phosphate pathway (non-oxidative branch) | 0.68 | 0.97 | 1 | 0.37 | 0.91 | 1 | 0.017 | 0.017 | 0.017 | 0.017 | 0.018 | 0.0042 | 0.017 | 0.015 | 0.0048 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5723~Rubisco shunt | 0.7 | 0.97 | 1 | 0.41 | 0.91 | 1 | 0.01 | 0.01 | 0.01 | 0.011 | 0.01 | 0.0032 | 0.01 | 0.011 | 0.0033 | -0.14 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6386~UDP-N-acetylmuramoyl-pentapeptide biosynthesis II (lysine-containing) | 0.95 | 0.99 | 1 | 0.63 | 0.96 | 1 | 0.015 | 0.015 | 0.014 | 0.015 | 0.014 | 0.0036 | 0.015 | 0.013 | 0.0054 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| P125-PWY~superpathway of (RR)-butanediol biosynthesis | 0.58 | 0.97 | 1 | 0.67 | 0.96 | 1 | 0.00032 | 3.4e-05 | 0 | 5.3e-05 | 0 | 2e-04 | 1.5e-05 | 0 | 4.9e-05 | -1.8 | 4 / 38 (11%) | 2 / 19 (11%) | 2 / 19 (11%) | 0.105 | 0.105 |
| PWY-6780~hydrogen production VI | 0.18 | 0.97 | 1 | 0.23 | 0.91 | 1 | 0.008 | 0.008 | 0.007 | 0.0075 | 0.0068 | 0.0044 | 0.0085 | 0.0079 | 0.0039 | 0.18 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-7344~UDP-α-D-galactose biosynthesis | 0.75 | 0.97 | 1 | 0.71 | 0.97 | 1 | 0.033 | 0.033 | 0.031 | 0.033 | 0.031 | 0.008 | 0.034 | 0.031 | 0.01 | 0.043 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| CYANCAT-PWY~cyanate degradation | 0.9 | 0.98 | 1 | 0.53 | 0.93 | 1 | 0.0034 | 0.0034 | 0.0027 | 0.0033 | 0.0029 | 0.002 | 0.0035 | 0.0027 | 0.0039 | 0.085 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| HOMOSER-METSYN-PWY~L-methionine biosynthesis I | 0.93 | 0.98 | 1 | 0.95 | 0.99 | 1 | 0.00062 | 0.00029 | 0 | 3e-04 | 0 | 0.00056 | 0.00029 | 0 | 0.00049 | -0.049 | 18 / 38 (47%) | 9 / 19 (47%) | 9 / 19 (47%) | 0.474 | 0.474 |
| ACETOACETATE-DEG-PWY~acetoacetate degradation (to acetyl CoA) | 0.0055 | 0.5 | 1 | 0.0059 | 0.62 | 1 | 7e-04 | 0.00057 | 0.00059 | 0.00075 | 0.00066 | 0.00053 | 0.00038 | 0.00037 | 0.00036 | -0.98 | 31 / 38 (82%) | 18 / 19 (95%) | 13 / 19 (68%) | 0.947 | 0.684 |
| PWY-6629~superpathway of L-tryptophan biosynthesis | 0.072 | 0.97 | 1 | 0.14 | 0.91 | 1 | 0.0022 | 0.002 | 0.002 | 0.0023 | 0.0021 | 0.0013 | 0.0017 | 0.0016 | 0.0011 | -0.44 | 34 / 38 (89%) | 18 / 19 (95%) | 16 / 19 (84%) | 0.947 | 0.842 |
| PWY0-1507~biotin biosynthesis from 8-amino-7-oxononanoate I | 0.91 | 0.98 | 1 | 0.47 | 0.91 | 1 | 0.0027 | 0.0026 | 0.0022 | 0.0027 | 0.0026 | 0.0018 | 0.0025 | 0.0021 | 0.0013 | -0.11 | 37 / 38 (97%) | 18 / 19 (95%) | 19 / 19 (100%) | 0.947 | 1 |
| PPGPPMET-PWY~ppGpp biosynthesis | 0.32 | 0.97 | 1 | 0.96 | 0.99 | 1 | 0.0031 | 0.0029 | 0.0016 | 0.0032 | 0.0023 | 0.0037 | 0.0026 | 0.0015 | 0.0024 | -0.3 | 35 / 38 (92%) | 17 / 19 (89%) | 18 / 19 (95%) | 0.895 | 0.947 |
| PWY-7196~superpathway of pyrimidine ribonucleosides salvage | 0.4 | 0.97 | 1 | 0.68 | 0.96 | 1 | 0.00075 | 0.00028 | 0 | 0.00038 | 0 | 9e-04 | 0.00018 | 0 | 0.00033 | -1.1 | 14 / 38 (37%) | 7 / 19 (37%) | 7 / 19 (37%) | 0.368 | 0.368 |
| PWY0-1297~superpathway of purine deoxyribonucleosides degradation | 0.25 | 0.97 | 1 | 0.23 | 0.91 | 1 | 0.003 | 0.003 | 0.0024 | 0.0028 | 0.0022 | 0.0016 | 0.0032 | 0.0024 | 0.0019 | 0.19 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY66-422~D-galactose degradation V (Leloir pathway) | 0.75 | 0.97 | 1 | 0.46 | 0.91 | 1 | 0.022 | 0.022 | 0.022 | 0.022 | 0.023 | 0.0054 | 0.022 | 0.022 | 0.0081 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| ASPARAGINE-BIOSYNTHESIS~L-asparagine biosynthesis I | 0.85 | 0.97 | 1 | 0.63 | 0.96 | 1 | 0.019 | 0.019 | 0.019 | 0.018 | 0.019 | 0.0046 | 0.019 | 0.018 | 0.0079 | 0.078 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| P461-PWY~hexitol fermentation to lactate formate ethanol and acetate | 0.029 | 0.97 | 1 | 0.083 | 0.91 | 1 | 0.0026 | 0.002 | 0.0018 | 0.0025 | 0.0022 | 0.002 | 0.0014 | 0.0014 | 0.0012 | -0.84 | 29 / 38 (76%) | 16 / 19 (84%) | 13 / 19 (68%) | 0.842 | 0.684 |
| PWY-7966~5-methoxy-6-methylbenzimidazolyl adenosylcobamide biosynthesis from adenosylcobinamide-GDP | 0.59 | 0.97 | 1 | 0.39 | 0.91 | 1 | 0.0049 | 0.0049 | 0.0047 | 0.005 | 0.0047 | 0.0018 | 0.0047 | 0.0046 | 0.0014 | -0.089 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5816~all trans undecaprenyl diphosphate biosynthesis | 0.81 | 0.97 | 1 | 0.52 | 0.93 | 1 | 0.0012 | 0.0011 | 0.00077 | 0.0011 | 0.00084 | 0.0012 | 0.0011 | 0.00076 | 0.00091 | 0 | 33 / 38 (87%) | 16 / 19 (84%) | 17 / 19 (89%) | 0.842 | 0.895 |
| PWY-5120~geranylgeranyl diphosphate biosynthesis | 0.63 | 0.97 | 1 | 0.75 | 0.98 | 1 | 0.0028 | 0.0028 | 0.002 | 0.0028 | 0.0023 | 0.0023 | 0.0028 | 0.002 | 0.0044 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5485~pyruvate fermentation to acetate IV | 0.96 | 0.99 | 1 | 0.72 | 0.97 | 1 | 0.018 | 0.018 | 0.018 | 0.018 | 0.018 | 0.0049 | 0.018 | 0.017 | 0.0063 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6270~isoprene biosynthesis I | 0.92 | 0.98 | 1 | 0.65 | 0.96 | 1 | 0.014 | 0.014 | 0.014 | 0.014 | 0.014 | 0.0034 | 0.014 | 0.013 | 0.0046 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-7343~UDP-α-D-glucose biosynthesis I | 0.24 | 0.97 | 1 | 0.55 | 0.93 | 1 | 0.0088 | 0.0088 | 0.0079 | 0.008 | 0.0076 | 0.0038 | 0.0096 | 0.0084 | 0.0071 | 0.26 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| ARGASEDEG-PWY~L-arginine degradation I (arginase pathway) | 0.76 | 0.97 | 1 | 0.82 | 0.99 | 1 | 0.0014 | 0.00033 | 0 | 0.00042 | 0 | 0.0012 | 0.00024 | 0 | 0.00062 | -0.81 | 9 / 38 (24%) | 4 / 19 (21%) | 5 / 19 (26%) | 0.211 | 0.263 |
| PHENOLDEG-PWY~phenol degradation II (anaerobic) | 0.14 | 0.97 | 1 | 0.05 | 0.91 | 1 | 0.00065 | 0.00021 | 0 | 0.00024 | 0 | 0.00053 | 0.00017 | 0 | 5e-04 | -0.5 | 12 / 38 (32%) | 9 / 19 (47%) | 3 / 19 (16%) | 0.474 | 0.158 |
| PWY3O-246~(RR)-butanediol degradation | 0.62 | 0.97 | 1 | 0.79 | 0.99 | 1 | 0.00014 | 2.9e-05 | 0 | 2.1e-05 | 0 | 5.4e-05 | 3.7e-05 | 0 | 9.4e-05 | 0.82 | 8 / 38 (21%) | 4 / 19 (21%) | 4 / 19 (21%) | 0.211 | 0.211 |
| PYRIDNUCSYN-PWY~NAD de novo biosynthesis I (from aspartate) | 0.38 | 0.97 | 1 | 0.25 | 0.91 | 1 | 0.01 | 0.01 | 0.01 | 0.011 | 0.01 | 0.0026 | 0.01 | 0.01 | 0.0026 | -0.14 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6963~ammonia assimilation cycle I | 0.44 | 0.97 | 1 | 0.25 | 0.91 | 1 | 0.034 | 0.034 | 0.035 | 0.035 | 0.035 | 0.0098 | 0.033 | 0.034 | 0.01 | -0.085 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5096~pyruvate fermentation to acetate and alanine | 0.83 | 0.97 | 1 | 0.55 | 0.93 | 1 | 0.0025 | 0.0012 | 0 | 0.0012 | 0.00046 | 0.0016 | 0.0012 | 0 | 0.0019 | 0 | 18 / 38 (47%) | 10 / 19 (53%) | 8 / 19 (42%) | 0.526 | 0.421 |
| ALANINE-VALINESYN-PWY~L-alanine biosynthesis I | 0.31 | 0.97 | 1 | 0.4 | 0.91 | 1 | 0.00078 | 0.00018 | 0 | 0.00015 | 0 | 0.00051 | 0.00022 | 0 | 0.00055 | 0.55 | 9 / 38 (24%) | 4 / 19 (21%) | 5 / 19 (26%) | 0.211 | 0.263 |
| PWY-7967~5-methoxybenzimidazolyl adenosylcobamide biosynthesis from adenosylcobinamide-GDP | 0.59 | 0.97 | 1 | 0.39 | 0.91 | 1 | 0.0049 | 0.0049 | 0.0047 | 0.005 | 0.0047 | 0.0018 | 0.0047 | 0.0046 | 0.0014 | -0.089 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6707~gallate biosynthesis | 0.96 | 0.99 | 1 | 0.88 | 0.99 | 1 | 0.012 | 0.012 | 0.012 | 0.012 | 0.012 | 0.0025 | 0.012 | 0.012 | 0.0019 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| P42-PWY~incomplete reductive TCA cycle | 0.71 | 0.97 | 1 | 0.54 | 0.93 | 1 | 0.0028 | 0.0012 | 0 | 0.0016 | 0 | 0.0033 | 0.00085 | 0.00039 | 0.0014 | -0.91 | 17 / 38 (45%) | 7 / 19 (37%) | 10 / 19 (53%) | 0.368 | 0.526 |
| MENAQUINONESYN-PWY~menaquinol-8 biosynthesis | 0.83 | 0.97 | 1 | 0.96 | 0.99 | 1 | 0.0057 | 0.0057 | 0.0056 | 0.0056 | 0.0056 | 0.0021 | 0.0057 | 0.0055 | 0.0018 | 0.026 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| SERDEG-PWY~L-serine degradation | 0.77 | 0.97 | 1 | 0.82 | 0.99 | 1 | 0.00018 | 0.00011 | 8.4e-05 | 0.00011 | 7.5e-05 | 0.00016 | 0.00011 | 8.7e-05 | 0.00016 | 0 | 23 / 38 (61%) | 11 / 19 (58%) | 12 / 19 (63%) | 0.579 | 0.632 |
| PWY-6751~superpathway of hydrogen production | 0.76 | 0.97 | 1 | 0.9 | 0.99 | 1 | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 | 0.009 | 0.03 | 0.031 | 0.008 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5600~pyruvate fermentation to acetate VII | 0.23 | 0.97 | 1 | 0.16 | 0.91 | 1 | 0.016 | 0.016 | 0.017 | 0.017 | 0.018 | 0.0043 | 0.016 | 0.016 | 0.0038 | -0.087 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6517~N-acetylglucosamine degradation II | 0.41 | 0.97 | 1 | 0.55 | 0.93 | 1 | 0.0015 | 0.0015 | 0.0014 | 0.0017 | 0.0015 | 0.0012 | 0.0014 | 0.0014 | 0.00075 | -0.28 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| SALVPURINE2-PWY~xanthine and xanthosine salvage | 0.96 | 0.99 | 1 | 0.73 | 0.97 | 1 | 0.0086 | 0.0086 | 0.0086 | 0.0086 | 0.0084 | 0.0021 | 0.0086 | 0.0086 | 0.0021 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5143~long-chain fatty acid activation | 0.41 | 0.97 | 1 | 0.21 | 0.91 | 1 | 0.031 | 0.031 | 0.024 | 0.031 | 0.029 | 0.016 | 0.031 | 0.023 | 0.026 | 0 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| CYSTSYN-PWY~L-cysteine biosynthesis I | 0.13 | 0.97 | 1 | 0.055 | 0.91 | 1 | 0.013 | 0.013 | 0.012 | 0.013 | 0.014 | 0.0027 | 0.012 | 0.011 | 0.0038 | -0.12 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| COLANSYN-PWY~colanic acid building blocks biosynthesis | 0.66 | 0.97 | 1 | 0.76 | 0.98 | 1 | 0.0028 | 0.0021 | 0.0017 | 0.0024 | 0.0015 | 0.0028 | 0.0019 | 0.0019 | 0.0017 | -0.34 | 29 / 38 (76%) | 15 / 19 (79%) | 14 / 19 (74%) | 0.789 | 0.737 |
| P161-PWY~acetylene degradation | 0.29 | 0.97 | 1 | 0.25 | 0.91 | 1 | 0.0031 | 0.003 | 0.002 | 0.0025 | 0.0018 | 0.0022 | 0.0035 | 0.002 | 0.0047 | 0.49 | 37 / 38 (97%) | 18 / 19 (95%) | 19 / 19 (100%) | 0.947 | 1 |
| PWY-7675~Kdo transfer to lipid IVA II | 0.85 | 0.97 | 1 | 0.36 | 0.91 | 1 | 0.00074 | 0.00018 | 0 | 0.00022 | 0 | 0.00064 | 0.00013 | 0 | 0.00026 | -0.76 | 9 / 38 (24%) | 3 / 19 (16%) | 6 / 19 (32%) | 0.158 | 0.316 |
| PWY-5667~CDP-diacylglycerol biosynthesis I | 0.91 | 0.98 | 1 | 0.54 | 0.93 | 1 | 0.00093 | 0.00083 | 0.00068 | 0.00084 | 0.00071 | 0.00069 | 0.00082 | 0.00055 | 0.00073 | -0.035 | 34 / 38 (89%) | 16 / 19 (84%) | 18 / 19 (95%) | 0.842 | 0.947 |
| PWY-6123~inosine-5-phosphate biosynthesis I | 0.43 | 0.97 | 1 | 0.27 | 0.91 | 1 | 0.0099 | 0.0099 | 0.0095 | 0.01 | 0.01 | 0.0031 | 0.0097 | 0.0087 | 0.0047 | -0.044 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-1861~formaldehyde assimilation II (assimilatory RuMP Cycle) | 0.0046 | 0.5 | 1 | 0.0061 | 0.62 | 1 | 0.0028 | 0.0027 | 0.0021 | 0.0033 | 0.0029 | 0.0023 | 0.0021 | 0.0015 | 0.0017 | -0.65 | 36 / 38 (95%) | 18 / 19 (95%) | 18 / 19 (95%) | 0.947 | 0.947 |
| PWY-I9~L-cysteine biosynthesis VI (from L-methionine) | 0.99 | 0.99 | 1 | 0.39 | 0.91 | 1 | 0.003 | 0.002 | 0.00084 | 0.0017 | 0.0011 | 0.0022 | 0.0024 | 8e-04 | 0.0053 | 0.5 | 26 / 38 (68%) | 14 / 19 (74%) | 12 / 19 (63%) | 0.737 | 0.632 |
| PWY-7645~hyaluronan degradation | 0.63 | 0.97 | 1 | 0.4 | 0.91 | 1 | 0.00031 | 5.8e-05 | 0 | 5.4e-05 | 0 | 0.00013 | 6.2e-05 | 0 | 2e-04 | 0.2 | 7 / 38 (18%) | 4 / 19 (21%) | 3 / 19 (16%) | 0.211 | 0.158 |
| DETOX1-PWY~superoxide radicals degradation | 0.97 | 0.99 | 1 | 0.96 | 0.99 | 1 | 0.0011 | 0.00046 | 0 | 0.00048 | 0 | 0.00099 | 0.00045 | 0 | 0.00091 | -0.093 | 16 / 38 (42%) | 8 / 19 (42%) | 8 / 19 (42%) | 0.421 | 0.421 |
| PWY-6952~glycerophosphodiester degradation | 0.42 | 0.97 | 1 | 0.39 | 0.91 | 1 | 0.0011 | 5e-04 | 0 | 0.00056 | 0.00013 | 0.00091 | 0.00044 | 0 | 8e-04 | -0.35 | 18 / 38 (47%) | 10 / 19 (53%) | 8 / 19 (42%) | 0.526 | 0.421 |
| THRESYN-PWY~superpathway of L-threonine biosynthesis | 0.22 | 0.97 | 1 | 0.11 | 0.91 | 1 | 0.017 | 0.017 | 0.016 | 0.017 | 0.018 | 0.0038 | 0.016 | 0.016 | 0.0065 | -0.087 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY66-380~estradiol biosynthesis I (via estrone) | 0.56 | 0.97 | 1 | 0.61 | 0.96 | 1 | 0.0015 | 0.00085 | 0.00012 | 6e-04 | 0.00022 | 0.00087 | 0.0011 | 0 | 0.0029 | 0.87 | 21 / 38 (55%) | 12 / 19 (63%) | 9 / 19 (47%) | 0.632 | 0.474 |
| AMMASSIM-PWY~ammonia assimilation cycle III | 0.58 | 0.97 | 1 | 0.35 | 0.91 | 1 | 0.067 | 0.067 | 0.067 | 0.068 | 0.066 | 0.01 | 0.066 | 0.069 | 0.016 | -0.043 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-6754~S-methyl-5-thioadenosine degradation I | 0.42 | 0.97 | 1 | 0.34 | 0.91 | 1 | 0.00065 | 5.1e-05 | 0 | 5.9e-05 | 0 | 0.00018 | 4.3e-05 | 0 | 0.00019 | -0.46 | 3 / 38 (7.9%) | 2 / 19 (11%) | 1 / 19 (5.3%) | 0.105 | 0.0526 |
| PWY-5480~pyruvate fermentation to ethanol I | 0.62 | 0.97 | 1 | 0.83 | 0.99 | 1 | 0.0038 | 0.0038 | 0.0023 | 0.0033 | 0.0023 | 0.0026 | 0.0042 | 0.0023 | 0.0057 | 0.35 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5004~superpathway of L-citrulline metabolism | 0.47 | 0.97 | 1 | 0.71 | 0.97 | 1 | 0.0016 | 0.00026 | 0 | 0.00035 | 0 | 0.001 | 0.00016 | 0 | 0.00045 | -1.1 | 6 / 38 (16%) | 3 / 19 (16%) | 3 / 19 (16%) | 0.158 | 0.158 |
| PWY-7586~β-14-D-mannosyl-N-acetyl-D-glucosamine degradation | 0.46 | 0.97 | 1 | 0.62 | 0.96 | 1 | 0.0012 | 0.00018 | 0 | 0.00024 | 0 | 0.00064 | 0.00013 | 0 | 0.00035 | -0.88 | 6 / 38 (16%) | 3 / 19 (16%) | 3 / 19 (16%) | 0.158 | 0.158 |
| PWY-4983~nitric oxide biosynthesis II (mammals) | 0.15 | 0.97 | 1 | 0.17 | 0.91 | 1 | 0.00052 | 0.00026 | 3.9e-05 | 0.00019 | 0 | 0.00031 | 0.00033 | 0.00016 | 0.00044 | 0.8 | 19 / 38 (50%) | 8 / 19 (42%) | 11 / 19 (58%) | 0.421 | 0.579 |
| CENTFERM-PWY~pyruvate fermentation to butanoate | 0.62 | 0.97 | 1 | 0.42 | 0.91 | 1 | 0.00064 | 0.00039 | 0.00033 | 0.00039 | 0.00034 | 0.00038 | 0.00038 | 3e-04 | 0.00052 | -0.037 | 23 / 38 (61%) | 13 / 19 (68%) | 10 / 19 (53%) | 0.684 | 0.526 |
| PWY-5509~adenosylcobalamin biosynthesis from adenosylcobinamide-GDP I | 0.59 | 0.97 | 1 | 0.39 | 0.91 | 1 | 0.0049 | 0.0049 | 0.0047 | 0.005 | 0.0047 | 0.0018 | 0.0047 | 0.0046 | 0.0014 | -0.089 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| MANNIDEG-PWY~mannitol degradation I | 0.046 | 0.97 | 1 | 0.18 | 0.91 | 1 | 0.0025 | 0.0025 | 0.0022 | 0.003 | 0.0025 | 0.0019 | 0.0021 | 0.002 | 0.001 | -0.51 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5340~sulfate activation for sulfonation | 0.8 | 0.97 | 1 | 0.39 | 0.91 | 1 | 0.0082 | 0.0082 | 0.0077 | 0.0082 | 0.0078 | 0.0031 | 0.0081 | 0.0077 | 0.0037 | -0.018 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5155~β-alanine biosynthesis III | 0.2 | 0.97 | 1 | 0.39 | 0.91 | 1 | 0.0031 | 0.0031 | 0.003 | 0.0034 | 0.0034 | 0.0015 | 0.0028 | 0.0027 | 0.00094 | -0.28 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5971~palmitate biosynthesis II (bacteria and plants) | 0.1 | 0.97 | 1 | 0.2 | 0.91 | 1 | 0.0047 | 0.0047 | 0.0035 | 0.0051 | 0.0042 | 0.0041 | 0.0043 | 0.0029 | 0.0045 | -0.25 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-3341~L-proline biosynthesis III | 0.99 | 0.99 | 1 | 0.61 | 0.96 | 1 | 0.00083 | 0.00026 | 0 | 0.00031 | 0 | 0.00077 | 0.00021 | 0 | 0.00041 | -0.56 | 12 / 38 (32%) | 5 / 19 (26%) | 7 / 19 (37%) | 0.263 | 0.368 |
| PWY-7180~2-deoxy-α-D-ribose 1-phosphate degradation | 0.18 | 0.97 | 1 | 0.24 | 0.91 | 1 | 0.00059 | 0.00017 | 0 | 1e-04 | 0 | 0.00025 | 0.00024 | 0 | 0.00037 | 1.3 | 11 / 38 (29%) | 4 / 19 (21%) | 7 / 19 (37%) | 0.211 | 0.368 |
| GLUTAMINEFUM-PWY~L-glutamine degradation II | 0.8 | 0.97 | 1 | 0.48 | 0.91 | 1 | 0.091 | 0.091 | 0.093 | 0.091 | 0.092 | 0.014 | 0.09 | 0.094 | 0.021 | -0.016 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY0-862~(5Z)-dodecenoate biosynthesis I | 0.092 | 0.97 | 1 | 0.16 | 0.91 | 1 | 0.0046 | 0.0046 | 0.0033 | 0.0051 | 0.0041 | 0.004 | 0.0042 | 0.0029 | 0.0044 | -0.28 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-4984~urea cycle | 0.12 | 0.97 | 1 | 0.24 | 0.91 | 1 | 0.002 | 0.0011 | 0.00029 | 0.0014 | 0.00072 | 0.0018 | 0.00066 | 0 | 0.00097 | -1.1 | 20 / 38 (53%) | 11 / 19 (58%) | 9 / 19 (47%) | 0.579 | 0.474 |
| LCYSDEG-PWY~L-cysteine degradation II | 0.77 | 0.97 | 1 | 0.82 | 0.99 | 1 | 0.00018 | 0.00011 | 8.4e-05 | 0.00011 | 7.5e-05 | 0.00016 | 0.00011 | 8.7e-05 | 0.00016 | 0 | 23 / 38 (61%) | 11 / 19 (58%) | 12 / 19 (63%) | 0.579 | 0.632 |
| PWY-5839~menaquinol-7 biosynthesis | 0.24 | 0.97 | 1 | 0.48 | 0.91 | 1 | 0.0015 | 0.00073 | 6.1e-05 | 0.00093 | 0.00012 | 0.0015 | 0.00052 | 0 | 0.00089 | -0.84 | 19 / 38 (50%) | 10 / 19 (53%) | 9 / 19 (47%) | 0.526 | 0.474 |
| NAGLIPASYN-PWY~lipid IVA biosynthesis | 0.086 | 0.97 | 1 | 0.24 | 0.91 | 1 | 0.0016 | 0.00078 | 0 | 0.0012 | 9.7e-05 | 0.0019 | 0.00037 | 0 | 0.00057 | -1.7 | 18 / 38 (47%) | 10 / 19 (53%) | 8 / 19 (42%) | 0.526 | 0.421 |
| HEMESYN2-PWY~heme b biosynthesis II (anaerobic) | 0.61 | 0.97 | 1 | 0.98 | 0.99 | 1 | 0.00051 | 5.4e-05 | 0 | 4.5e-05 | 0 | 0.00018 | 6.3e-05 | 0 | 0.00026 | 0.49 | 4 / 38 (11%) | 2 / 19 (11%) | 2 / 19 (11%) | 0.105 | 0.105 |
| PWY-5844~menaquinol-9 biosynthesis | 0.83 | 0.97 | 1 | 0.96 | 0.99 | 1 | 0.0057 | 0.0057 | 0.0056 | 0.0056 | 0.0056 | 0.0021 | 0.0057 | 0.0055 | 0.0018 | 0.026 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5484~glycolysis II (from fructose 6-phosphate) | 0.12 | 0.97 | 1 | 0.019 | 0.91 | 1 | 0.011 | 0.011 | 0.012 | 0.012 | 0.013 | 0.0031 | 0.011 | 0.011 | 0.0029 | -0.13 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5766~L-glutamate degradation X | 0.29 | 0.97 | 1 | 0.98 | 0.99 | 1 | 0.00091 | 0.00048 | 5.2e-05 | 0.00068 | 0 | 0.0014 | 0.00028 | 8e-05 | 0.00046 | -1.3 | 20 / 38 (53%) | 9 / 19 (47%) | 11 / 19 (58%) | 0.474 | 0.579 |
| PWY-6599~guanine and guanosine salvage II | 0.86 | 0.97 | 1 | 0.46 | 0.91 | 1 | 0.0018 | 0.0017 | 0.0014 | 0.0015 | 0.0017 | 0.0011 | 0.0019 | 0.0012 | 0.002 | 0.34 | 35 / 38 (92%) | 17 / 19 (89%) | 18 / 19 (95%) | 0.895 | 0.947 |
| PWY-841~superpathway of purine nucleotides de novo biosynthesis I | 0.7 | 0.97 | 1 | 0.72 | 0.97 | 1 | 0.0041 | 0.004 | 0.0034 | 0.0042 | 0.0032 | 0.003 | 0.0038 | 0.0038 | 0.0023 | -0.14 | 37 / 38 (97%) | 18 / 19 (95%) | 19 / 19 (100%) | 0.947 | 1 |
| PHESYN~L-phenylalanine biosynthesis I | 0.94 | 0.99 | 1 | 0.7 | 0.97 | 1 | 0.0046 | 0.0046 | 0.0029 | 0.0042 | 0.0047 | 0.0032 | 0.005 | 0.0025 | 0.0069 | 0.25 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5897~superpathway of menaquinol-11 biosynthesis | 0.27 | 0.97 | 1 | 0.14 | 0.91 | 1 | 0.00069 | 7.3e-05 | 0 | 9.2e-05 | 0 | 0.00026 | 5.4e-05 | 0 | 0.00023 | -0.77 | 4 / 38 (11%) | 3 / 19 (16%) | 1 / 19 (5.3%) | 0.158 | 0.0526 |
| PWY-6471~peptidoglycan biosynthesis IV (Enterococcus faecium) | 0.64 | 0.97 | 1 | 0.73 | 0.97 | 1 | 0.0053 | 0.0035 | 0.0011 | 0.0028 | 0.00085 | 0.0036 | 0.0041 | 0.0012 | 0.0094 | 0.55 | 25 / 38 (66%) | 13 / 19 (68%) | 12 / 19 (63%) | 0.684 | 0.632 |
| PWY-1263~taurine degradation I | 0.65 | 0.97 | 1 | 0.98 | 0.99 | 1 | 0.00081 | 0.00057 | 0.00027 | 0.00053 | 0.00023 | 0.00058 | 0.00061 | 0.00048 | 0.00069 | 0.2 | 27 / 38 (71%) | 14 / 19 (74%) | 13 / 19 (68%) | 0.737 | 0.684 |
| PWY-5505~L-glutamate and L-glutamine biosynthesis | 0.38 | 0.97 | 1 | 0.26 | 0.91 | 1 | 0.047 | 0.047 | 0.048 | 0.048 | 0.05 | 0.0066 | 0.046 | 0.046 | 0.011 | -0.061 | 38 / 38 (100%) | 19 / 19 (100%) | 19 / 19 (100%) | 1 | 1 |
| PWY-5274~sulfide oxidation II (sulfide dehydrogenase) | 0.59 | 0.97 | 1 | 0.56 | 0.93 | 1 | 9.5e-05 | 7.5e-06 | 0 | 1.2e-05 | 0 | 5.4e-05 | 2.6e-06 | 0 | 8.2e-06 | -2.2 | 3 / 38 (7.9%) | 1 / 19 (5.3%) | 2 / 19 (11%) | 0.0526 | 0.105 |
| PWY66-423~fructose 26-bisphosphate biosynthesis | 0.78 | 0.97 | 1 | 0.93 | 0.99 | 1 | 0.0016 | 0.0013 | 6e-04 | 0.0012 | 0.00072 | 0.0015 | 0.0015 | 0.00059 | 0.0033 | 0.32 | 32 / 38 (84%) | 15 / 19 (79%) | 17 / 19 (89%) | 0.789 | 0.895 |
| ARG-PRO-PWY~L-arginine degradation VI (arginase 2 pathway) | 0.82 | 0.97 | 1 | 0.67 | 0.96 | 1 | 0.001 | 0.00022 | 0 | 0.00028 | 0 | 0.00081 | 0.00016 | 0 | 0.00044 | -0.81 | 8 / 38 (21%) | 3 / 19 (16%) | 5 / 19 (26%) | 0.158 | 0.263 |
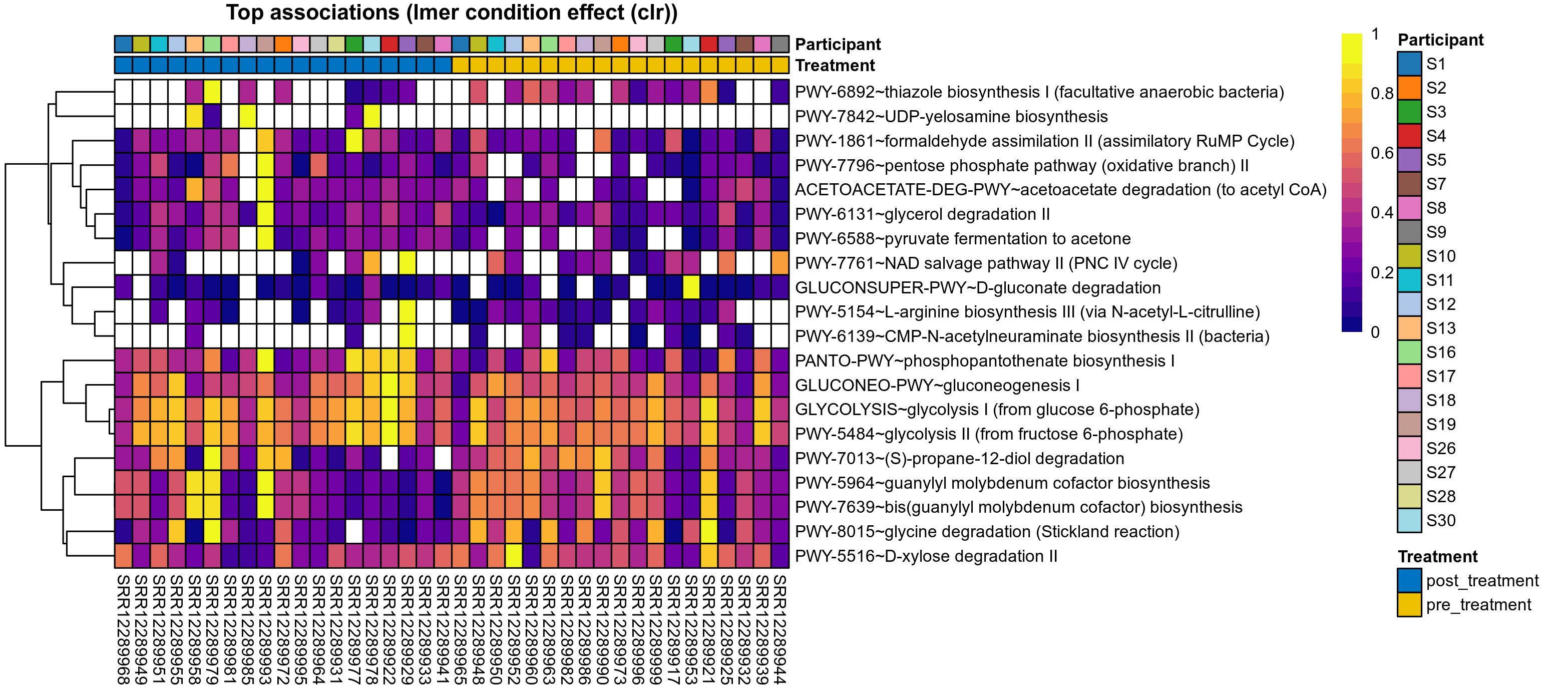
Click here to open full-sized image in new window.

Click here to open full-sized image in new window.
P lmer condition effect (sqrt): Differentially abundant functions were identified by linear mixed effect regression without confounders. Random effects: Participant (intercept). Fixed effects: Treatment. Effect tested: Treatment.
P lmer condition effect (clr): Differentially abundant functions were identified by linear mixed effect regression without confounders. Random effects: Participant (intercept). Fixed effects: Treatment. Effect tested: Treatment.
The following plots present the distribution of the top most differentially abundant microbial functions across all applied statistical analysis. Plots are ordered alphabetically.