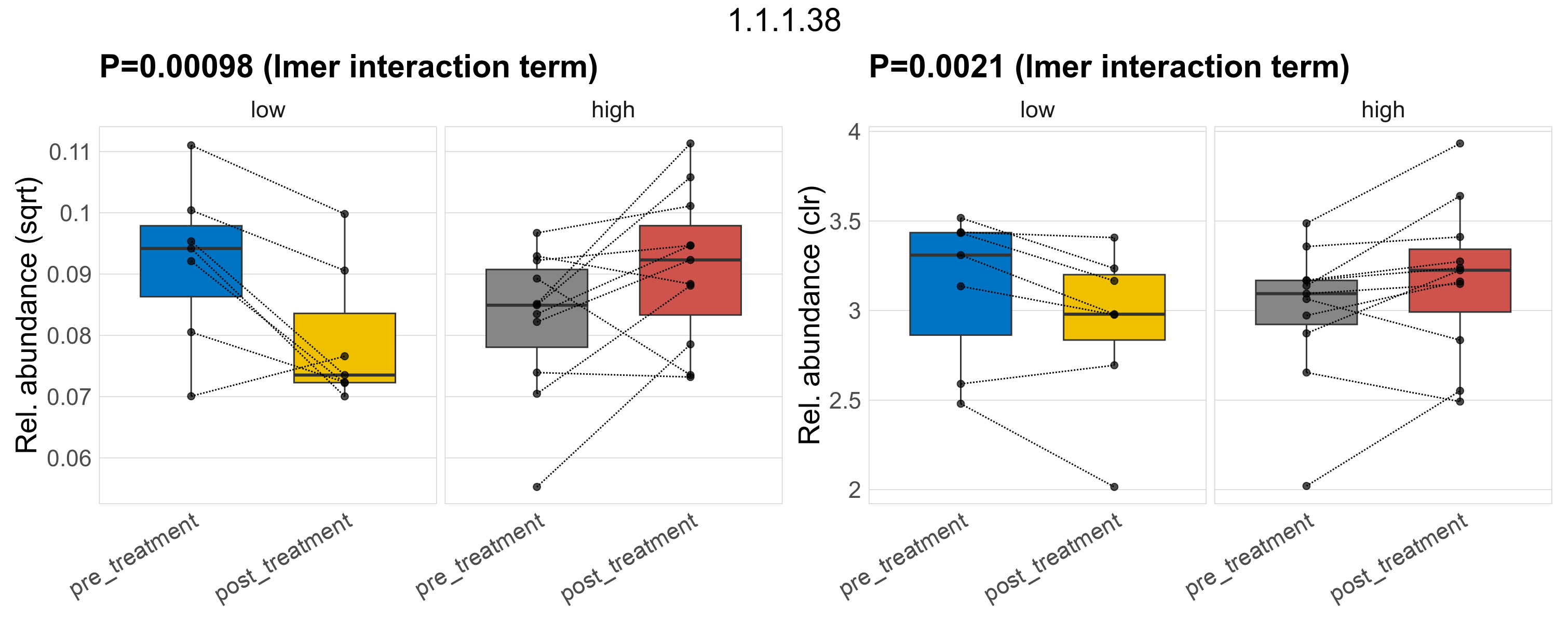
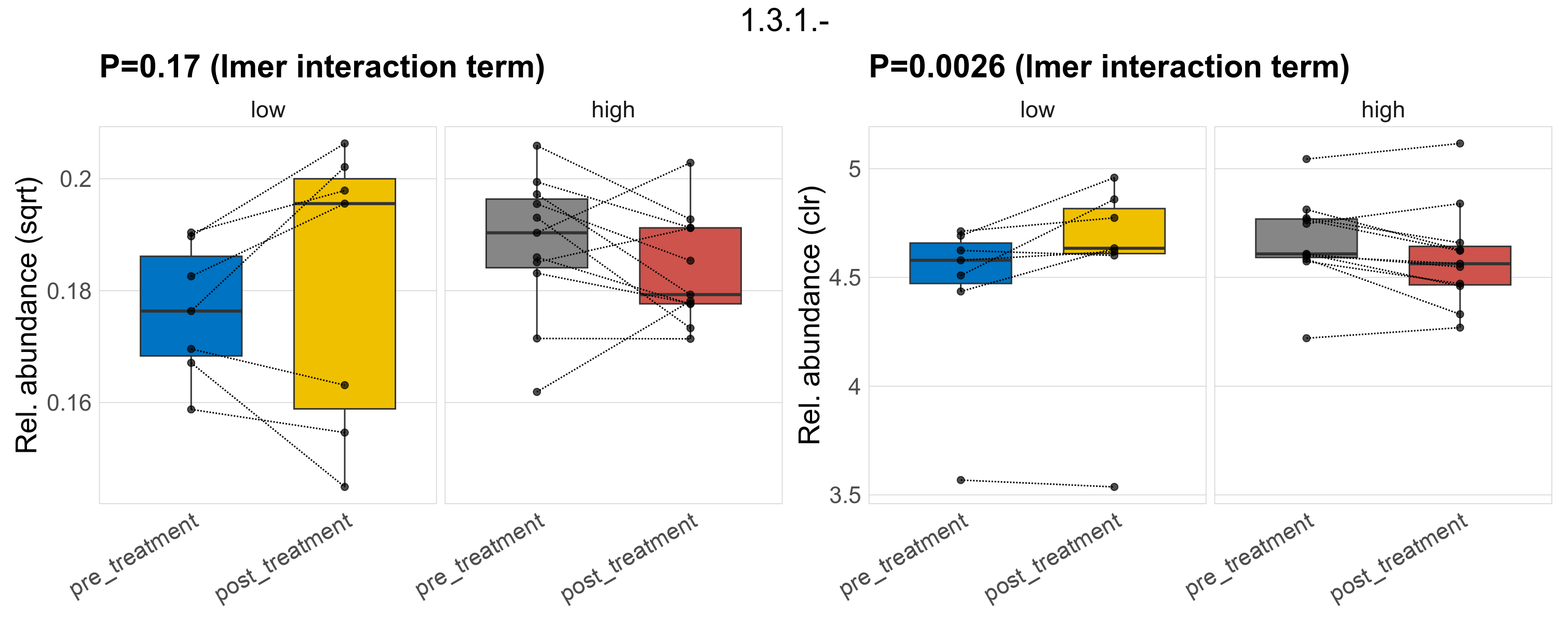
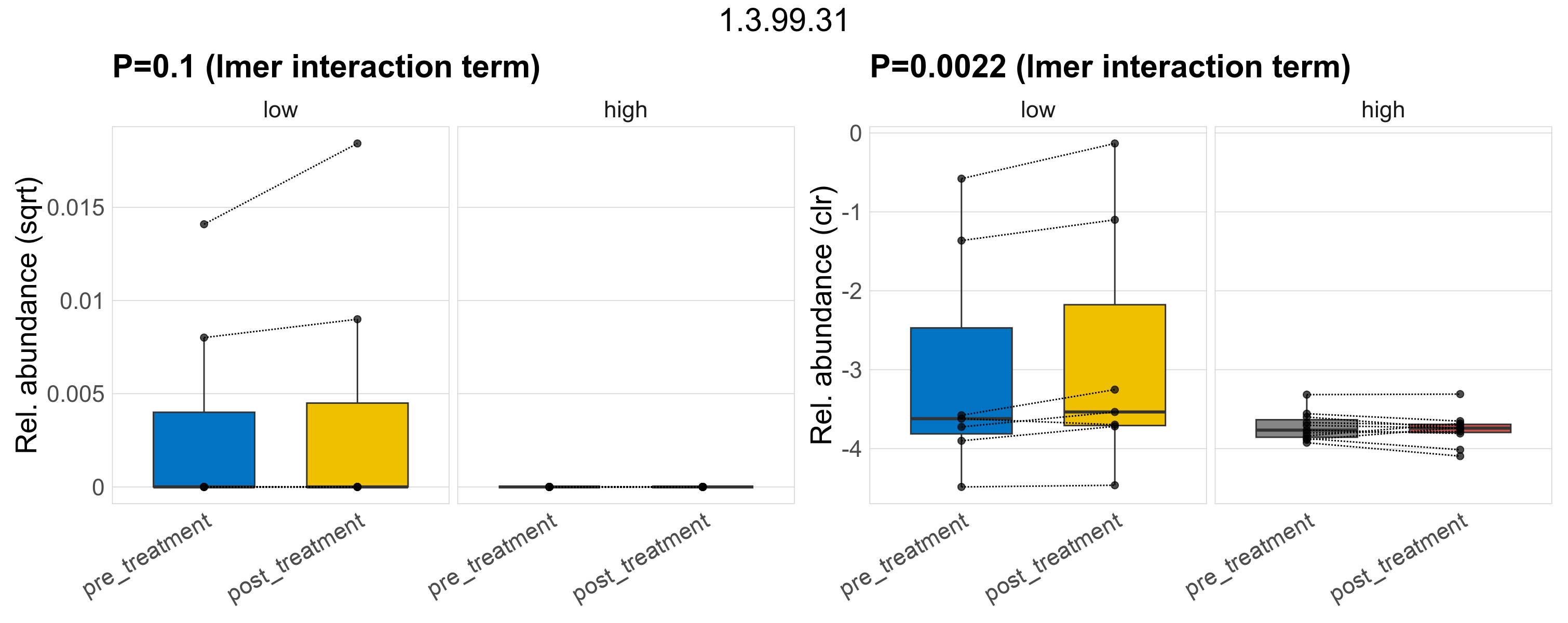
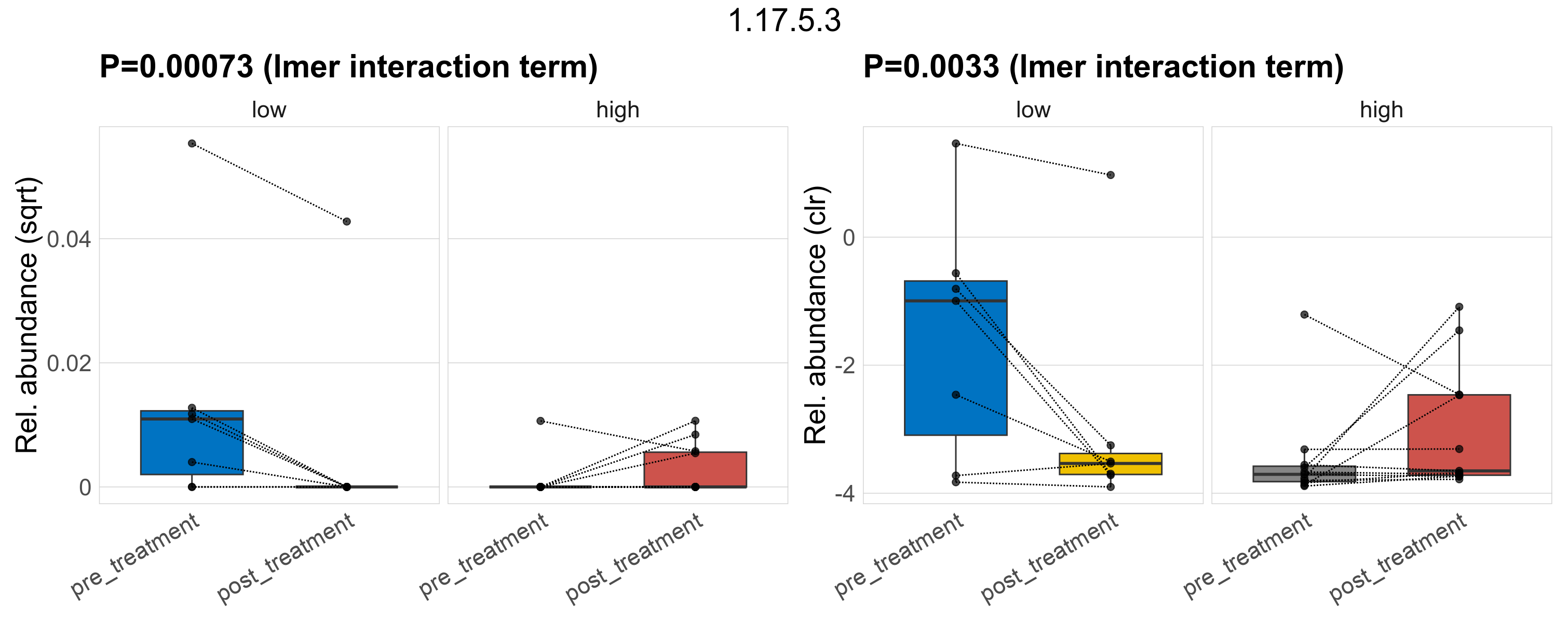
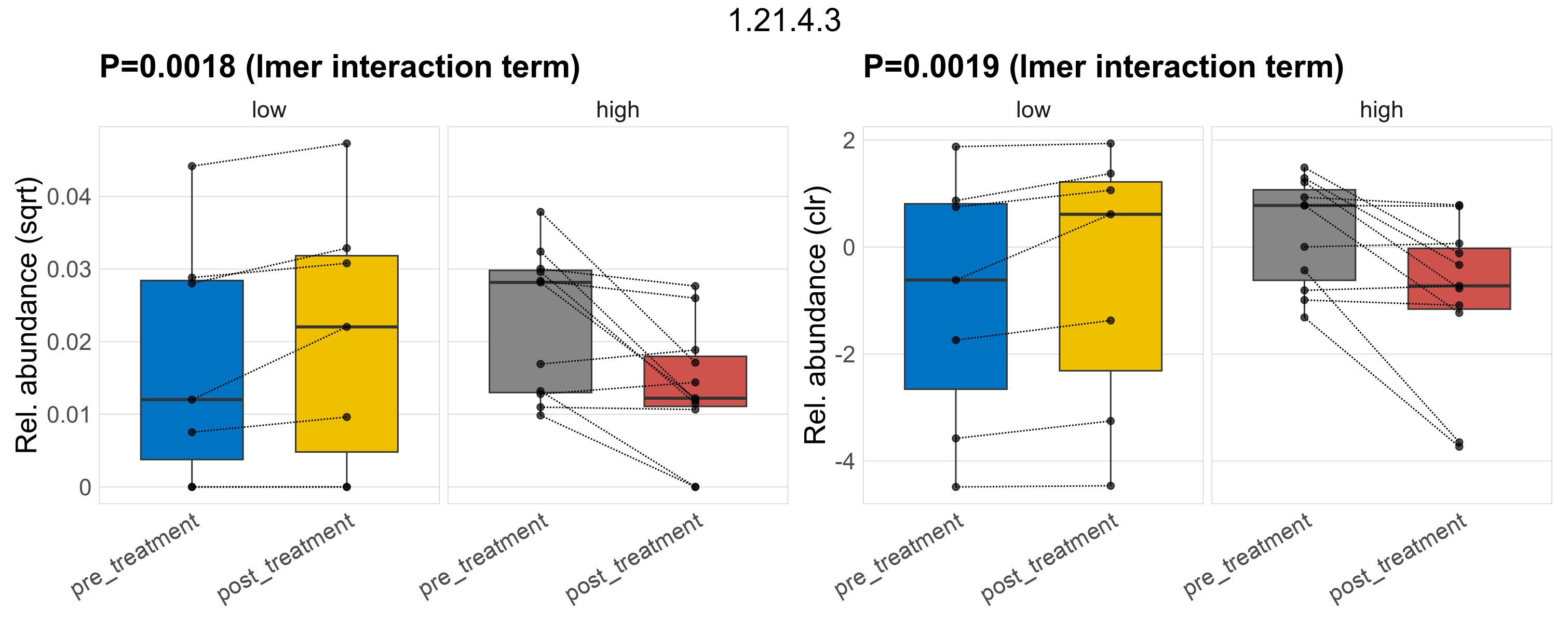
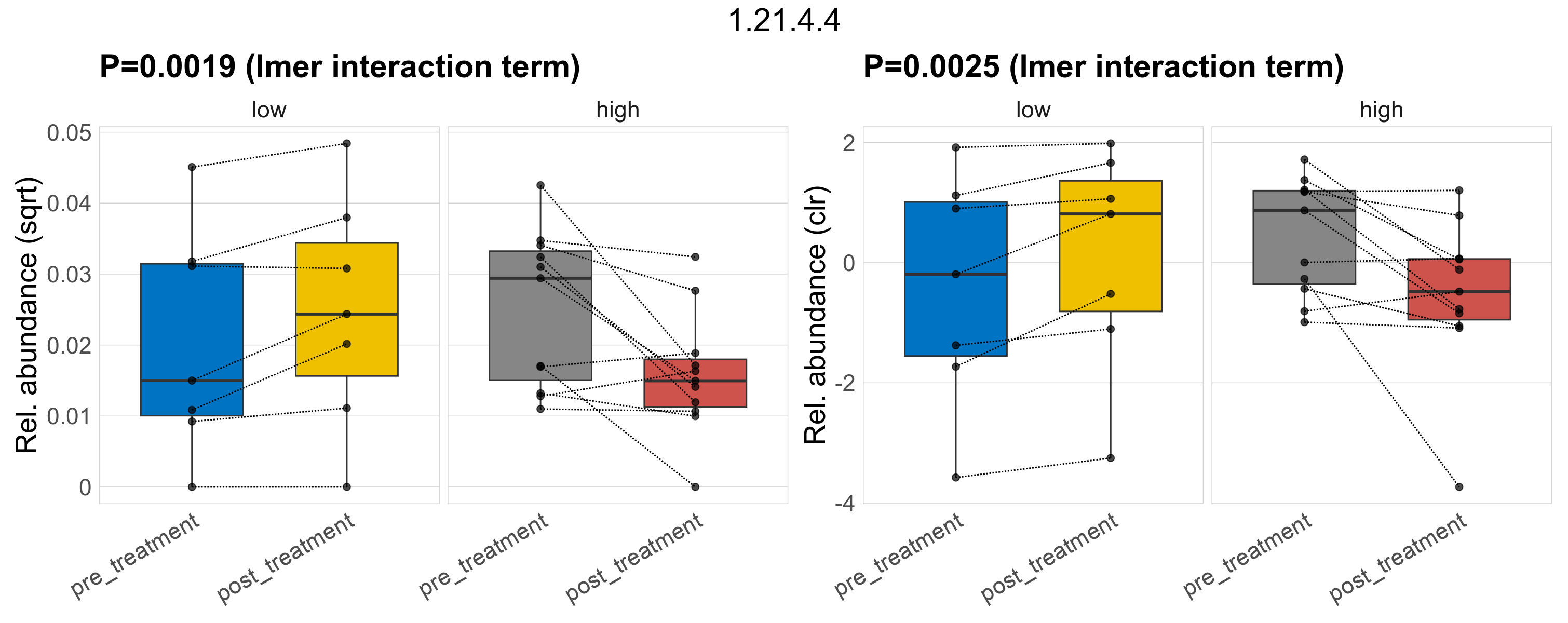
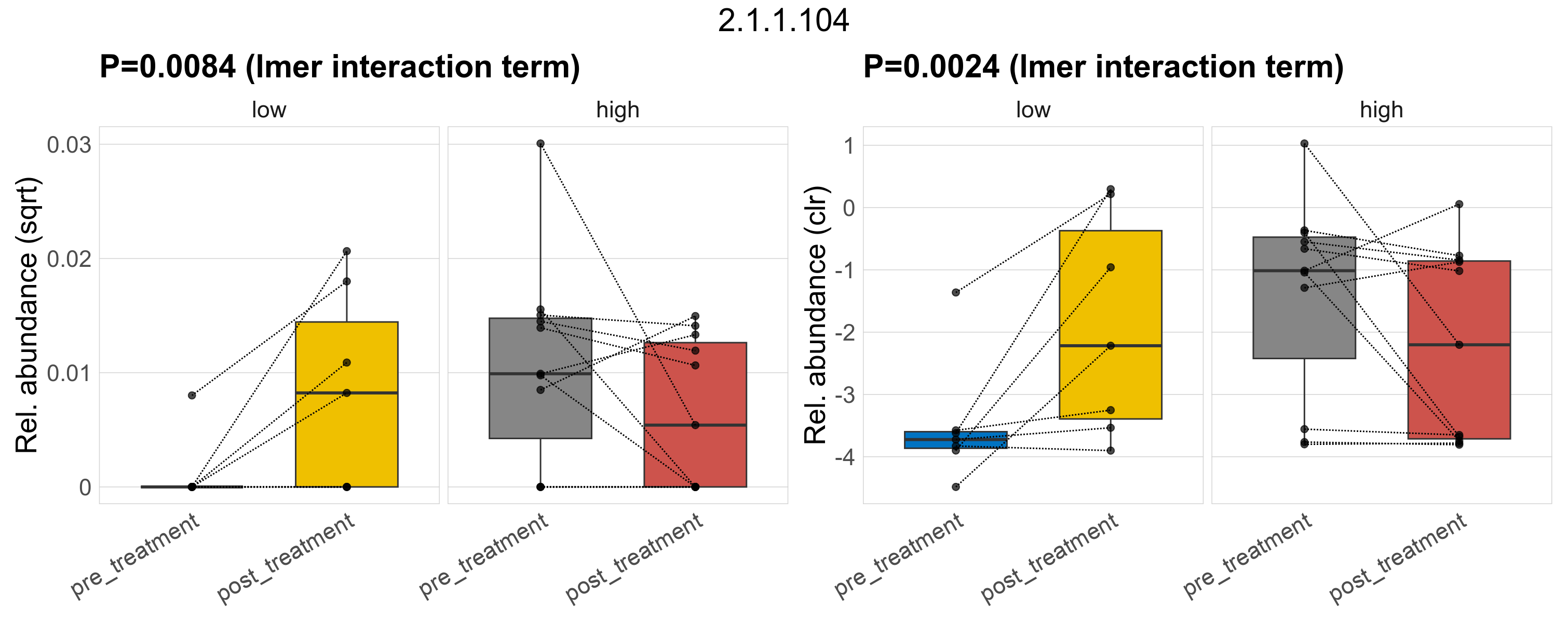
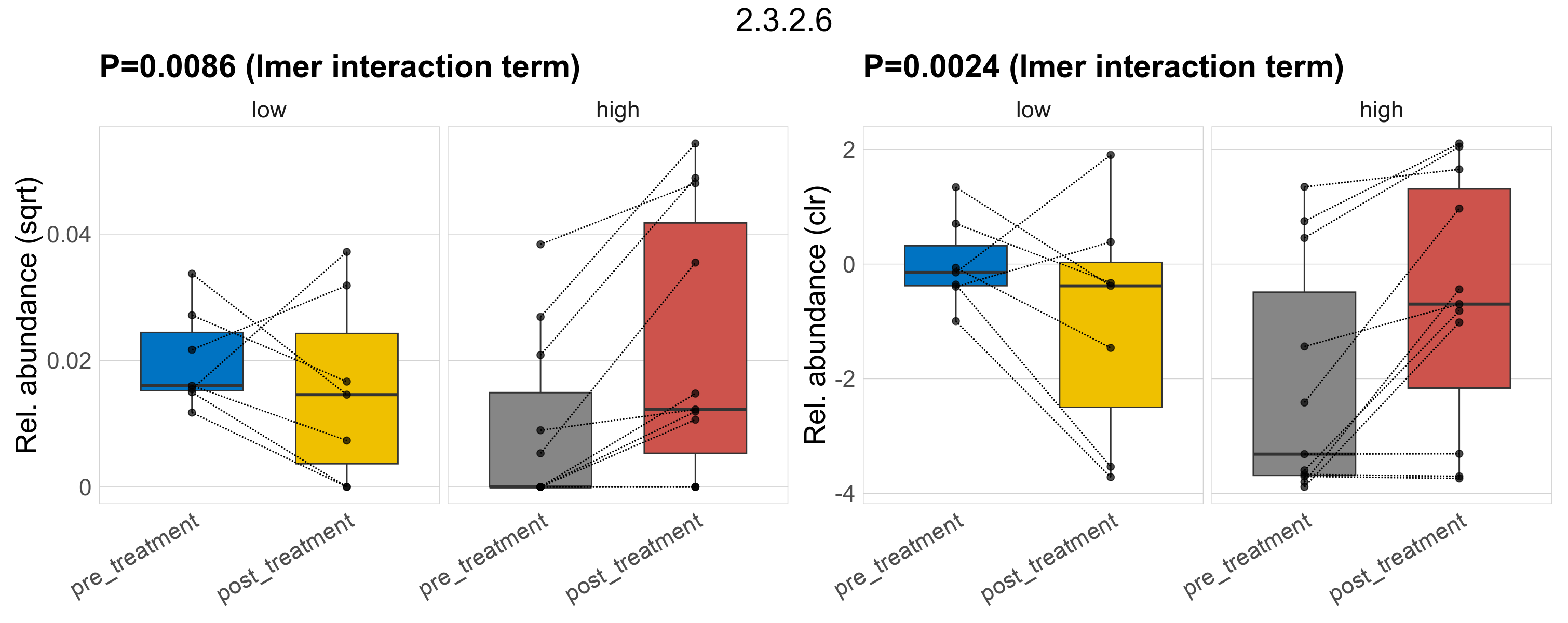
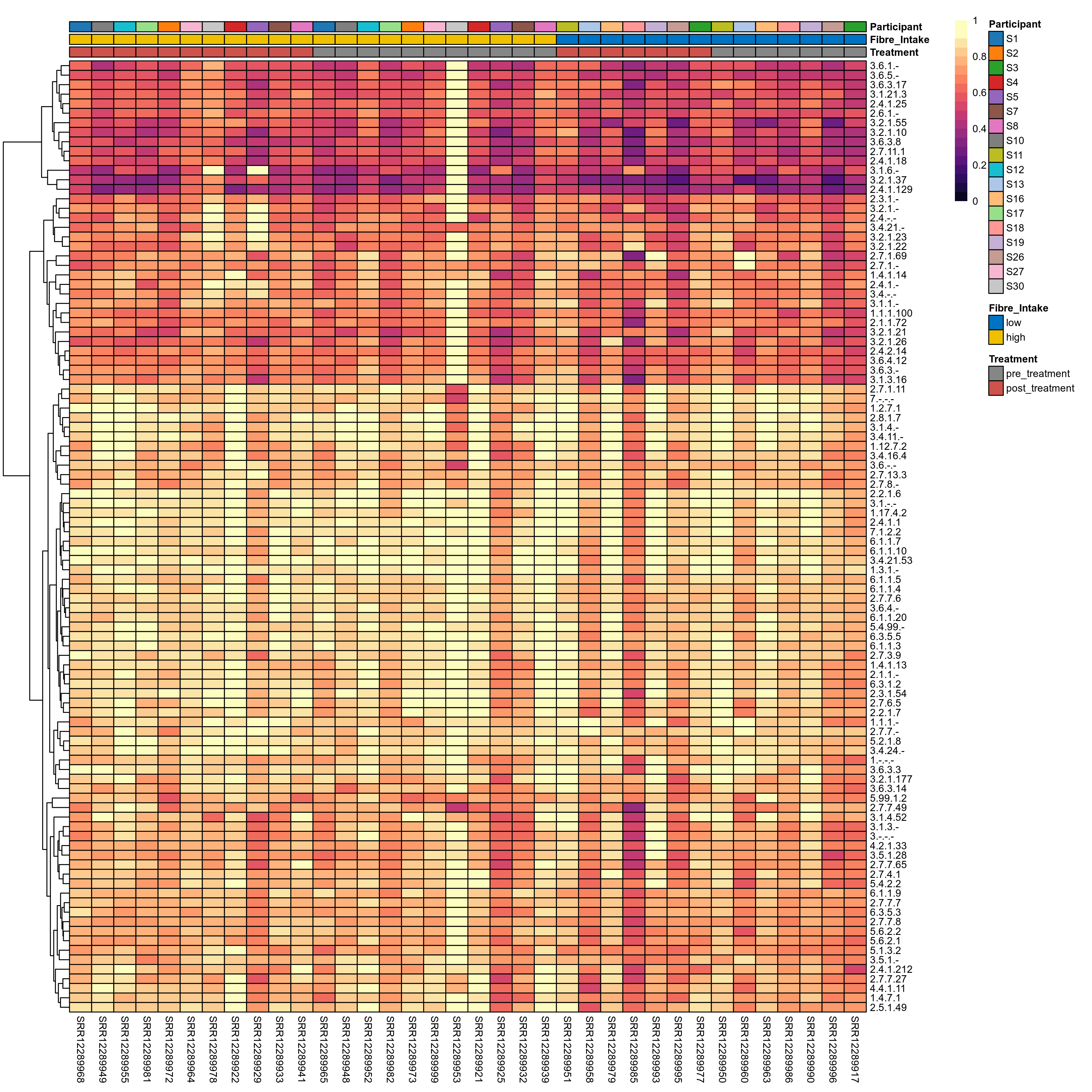
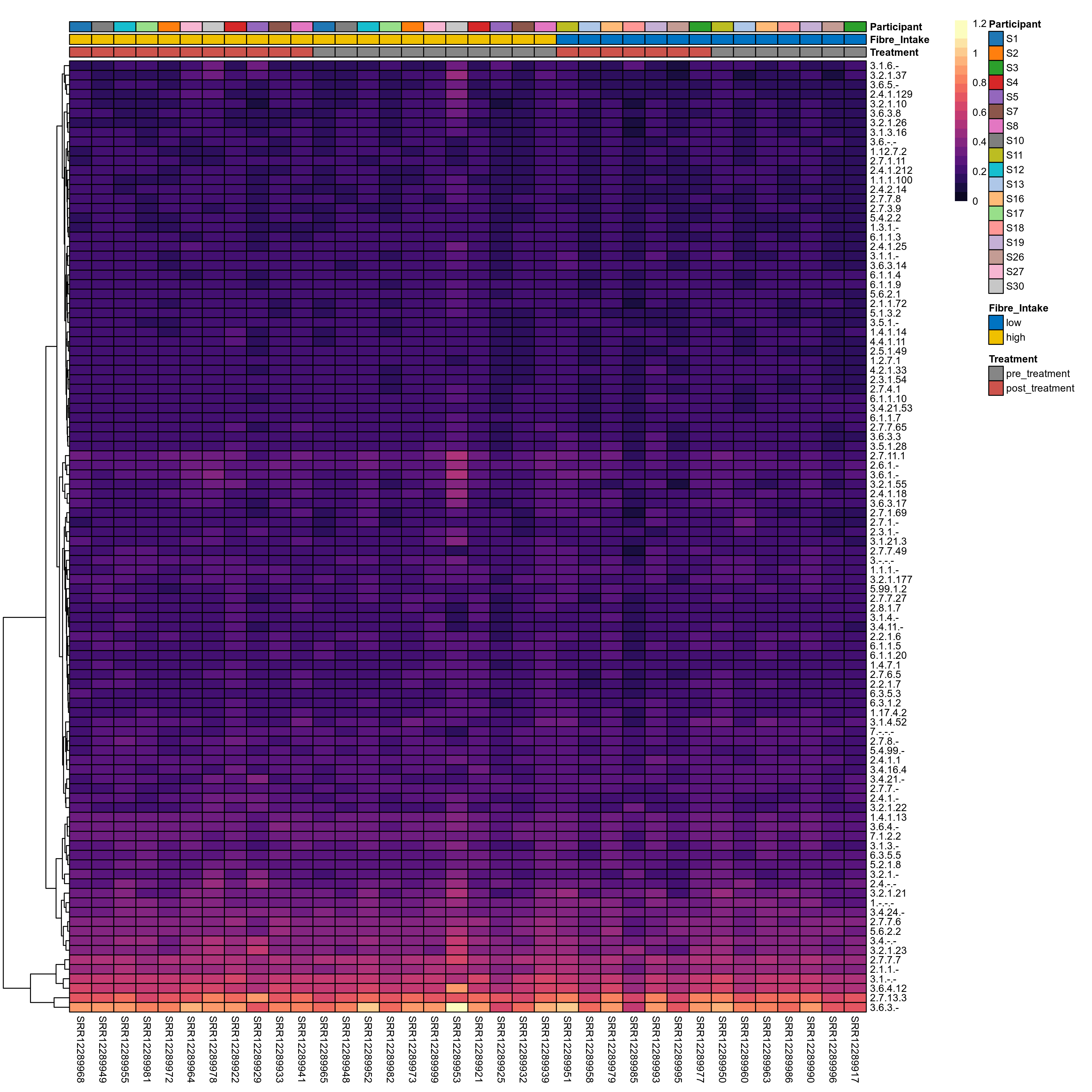
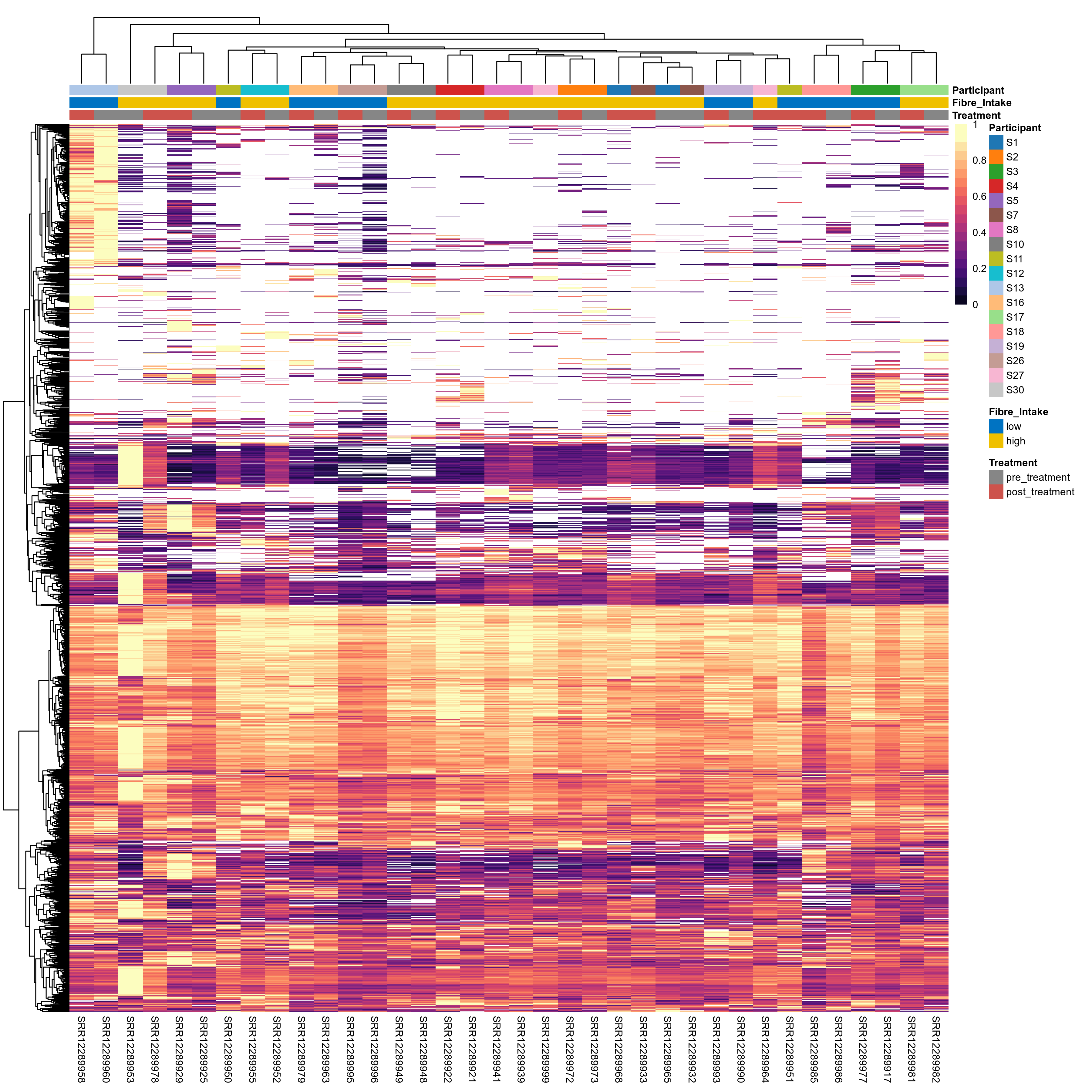
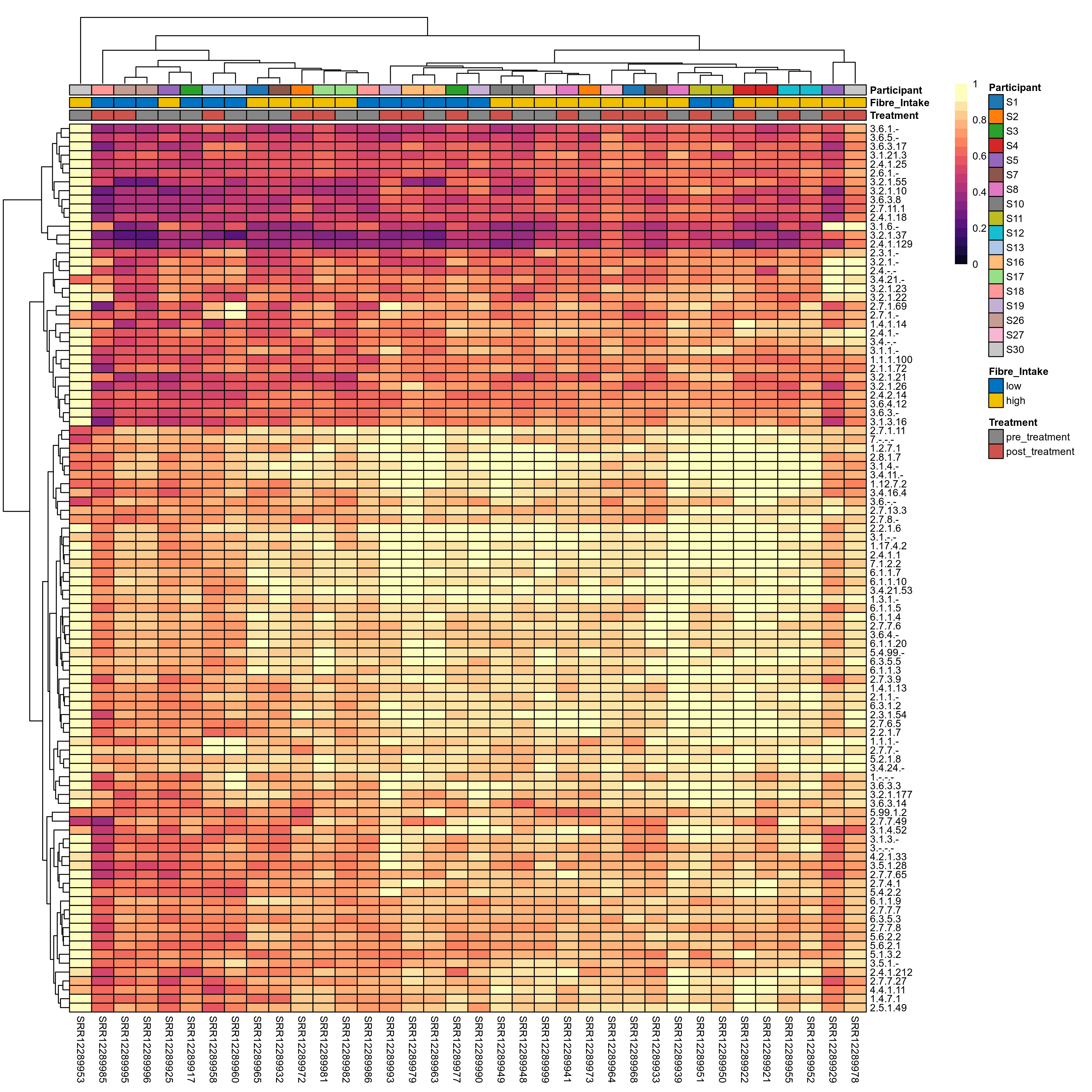
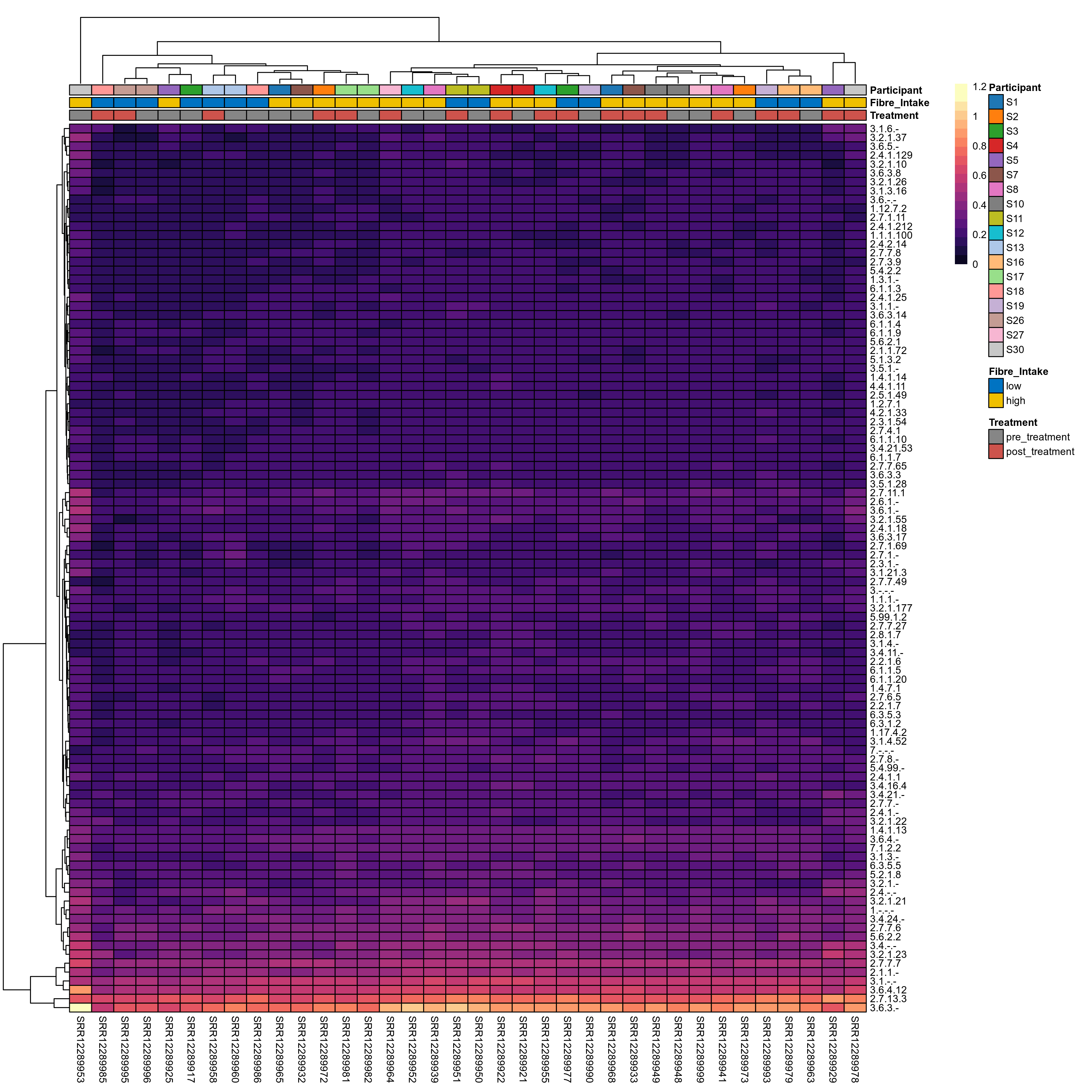
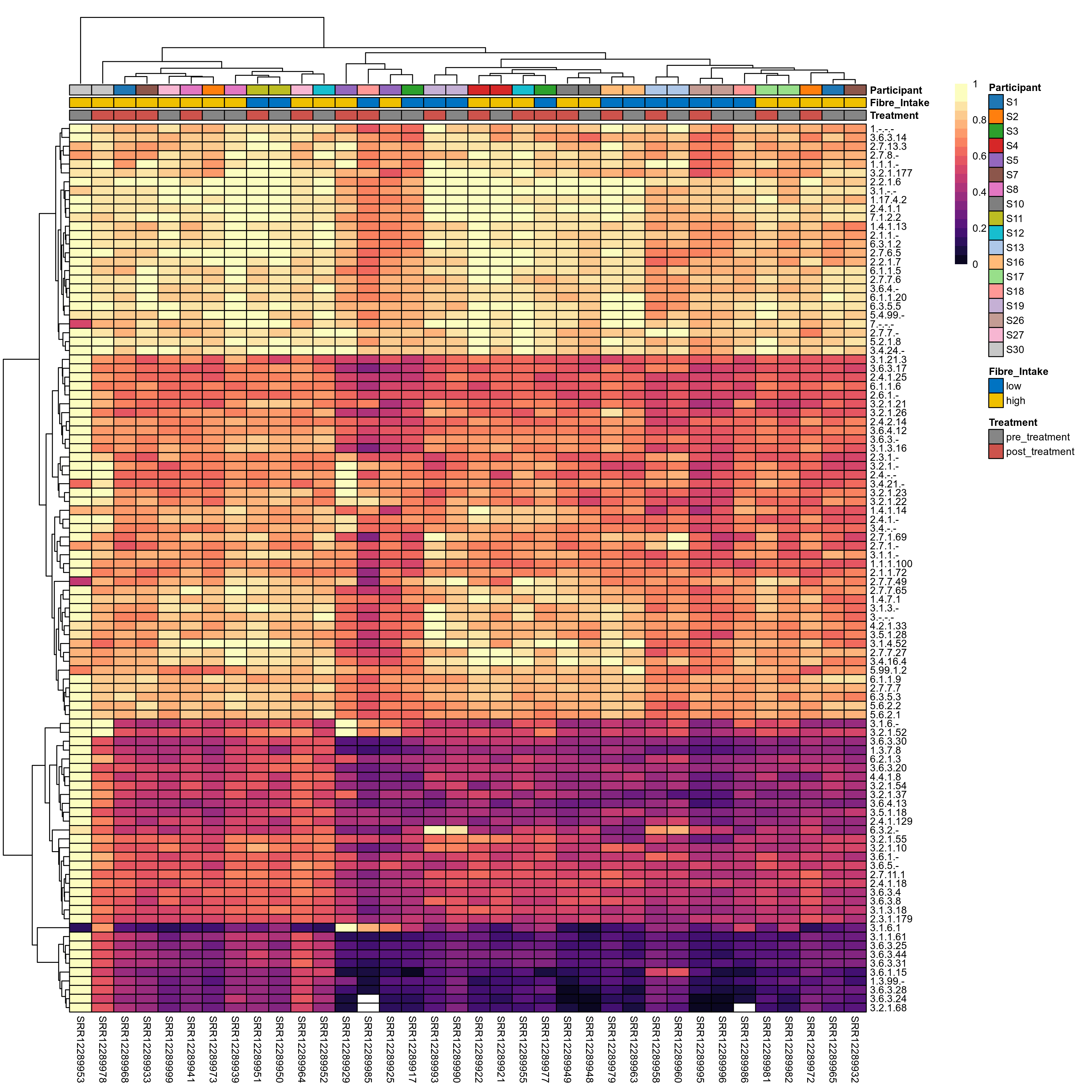
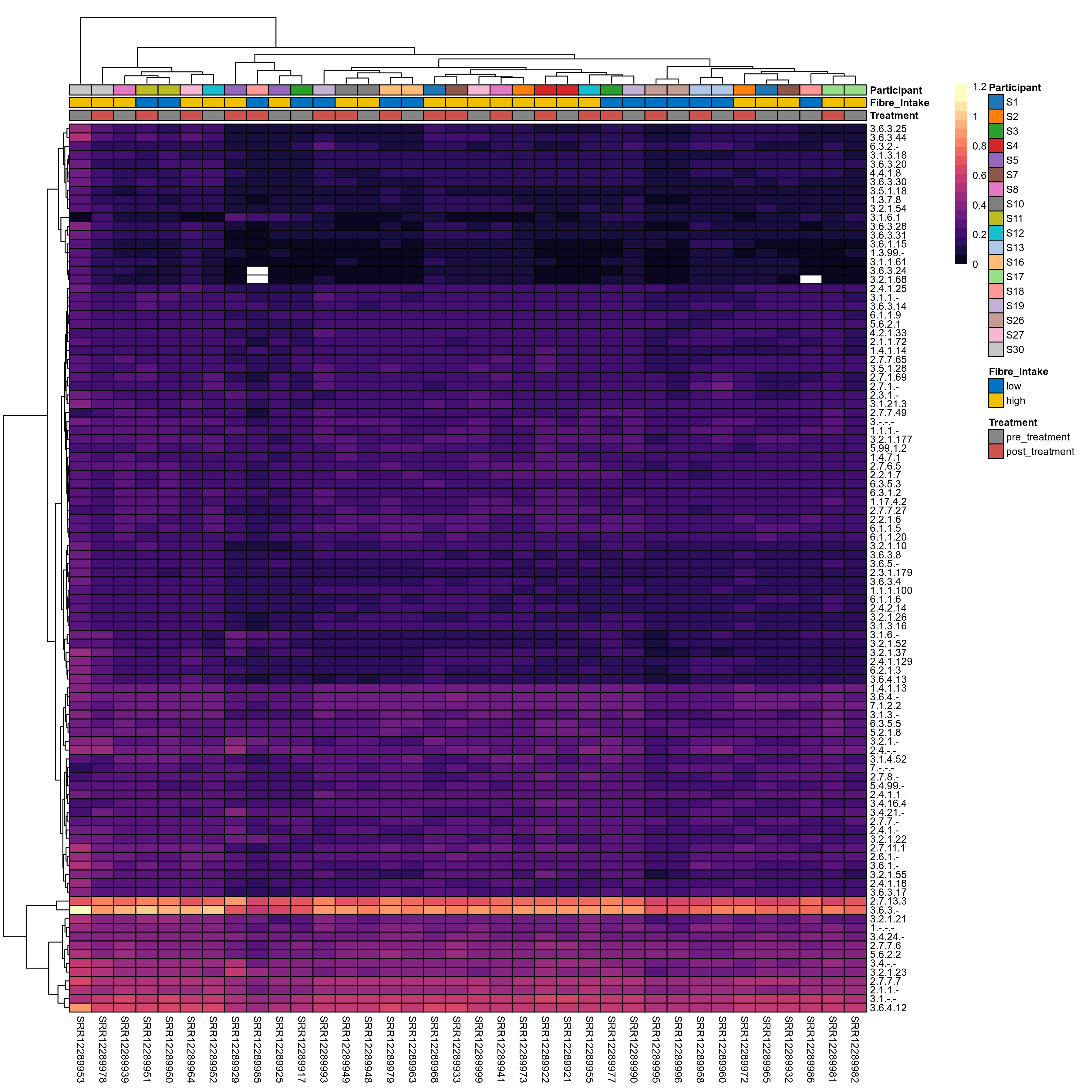
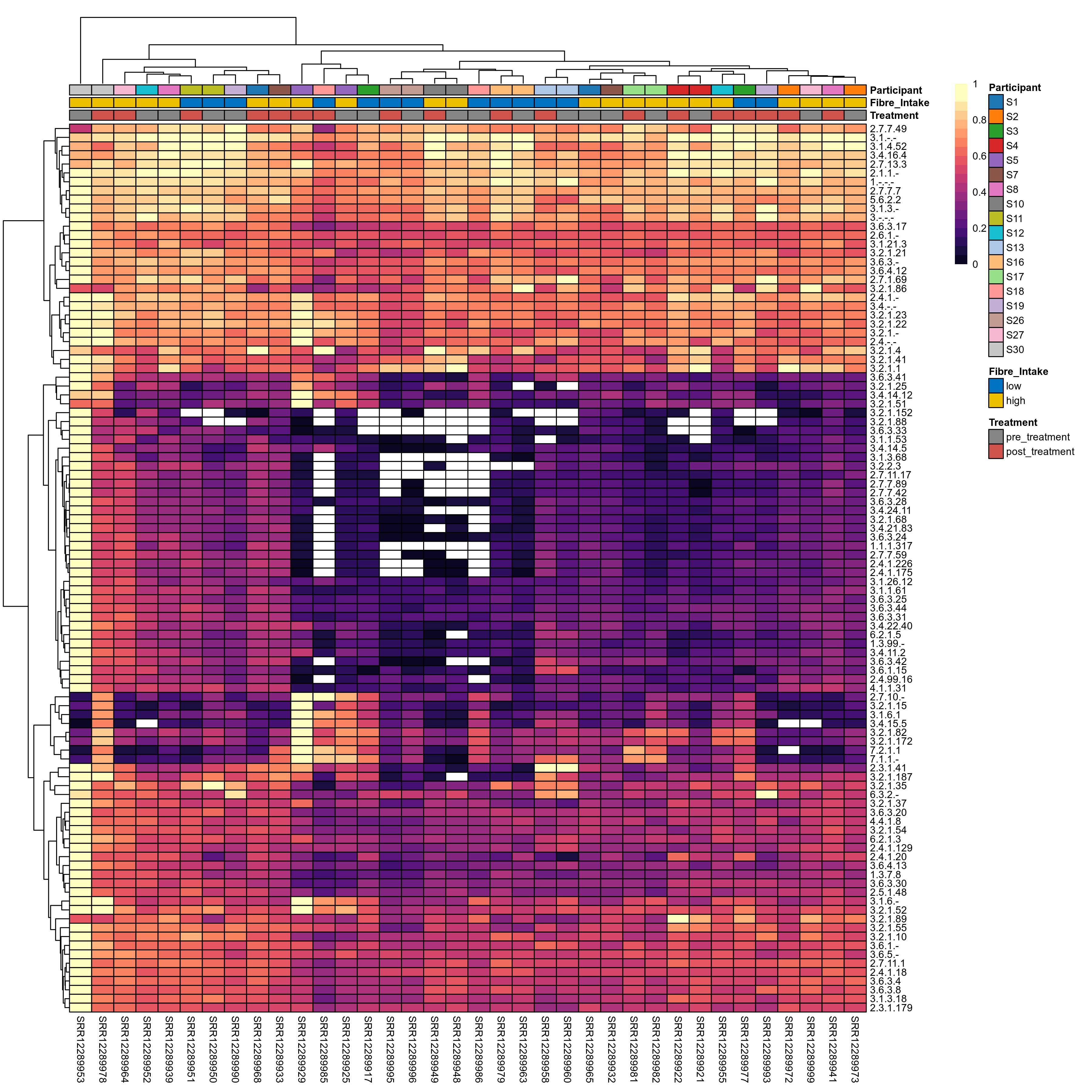
Quantitative visualisation of the top most abundant microbial functions identified in the analysed samples.
Click here to open full-sized image in new window.
Click here to open full-sized image in new window.
Click here to open full-sized image in new window.
Click here to open full-sized image in new window.
Click here to open full-sized image in new window.
Click here to open full-sized image in new window.
Click here to open full-sized image in new window.
Click here to open full-sized image in new window.
Click here to open full-sized image in new window.
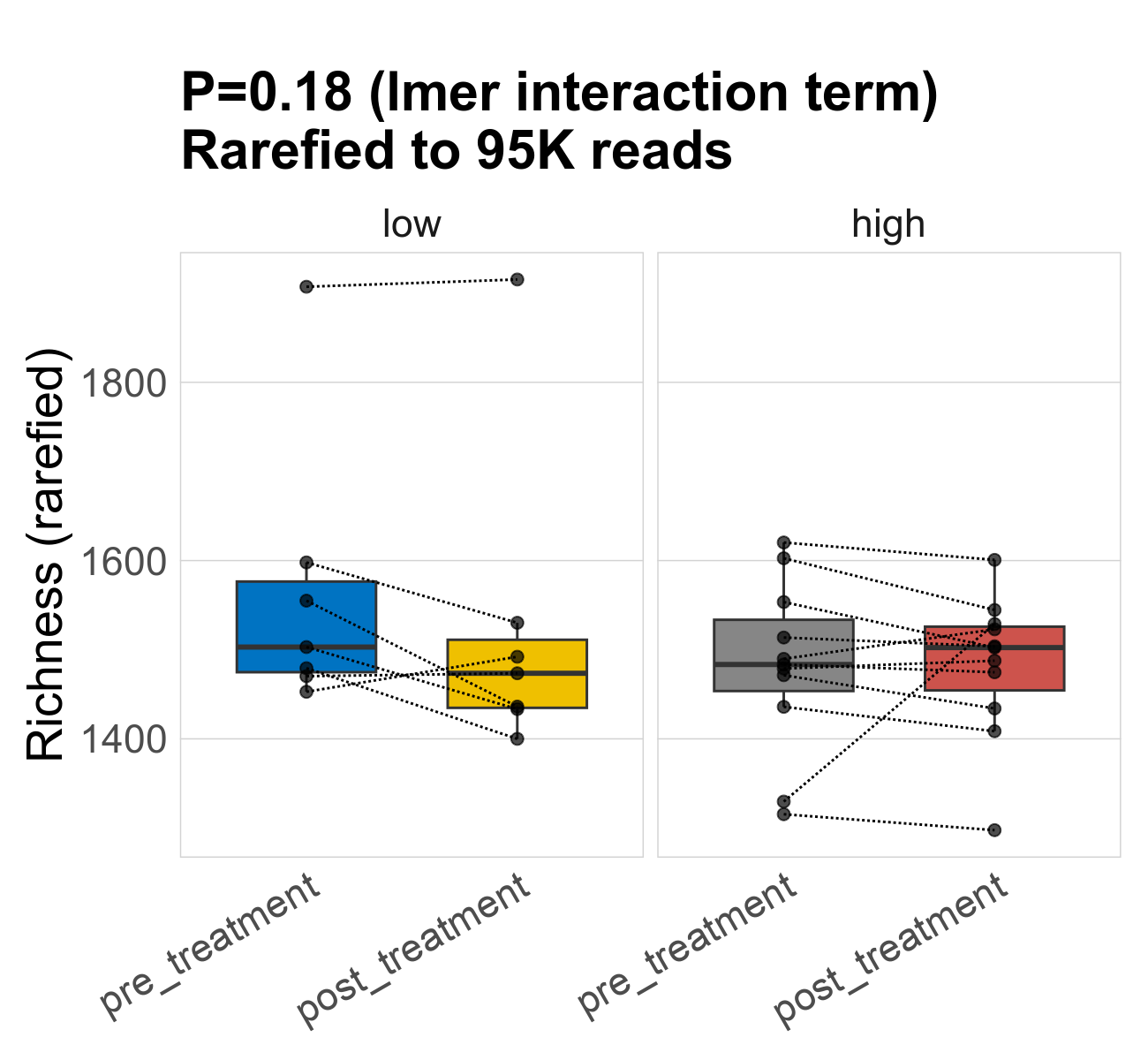
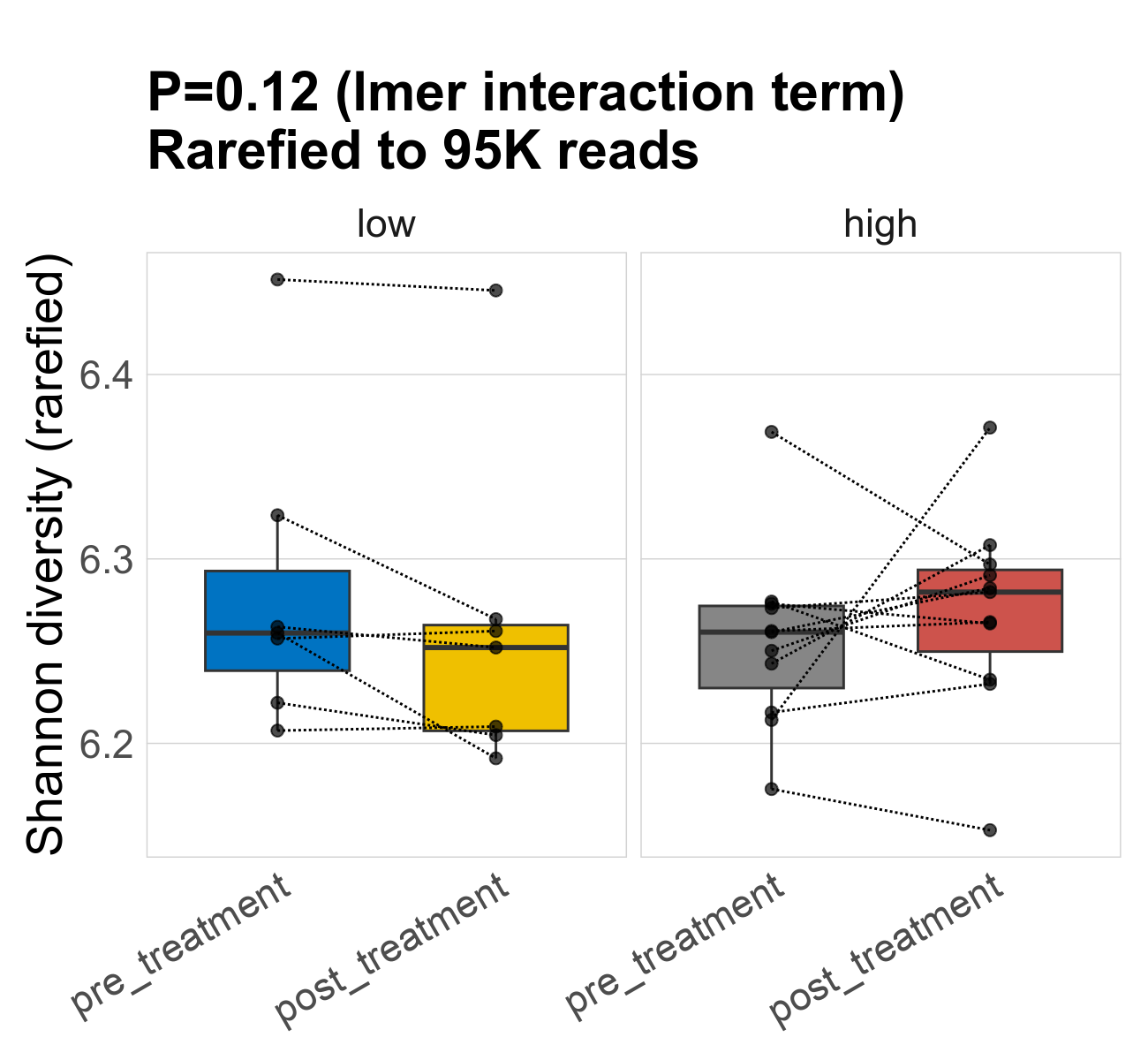
This page provides an overview of the functional alpha diversity of the analysed sample. Alpha diversity was measured using the Shannon index and functional richnes. Richness measures the total number of gene functions present in each sample. Shannon index combines richness and evenness.
| Index | rarefiedTo | P lmer treatment effect (interaction) | Mean Pos | Mean Abundance | Median Abundance | Mean Fibre_Intakelow:Treatmentpre_treatment | Median Fibre_Intakelow:Treatmentpre_treatment | SD Fibre_Intakelow:Treatmentpre_treatment | Mean Fibre_Intakelow:Treatmentpost_treatment | Median Fibre_Intakelow:Treatmentpost_treatment | SD Fibre_Intakelow:Treatmentpost_treatment | Fold Change Log2(Fibre_Intakelow:Treatmentpost_treatment/Fibre_Intakelow:Treatmentpre_treatment) | Positive samples | Positive Fibre_Intakelow:Treatmentpre_treatment | Positive Fibre_Intakelow:Treatmentpost_treatment | Positive_Fibre_Intakelow:Treatmentpre_treatment_percent | Positive_Fibre_Intakelow:Treatmentpost_treatment_percent | Mean Fibre_Intakehigh:Treatmentpre_treatment | Median Fibre_Intakehigh:Treatmentpre_treatment | SD Fibre_Intakehigh:Treatmentpre_treatment | Mean Fibre_Intakehigh:Treatmentpost_treatment | Median Fibre_Intakehigh:Treatmentpost_treatment | SD Fibre_Intakehigh:Treatmentpost_treatment | Fold Change Log2(Fibre_Intakehigh:Treatmentpost_treatment/Fibre_Intakehigh:Treatmentpre_treatment) | Positive Fibre_Intakehigh:Treatmentpre_treatment | Positive Fibre_Intakehigh:Treatmentpost_treatment | Positive_Fibre_Intakehigh:Treatmentpre_treatment_percent | Positive_Fibre_Intakehigh:Treatmentpost_treatment_percent |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Shannon | 95287 | 0.12 | 6.3 | 6.3 | 6.3 | 6.3 | 6.3 | 0.083 | 6.3 | 6.3 | 0.086 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 6.3 | 6.3 | 0.049 | 6.3 | 6.3 | 0.054 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| Richness | 95287 | 0.18 | 1500 | 1500 | 1500 | 1600 | 1500 | 160 | 1500 | 1500 | 180 | -0.093 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 1500 | 1500 | 96 | 1500 | 1500 | 80 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
Microbial functions were analyzed using supervised and unsupervised multivariate techniques. Functional profiles were ordinated using the unsupervised methods Principal Coordinates Analysis (PCoA), Non-Metric Multidimensional Scaling (NMDS) and Principal Component Analysis (PCA). The supervised methods Adonis and Redundancy analysis (RDA) were used to assess if variance in the functional profiles was significantly associated with the study condition. A guide explaining these methods can be found here. Sparse Partial Least Square Discriminant Analysis (sPLS-DA) from the MixMc package was additionaly used to extract features associated with the condition of interest.
Differentially abundant microbial functions were identified using the univariate methods ANOVA or LMER (linear mixed effect regression) on clr transformed relative abundance data, Fisher's exact test, and Aldex2 (ANOVA-like Differential Expression). Aldex2 was run on read count data. Fisher's exact test was used to test for diffrerences in the presence and absence (detection rate) of microbial functions across study groups.
LMER is used for repeated measures data, using random effects to control for correlation between samples from the same subject. Fixed effects are included for treatment groups, time, and treatment over time, where appropriate. The LMER P values correspond to a nested model test of the significance of including the corresponding fixed effect.
ALDEx2 uses subsampling (Bayesian sampling) to estimate the underlying technical variation. For each subsample instance, center log-ratio transformed data is statistically compared across study groups and computed P values are corrected for multiple testing using the Benjamini–Hochberg procedure. The expected P value (mean P value) is reported, which are those that would likely have been observed if the same samples had been run multiple times. The expected values are reported for both the distribution of P values and for the distribution of Benjamini–Hochberg corrected values.
| Function | Annotation | P lmer study group effect (sqrt) | FDR lmer study group effect (sqrt) | Pbonf lmer study group effect (sqrt) | P lmer time effect (sqrt) | FDR lmer time effect (sqrt) | Pbonf lmer time effect (sqrt) | P lmer treatment effect (interaction) (sqrt) | FDR lmer treatment effect (interaction) (sqrt) | Pbonf lmer treatment effect (interaction) (sqrt) | P lmer study group effect (clr) | FDR lmer study group effect (clr) | Pbonf lmer study group effect (clr) | P lmer time effect (clr) | FDR lmer time effect (clr) | Pbonf lmer time effect (clr) | P lmer treatment effect (interaction) (clr) | FDR lmer treatment effect (interaction) (clr) | Pbonf lmer treatment effect (interaction) (clr) | Mean Pos | Mean Abundance | Median Abundance | Mean Fibre_Intakelow:Treatmentpre_treatment | Median Fibre_Intakelow:Treatmentpre_treatment | SD Fibre_Intakelow:Treatmentpre_treatment | Mean Fibre_Intakelow:Treatmentpost_treatment | Median Fibre_Intakelow:Treatmentpost_treatment | SD Fibre_Intakelow:Treatmentpost_treatment | Fold Change Log2(Fibre_Intakelow:Treatmentpost_treatment/Fibre_Intakelow:Treatmentpre_treatment) | Positive samples | Positive Fibre_Intakelow:Treatmentpre_treatment | Positive Fibre_Intakelow:Treatmentpost_treatment | Positive_Fibre_Intakelow:Treatmentpre_treatment_percent | Positive_Fibre_Intakelow:Treatmentpost_treatment_percent | Mean Fibre_Intakehigh:Treatmentpre_treatment | Median Fibre_Intakehigh:Treatmentpre_treatment | SD Fibre_Intakehigh:Treatmentpre_treatment | Mean Fibre_Intakehigh:Treatmentpost_treatment | Median Fibre_Intakehigh:Treatmentpost_treatment | SD Fibre_Intakehigh:Treatmentpost_treatment | Fold Change Log2(Fibre_Intakehigh:Treatmentpost_treatment/Fibre_Intakehigh:Treatmentpre_treatment) | Positive Fibre_Intakehigh:Treatmentpre_treatment | Positive Fibre_Intakehigh:Treatmentpost_treatment | Positive_Fibre_Intakehigh:Treatmentpre_treatment_percent | Positive_Fibre_Intakehigh:Treatmentpost_treatment_percent |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2.7.1.73 | Inosine kinase. | 0.11 | 0.5 | 1 | 0.87 | 1 | 1 | 0.56 | 0.96 | 1 | 0.11 | 0.47 | 1 | 0.45 | 0.92 | 1 | 0.91 | 0.98 | 1 | 5e-04 | 7e-05 | 0 | 2e-04 | 0 | 0.00053 | 0.00014 | 0 | 0.00033 | -0.51 | 5 / 36 (14%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0 | 0 | 0 | 8.1e-06 | 0 | 2.7e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 6.3.4.3 | Formate--tetrahydrofolate ligase. | 0.079 | 0.49 | 1 | 0.62 | 1 | 1 | 1 | 1 | 1 | 0.14 | 0.47 | 1 | 0.44 | 0.91 | 1 | 0.19 | 0.72 | 1 | 0.027 | 0.027 | 0.026 | 0.025 | 0.025 | 0.0042 | 0.024 | 0.024 | 0.0047 | -0.059 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.029 | 0.029 | 0.0064 | 0.027 | 0.027 | 0.0036 | -0.1 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.8.1.5 | Haloalkane dehalogenase. | 0.25 | 0.55 | 1 | 0.52 | 1 | 1 | 0.18 | 0.85 | 1 | 0.11 | 0.47 | 1 | 0.21 | 0.87 | 1 | 0.037 | 0.63 | 1 | 0.00066 | 0.00052 | 0.00029 | 0.00039 | 0.00028 | 0.00051 | 4e-04 | 0.00025 | 0.00039 | 0.037 | 28 / 36 (78%) | 4 / 7 (57%) | 6 / 7 (86%) | 0.571 | 0.857 | 0.00065 | 0.00032 | 0.00083 | 0.00053 | 0.00023 | 0.00064 | -0.29 | 10 / 11 (91%) | 8 / 11 (73%) | 0.909 | 0.727 |
| 1.3.5.4 | Fumarate reductase (quinol). | 0.39 | 0.65 | 1 | 0.99 | 1 | 1 | 0.62 | 0.96 | 1 | 0.28 | 0.57 | 1 | 0.84 | 0.98 | 1 | 0.73 | 0.94 | 1 | 0.0017 | 0.0017 | 0.00075 | 0.0017 | 0.0011 | 0.0013 | 0.0021 | 0.0012 | 0.0024 | 0.3 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0012 | 0.00039 | 0.0018 | 0.002 | 0.00071 | 0.0033 | 0.74 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.4.16 | Thiamine-phosphate kinase. | 0.32 | 0.6 | 1 | 0.66 | 1 | 1 | 0.18 | 0.86 | 1 | 0.11 | 0.47 | 1 | 0.57 | 0.93 | 1 | 0.18 | 0.72 | 1 | 0.0016 | 0.0015 | 0.00095 | 0.0014 | 0.00099 | 0.00081 | 0.0015 | 0.0015 | 0.0017 | 0.1 | 33 / 36 (92%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0012 | 4e-04 | 0.0016 | 0.0017 | 0.0013 | 0.0021 | 0.5 | 9 / 11 (82%) | 10 / 11 (91%) | 0.818 | 0.909 |
| 2.1.1.63 | Methylated-DNA--[protein]-cysteine S-methyltransferase. | 0.03 | 0.49 | 1 | 0.37 | 1 | 1 | 0.48 | 0.93 | 1 | 0.018 | 0.42 | 1 | 0.08 | 0.86 | 1 | 0.12 | 0.66 | 1 | 0.01 | 0.01 | 0.0098 | 0.0086 | 0.0084 | 0.0012 | 0.0094 | 0.0099 | 0.0024 | 0.13 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.0098 | 0.0028 | 0.011 | 0.01 | 0.001 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.6.3.2 | Transferred entry: 7.2.2.14. | 0.58 | 0.79 | 1 | 0.83 | 1 | 1 | 0.44 | 0.93 | 1 | 0.52 | 0.76 | 1 | 0.54 | 0.93 | 1 | 0.35 | 0.81 | 1 | 0.0026 | 0.0026 | 0.0022 | 0.0029 | 0.0026 | 0.0014 | 0.003 | 0.0031 | 0.0013 | 0.049 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0028 | 0.0016 | 0.0027 | 0.002 | 0.0019 | 0.0011 | -0.49 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.1.187 | Acetyl-S-ACP:malonate ACP transferase. | 0.98 | 1 | 1 | 0.34 | 1 | 1 | 0.58 | 0.96 | 1 | 0.99 | 1 | 1 | 0.079 | 0.86 | 1 | 0.29 | 0.78 | 1 | 0.00024 | 9.4e-05 | 0 | 8.1e-05 | 0 | 0.00014 | 7.2e-05 | 6.8e-05 | 5.9e-05 | -0.17 | 14 / 36 (39%) | 2 / 7 (29%) | 5 / 7 (71%) | 0.286 | 0.714 | 0.00012 | 0 | 0.00031 | 9.3e-05 | 0 | 0.00017 | -0.37 | 3 / 11 (27%) | 4 / 11 (36%) | 0.273 | 0.364 |
| 3.2.1.143 | Poly(ADP-ribose) glycohydrolase. | 0.19 | 0.53 | 1 | 0.83 | 1 | 1 | 0.48 | 0.93 | 1 | 0.19 | 0.5 | 1 | 0.63 | 0.95 | 1 | 0.62 | 0.92 | 1 | 0.0013 | 0.00021 | 0 | 1.6e-05 | 0 | 4.2e-05 | 0 | 0 | 0 | -Inf | 6 / 36 (17%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 0.00065 | 0 | 0.002 | 2.7e-05 | 0 | 7e-05 | -4.6 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 5.3.3.10 | 5-carboxymethyl-2-hydroxymuconate Delta-isomerase. | 0.12 | 0.5 | 1 | 0.88 | 1 | 1 | 0.43 | 0.93 | 1 | 0.19 | 0.5 | 1 | 0.47 | 0.92 | 1 | 0.64 | 0.92 | 1 | 0.002 | 0.0015 | 0.00046 | 0.00048 | 0.00024 | 0.00067 | 0.00079 | 0.00041 | 0.0014 | 0.72 | 28 / 36 (78%) | 6 / 7 (86%) | 4 / 7 (57%) | 0.857 | 0.571 | 0.0026 | 0.0011 | 0.0053 | 0.0016 | 0.0011 | 0.0019 | -0.7 | 10 / 11 (91%) | 8 / 11 (73%) | 0.909 | 0.727 |
| 2.4.1.4 | Amylosucrase. | 0.34 | 0.61 | 1 | 0.82 | 1 | 1 | 0.85 | 1 | 1 | 0.86 | 0.95 | 1 | 0.34 | 0.9 | 1 | 0.3 | 0.78 | 1 | 0.011 | 0.011 | 0.0098 | 0.012 | 0.013 | 0.0049 | 0.012 | 0.0098 | 0.0054 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0099 | 0.0092 | 0.0029 | 0.0099 | 0.0098 | 0.003 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
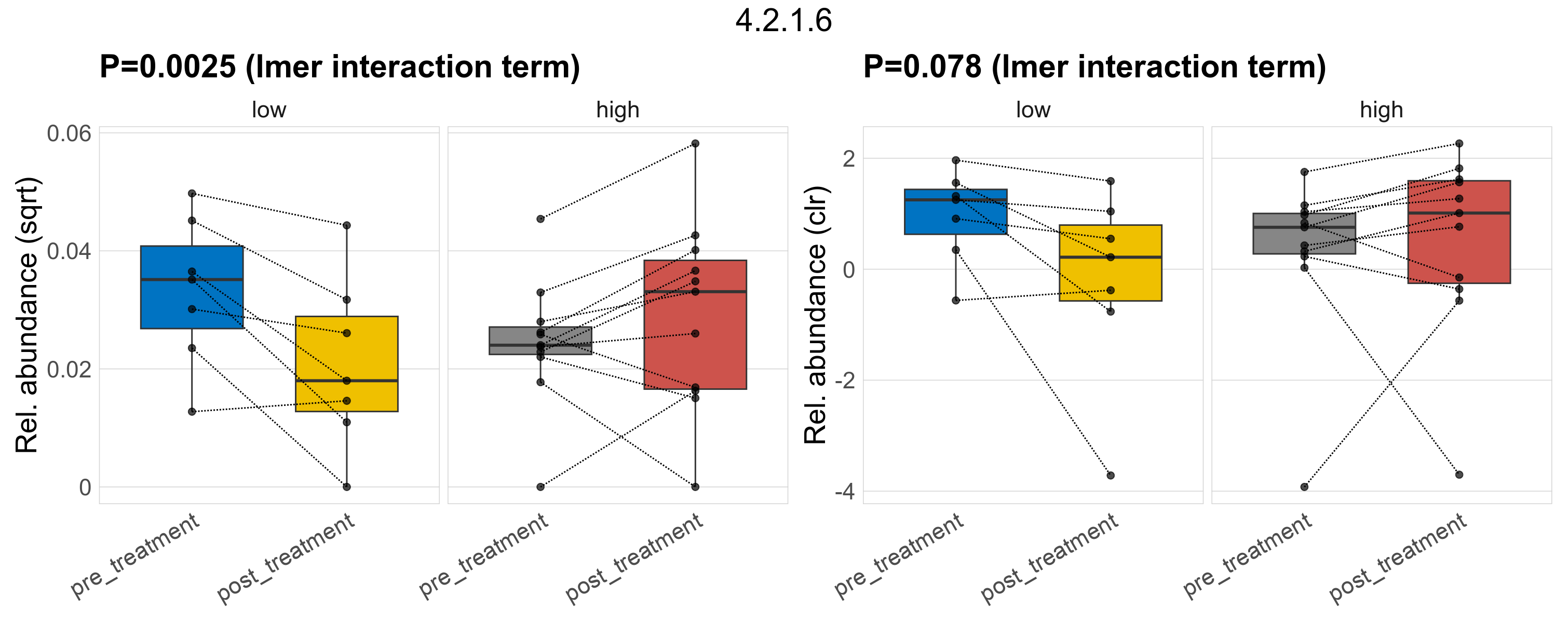
| 4.2.1.10 | 3-dehydroquinate dehydratase. | 0.68 | 0.85 | 1 | 0.41 | 1 | 1 | 0.4 | 0.92 | 1 | 0.39 | 0.66 | 1 | 0.91 | 0.99 | 1 | 0.83 | 0.96 | 1 | 0.013 | 0.013 | 0.013 | 0.013 | 0.013 | 0.0022 | 0.012 | 0.012 | 0.0034 | -0.12 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.013 | 0.013 | 0.0023 | 0.014 | 0.014 | 0.0026 | 0.11 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
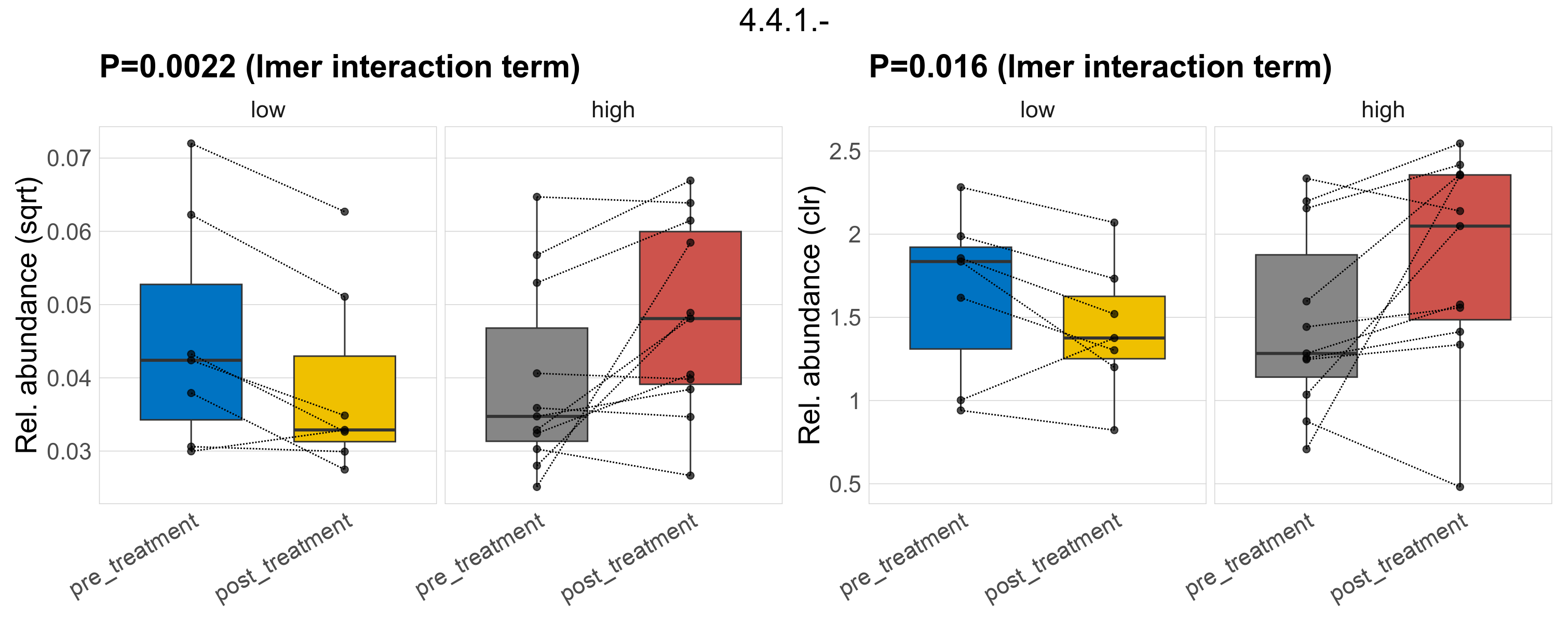
| 5.4.2.- | 0.77 | 0.89 | 1 | 0.83 | 1 | 1 | 0.48 | 0.93 | 1 | 0.46 | 0.71 | 1 | 0.63 | 0.95 | 1 | 0.94 | 0.98 | 1 | 0.0092 | 0.0092 | 0.0085 | 0.0086 | 0.0091 | 0.0029 | 0.0084 | 0.0084 | 0.0034 | -0.034 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0092 | 0.0075 | 0.0045 | 0.0099 | 0.011 | 0.0033 | 0.11 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 3.2.1.26 | Beta-fructofuranosidase. | 0.71 | 0.86 | 1 | 0.71 | 1 | 1 | 0.29 | 0.89 | 1 | 0.51 | 0.75 | 1 | 0.37 | 0.9 | 1 | 0.12 | 0.66 | 1 | 0.034 | 0.034 | 0.031 | 0.033 | 0.029 | 0.0094 | 0.036 | 0.04 | 0.016 | 0.13 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.036 | 0.037 | 0.017 | 0.031 | 0.031 | 0.0065 | -0.22 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.3.21 | Glycerol-3-phosphate oxidase. | 0.53 | 0.75 | 1 | 0.67 | 1 | 1 | 0.53 | 0.95 | 1 | 0.65 | 0.85 | 1 | 0.58 | 0.93 | 1 | 0.52 | 0.89 | 1 | 3e-04 | 0.00016 | 4.6e-05 | 0.00022 | 6.4e-05 | 0.00038 | 0.00026 | 0.00012 | 0.00049 | 0.24 | 19 / 36 (53%) | 4 / 7 (57%) | 5 / 7 (71%) | 0.571 | 0.714 | 0.00013 | 0 | 0.00023 | 8e-05 | 0 | 0.00014 | -0.7 | 5 / 11 (45%) | 5 / 11 (45%) | 0.455 | 0.455 |
| 1.3.5.2 | Dihydroorotate dehydrogenase (quinone). | 0.13 | 0.5 | 1 | 0.78 | 1 | 1 | 0.57 | 0.96 | 1 | 0.081 | 0.44 | 1 | 0.62 | 0.95 | 1 | 0.42 | 0.85 | 1 | 0.0029 | 0.0029 | 0.0021 | 0.0014 | 0.0012 | 0.001 | 0.0017 | 0.0016 | 0.0012 | 0.28 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0044 | 0.0025 | 0.0072 | 0.0032 | 0.0021 | 0.0029 | -0.46 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.1.207 | tRNA (cytidine(34)-2'-O)-methyltransferase. | 0.019 | 0.49 | 1 | 0.87 | 1 | 1 | 0.26 | 0.89 | 1 | 0.029 | 0.43 | 1 | 0.35 | 0.9 | 1 | 0.066 | 0.66 | 1 | 0.0096 | 0.0096 | 0.0092 | 0.0081 | 0.0081 | 0.00091 | 0.0083 | 0.0079 | 0.0023 | 0.035 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.0099 | 0.0037 | 0.0099 | 0.0098 | 0.0023 | -0.15 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.2.1.3 | Long-chain-fatty-acid--CoA ligase. | 0.49 | 0.72 | 1 | 0.69 | 1 | 1 | 0.21 | 0.87 | 1 | 0.22 | 0.52 | 1 | 0.96 | 0.99 | 1 | 0.25 | 0.76 | 1 | 0.033 | 0.033 | 0.027 | 0.027 | 0.024 | 0.0094 | 0.026 | 0.024 | 0.011 | -0.054 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.036 | 0.025 | 0.034 | 0.039 | 0.038 | 0.017 | 0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.2.1 | Purine-nucleoside phosphorylase. | 0.19 | 0.53 | 1 | 0.76 | 1 | 1 | 0.59 | 0.96 | 1 | 0.74 | 0.9 | 1 | 0.56 | 0.93 | 1 | 0.83 | 0.96 | 1 | 0.015 | 0.015 | 0.015 | 0.016 | 0.016 | 0.0035 | 0.016 | 0.015 | 0.0024 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.014 | 0.015 | 0.004 | 0.015 | 0.014 | 0.0022 | 0.1 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.45 | 2-dehydro-3-deoxygluconokinase. | 0.074 | 0.49 | 1 | 0.24 | 1 | 1 | 0.031 | 0.71 | 1 | 0.15 | 0.48 | 1 | 0.39 | 0.9 | 1 | 0.082 | 0.66 | 1 | 0.0066 | 0.0066 | 0.0065 | 0.0076 | 0.0078 | 0.0027 | 0.0058 | 0.0064 | 0.0027 | -0.39 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0051 | 0.0043 | 0.0022 | 0.0079 | 0.0086 | 0.0039 | 0.63 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.1.246 | Mannosylfructose-phosphate synthase. | 0.22 | 0.54 | 1 | 0.56 | 1 | 1 | 0.42 | 0.92 | 1 | 0.32 | 0.59 | 1 | 0.84 | 0.98 | 1 | 0.9 | 0.98 | 1 | 0.002 | 0.002 | 0.0017 | 0.0023 | 0.0018 | 0.0013 | 0.0021 | 0.0016 | 0.0011 | -0.13 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0017 | 0.0018 | 0.00083 | 0.002 | 0.0015 | 0.0015 | 0.23 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.8.40 | UDP-N-acetylgalactosamine-undecaprenyl-phosphate | 0.17 | 0.5 | 1 | 0.037 | 0.97 | 1 | 0.2 | 0.87 | 1 | 0.16 | 0.48 | 1 | 0.056 | 0.82 | 1 | 0.27 | 0.77 | 1 | 0.00024 | 9.4e-05 | 0 | 0.00017 | 9.6e-05 | 2e-04 | 7.6e-05 | 0 | 0.00014 | -1.2 | 14 / 36 (39%) | 5 / 7 (71%) | 2 / 7 (29%) | 0.714 | 0.286 | 8.6e-05 | 0 | 0.00016 | 6.6e-05 | 0 | 0.00017 | -0.38 | 4 / 11 (36%) | 3 / 11 (27%) | 0.364 | 0.273 |
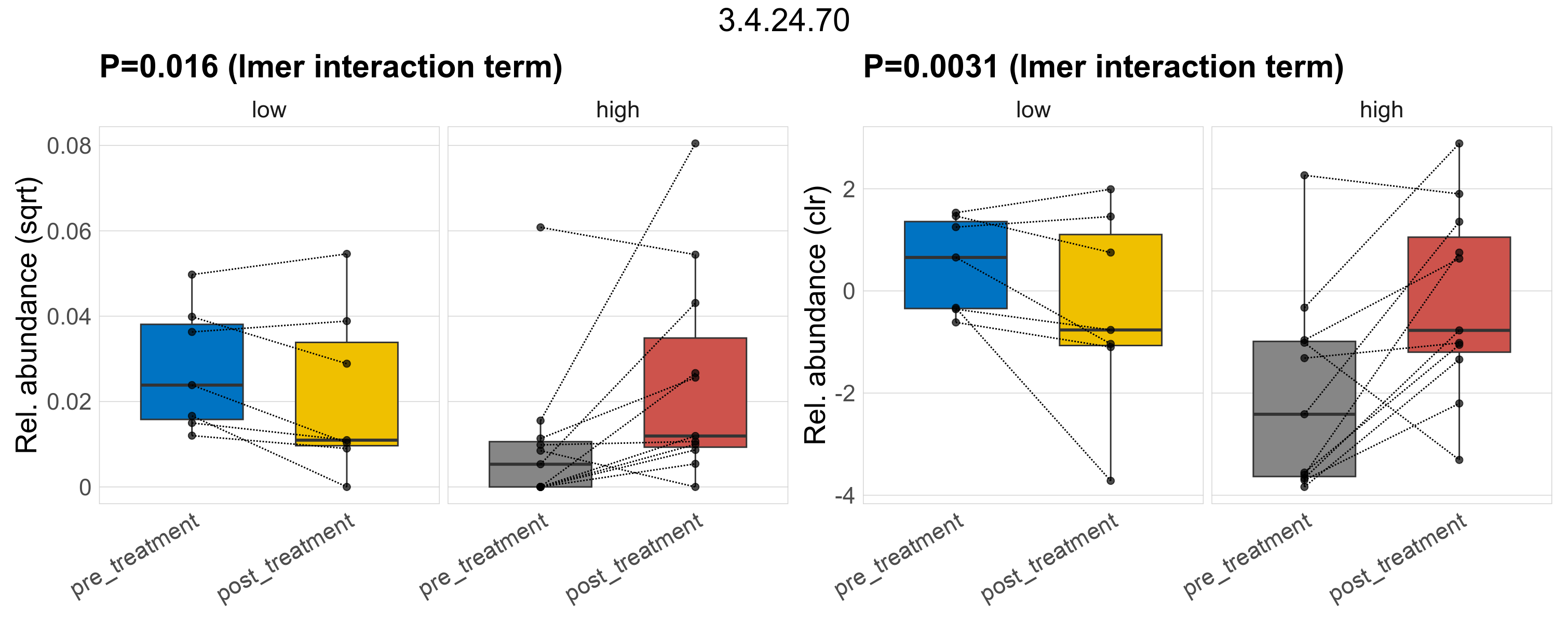
| 3.4.24.64 | Mitochondrial processing peptidase. | 0.13 | 0.5 | 1 | 0.039 | 0.97 | 1 | 0.073 | 0.73 | 1 | 0.24 | 0.53 | 1 | 0.18 | 0.87 | 1 | 0.24 | 0.76 | 1 | 0.00027 | 5.2e-05 | 0 | 0.00017 | 0 | 4e-04 | 4.8e-05 | 0 | 0.00013 | -1.8 | 7 / 36 (19%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 1.4e-05 | 0 | 3.7e-05 | 1.8e-05 | 0 | 4.1e-05 | 0.36 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 2.7.2.1 | Acetate kinase. | 0.092 | 0.49 | 1 | 0.77 | 1 | 1 | 0.91 | 1 | 1 | 0.13 | 0.47 | 1 | 0.42 | 0.91 | 1 | 0.23 | 0.75 | 1 | 0.022 | 0.022 | 0.022 | 0.02 | 0.019 | 0.0031 | 0.02 | 0.02 | 0.0039 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.023 | 0.023 | 0.0047 | 0.023 | 0.022 | 0.003 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.1.1 | Glycogen phosphorylase. | 0.4 | 0.66 | 1 | 0.71 | 1 | 1 | 0.99 | 1 | 1 | 0.29 | 0.58 | 1 | 0.32 | 0.9 | 1 | 0.15 | 0.71 | 1 | 0.073 | 0.073 | 0.075 | 0.072 | 0.07 | 0.014 | 0.07 | 0.074 | 0.017 | -0.041 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.076 | 0.076 | 0.011 | 0.074 | 0.074 | 0.0076 | -0.038 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.3.99.2 | Transferred entry: 7.2.4.1. | 0.078 | 0.49 | 1 | 0.91 | 1 | 1 | 0.42 | 0.92 | 1 | 0.083 | 0.44 | 1 | 0.51 | 0.93 | 1 | 0.89 | 0.97 | 1 | 0.00051 | 2e-04 | 0 | 0.00031 | 8.1e-05 | 0.00041 | 0.00026 | 0.00021 | 0.00024 | -0.25 | 14 / 36 (39%) | 4 / 7 (57%) | 5 / 7 (71%) | 0.571 | 0.714 | 5.4e-05 | 0 | 0.00012 | 0.00023 | 0 | 0.00045 | 2.1 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 1.6.8.- | 0.22 | 0.54 | 1 | 0.99 | 1 | 1 | 0.41 | 0.92 | 1 | 0.24 | 0.53 | 1 | 0.72 | 0.95 | 1 | 0.63 | 0.92 | 1 | 0.00036 | 3e-05 | 0 | 6.7e-05 | 0 | 0.00018 | 6.8e-05 | 0 | 0.00018 | 0.021 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 1.3e-05 | 0 | 4.5e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 | |
| 1.5.1.30 | Flavin reductase (NADPH). | 0.14 | 0.5 | 1 | 0.54 | 1 | 1 | 0.68 | 0.98 | 1 | 0.13 | 0.47 | 1 | 0.51 | 0.93 | 1 | 0.75 | 0.95 | 1 | 0.00023 | 3.8e-05 | 0 | 7.2e-05 | 0 | 0.00018 | 9.4e-05 | 0 | 2e-04 | 0.38 | 6 / 36 (17%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0 | 0 | 0 | 1.8e-05 | 0 | 4e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 1.2.1.8 | Betaine-aldehyde dehydrogenase. | 0.77 | 0.89 | 1 | 0.91 | 1 | 1 | 0.99 | 1 | 1 | 0.74 | 0.9 | 1 | 0.72 | 0.95 | 1 | 0.99 | 1 | 1 | 7e-04 | 0.00053 | 0.00022 | 0.00054 | 0.00028 | 0.00062 | 0.00056 | 0.00064 | 0.00056 | 0.052 | 27 / 36 (75%) | 6 / 7 (86%) | 5 / 7 (71%) | 0.857 | 0.714 | 0.00046 | 0.00049 | 0.00047 | 0.00057 | 0.00011 | 0.001 | 0.31 | 8 / 11 (73%) | 8 / 11 (73%) | 0.727 | 0.727 |
| 3.2.2.23 | DNA-formamidopyrimidine glycosylase. | 0.12 | 0.5 | 1 | 0.27 | 1 | 1 | 0.28 | 0.89 | 1 | 0.23 | 0.53 | 1 | 0.37 | 0.9 | 1 | 0.34 | 0.8 | 1 | 0.001 | 0.001 | 0.00083 | 0.0014 | 0.00094 | 0.00094 | 0.0011 | 0.00084 | 0.00079 | -0.35 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00087 | 0.00072 | 0.00054 | 0.00089 | 0.00087 | 0.00032 | 0.033 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.8.1.4 | tRNA uracil 4-sulfurtransferase. | 0.58 | 0.79 | 1 | 0.96 | 1 | 1 | 0.83 | 1 | 1 | 0.44 | 0.69 | 1 | 0.42 | 0.91 | 1 | 0.26 | 0.76 | 1 | 0.017 | 0.017 | 0.018 | 0.016 | 0.016 | 0.0026 | 0.016 | 0.018 | 0.0044 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.017 | 0.018 | 0.0039 | 0.017 | 0.018 | 0.0038 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.14.14.51 | (S)-limonene 6-monooxygenase. | 0.12 | 0.5 | 1 | 0.38 | 1 | 1 | 0.62 | 0.96 | 1 | 0.12 | 0.47 | 1 | 0.46 | 0.92 | 1 | 0.67 | 0.93 | 1 | 0.00035 | 9.6e-05 | 0 | 0.00023 | 2.8e-05 | 0.00048 | 0.00016 | 0 | 0.00033 | -0.52 | 10 / 36 (28%) | 4 / 7 (57%) | 2 / 7 (29%) | 0.571 | 0.286 | 4.8e-05 | 0 | 0.00012 | 2.3e-05 | 0 | 5.6e-05 | -1.1 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 4.2.2.23 | Rhamnogalacturonan endolyase. | 0.7 | 0.86 | 1 | 0.85 | 1 | 1 | 0.37 | 0.9 | 1 | 0.64 | 0.84 | 1 | 0.98 | 0.99 | 1 | 0.47 | 0.87 | 1 | 0.0024 | 0.0023 | 0.0011 | 0.0022 | 0.0011 | 0.0023 | 0.002 | 0.0011 | 0.002 | -0.14 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0018 | 0.0011 | 0.002 | 0.0032 | 9e-04 | 0.0053 | 0.83 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 2.3.1.210 | dTDP-4-amino-4.6-dideoxy-D-galactose acyltransferase. | 0.091 | 0.49 | 1 | 0.13 | 1 | 1 | 0.59 | 0.96 | 1 | 0.22 | 0.52 | 1 | 0.42 | 0.91 | 1 | 0.75 | 0.95 | 1 | 0.00028 | 5.5e-05 | 0 | 0.00019 | 0 | 0.00033 | 7.2e-05 | 0 | 0.00018 | -1.4 | 7 / 36 (19%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 1.4e-05 | 0 | 3e-05 | 0 | 0 | 0 | -Inf | 3 / 11 (27%) | 0 / 11 (0%) | 0.273 | 0 |
| 2.5.1.90 | All-trans-octaprenyl-diphosphate synthase. | 0.38 | 0.65 | 1 | 0.98 | 1 | 1 | 0.85 | 1 | 1 | 0.42 | 0.69 | 1 | 0.65 | 0.95 | 1 | 0.77 | 0.95 | 1 | 0.0035 | 0.0035 | 0.0022 | 0.0023 | 0.0024 | 0.0013 | 0.0023 | 0.002 | 0.0013 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0045 | 0.0022 | 0.0069 | 0.004 | 0.003 | 0.003 | -0.17 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.6.5 | GTP diphosphokinase. | 0.38 | 0.65 | 1 | 0.57 | 1 | 1 | 0.58 | 0.96 | 1 | 0.26 | 0.55 | 1 | 0.57 | 0.93 | 1 | 0.55 | 0.9 | 1 | 0.052 | 0.052 | 0.054 | 0.049 | 0.052 | 0.013 | 0.048 | 0.054 | 0.016 | -0.03 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.054 | 0.057 | 0.013 | 0.054 | 0.054 | 0.0084 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.3.2.9 | UDP-N-acetylmuramoyl-L-alanine--D-glutamate ligase. | 0.14 | 0.5 | 1 | 0.81 | 1 | 1 | 0.9 | 1 | 1 | 0.13 | 0.47 | 1 | 0.37 | 0.9 | 1 | 0.32 | 0.79 | 1 | 0.023 | 0.023 | 0.022 | 0.021 | 0.019 | 0.0041 | 0.021 | 0.018 | 0.0048 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.024 | 0.022 | 0.0062 | 0.024 | 0.023 | 0.0041 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.3.3 | Citrate (Re)-synthase. | 0.11 | 0.5 | 1 | 0.52 | 1 | 1 | 0.51 | 0.95 | 1 | 0.12 | 0.47 | 1 | 0.61 | 0.95 | 1 | 0.5 | 0.88 | 1 | 0.00012 | 2.3e-05 | 0 | 1.1e-05 | 0 | 3e-05 | 0 | 0 | 0 | -Inf | 7 / 36 (19%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 5.6e-05 | 0 | 8.8e-05 | 1.2e-05 | 0 | 2.7e-05 | -2.2 | 4 / 11 (36%) | 2 / 11 (18%) | 0.364 | 0.182 |
| 3.3.1.- | 0.57 | 0.79 | 1 | 0.58 | 1 | 1 | 0.6 | 0.96 | 1 | 0.77 | 0.92 | 1 | 0.88 | 0.99 | 1 | 0.9 | 0.98 | 1 | 0.00036 | 4e-05 | 0 | 7.3e-05 | 0 | 0.00019 | 2.2e-05 | 0 | 5.8e-05 | -1.7 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.9e-05 | 0 | 9.5e-05 | 4.1e-05 | 0 | 0.00014 | 0.5 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 | |
| 3.6.1.41 | Bis(5'-nucleosyl)-tetraphosphatase (symmetrical). | 0.035 | 0.49 | 1 | 0.94 | 1 | 1 | 0.23 | 0.88 | 1 | 0.014 | 0.42 | 1 | 0.67 | 0.95 | 1 | 0.31 | 0.79 | 1 | 0.00062 | 0.00059 | 5e-04 | 0.00047 | 0.00034 | 0.00064 | 0.00042 | 0.00036 | 0.00033 | -0.16 | 34 / 36 (94%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00085 | 0.00065 | 0.00053 | 5e-04 | 5e-04 | 0.00022 | -0.77 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.4.2.8 | Phosphomannomutase. | 0.99 | 1 | 1 | 0.9 | 1 | 1 | 0.43 | 0.93 | 1 | 0.57 | 0.79 | 1 | 0.16 | 0.87 | 1 | 0.52 | 0.9 | 1 | 0.021 | 0.021 | 0.02 | 0.02 | 0.02 | 0.0074 | 0.02 | 0.019 | 0.0082 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.02 | 0.02 | 0.0044 | 0.022 | 0.022 | 0.0061 | 0.14 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.2.1.76 | Succinate-semialdehyde dehydrogenase (acetylating). | 0.31 | 0.59 | 1 | 0.9 | 1 | 1 | 0.48 | 0.93 | 1 | 0.89 | 0.96 | 1 | 0.48 | 0.92 | 1 | 0.18 | 0.72 | 1 | 0.0041 | 0.0041 | 0.0038 | 0.0051 | 0.0046 | 0.0027 | 0.0051 | 0.0041 | 0.0029 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0038 | 0.0039 | 0.0011 | 0.0032 | 0.0031 | 0.0011 | -0.25 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.208 | Protein-N(pi)-phosphohistidine--maltose phosphotransferase. | 0.5 | 0.73 | 1 | 0.44 | 1 | 1 | 0.32 | 0.89 | 1 | 0.68 | 0.87 | 1 | 0.75 | 0.97 | 1 | 0.68 | 0.93 | 1 | 0.00026 | 5e-05 | 0 | 8.6e-05 | 0 | 0.00022 | 7.7e-05 | 0 | 2e-04 | -0.16 | 7 / 36 (19%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.5e-05 | 0 | 7.2e-05 | 3.6e-05 | 0 | 1e-04 | 0.53 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 1.97.-.- | 0.9 | 0.96 | 1 | 0.25 | 1 | 1 | 0.11 | 0.82 | 1 | 0.88 | 0.96 | 1 | 0.44 | 0.91 | 1 | 0.26 | 0.76 | 1 | 0.0092 | 0.0092 | 0.0087 | 0.0089 | 0.0088 | 0.0011 | 0.0079 | 0.0079 | 0.0031 | -0.17 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.009 | 0.0084 | 0.0035 | 0.01 | 0.0095 | 0.0043 | 0.15 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 2.5.1.26 | Alkylglycerone-phosphate synthase. | 0.43 | 0.69 | 1 | 0.21 | 1 | 1 | 0.093 | 0.79 | 1 | 0.42 | 0.69 | 1 | 0.17 | 0.87 | 1 | 0.23 | 0.75 | 1 | 0.00061 | 0.00042 | 0.00013 | 0.00047 | 0.00021 | 0.00057 | 0.00025 | 0.00012 | 0.00035 | -0.91 | 25 / 36 (69%) | 6 / 7 (86%) | 4 / 7 (57%) | 0.857 | 0.571 | 0.00028 | 0.00011 | 0.00044 | 0.00065 | 1e-04 | 0.001 | 1.2 | 8 / 11 (73%) | 7 / 11 (64%) | 0.727 | 0.636 |
| 1.1.99.5 | Transferred entry: 1.1.5.3. | 0.48 | 0.72 | 1 | 0.48 | 1 | 1 | 0.99 | 1 | 1 | 0.58 | 0.8 | 1 | 0.61 | 0.95 | 1 | 0.94 | 0.99 | 1 | 0.00028 | 3.9e-05 | 0 | 5e-05 | 0 | 0.00013 | 0.00011 | 0 | 0.00028 | 1.1 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 5.1e-06 | 0 | 1.7e-05 | 2.3e-05 | 0 | 6.8e-05 | 2.2 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 4.2.1.112 | Acetylene hydratase. | 0.68 | 0.85 | 1 | 0.4 | 1 | 1 | 0.93 | 1 | 1 | 0.97 | 0.99 | 1 | 0.59 | 0.93 | 1 | 0.64 | 0.92 | 1 | 0.00012 | 2.3e-05 | 0 | 5.5e-05 | 0 | 0.00013 | 1.7e-05 | 0 | 4.5e-05 | -1.7 | 7 / 36 (19%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-05 | 0 | 4.6e-05 | 2.7e-06 | 0 | 8.9e-06 | -3.3 | 3 / 11 (27%) | 1 / 11 (9.1%) | 0.273 | 0.0909 |
| 2.1.1.37 | DNA (cytosine-5-)-methyltransferase. | 0.44 | 0.69 | 1 | 0.45 | 1 | 1 | 0.38 | 0.9 | 1 | 0.29 | 0.58 | 1 | 0.77 | 0.97 | 1 | 0.79 | 0.95 | 1 | 0.023 | 0.023 | 0.024 | 0.022 | 0.021 | 0.0052 | 0.021 | 0.02 | 0.0044 | -0.067 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.024 | 0.025 | 0.0047 | 0.025 | 0.025 | 0.006 | 0.059 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.21.53 | Endopeptidase La. | 0.21 | 0.53 | 1 | 0.45 | 1 | 1 | 0.94 | 1 | 1 | 0.24 | 0.53 | 1 | 0.49 | 0.92 | 1 | 0.1 | 0.66 | 1 | 0.046 | 0.046 | 0.047 | 0.044 | 0.042 | 0.0079 | 0.042 | 0.046 | 0.012 | -0.067 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.048 | 0.05 | 0.0064 | 0.046 | 0.046 | 0.0057 | -0.061 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.11.1.15 | Peroxiredoxin. | 0.47 | 0.72 | 1 | 0.83 | 1 | 1 | 0.14 | 0.83 | 1 | 0.5 | 0.74 | 1 | 0.7 | 0.95 | 1 | 0.35 | 0.81 | 1 | 0.014 | 0.014 | 0.012 | 0.014 | 0.013 | 0.0043 | 0.013 | 0.014 | 0.0041 | -0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.012 | 0.0095 | 0.0054 | 0.017 | 0.01 | 0.017 | 0.5 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.3.4.23 | Formate--phosphoribosylaminoimidazolecarboxamide ligase. | 0.62 | 0.82 | 1 | 0.59 | 1 | 1 | 0.21 | 0.87 | 1 | 0.8 | 0.92 | 1 | 0.84 | 0.98 | 1 | 0.35 | 0.81 | 1 | 0.00017 | 2.4e-05 | 0 | 2.8e-05 | 0 | 7.5e-05 | 8.8e-06 | 0 | 2.3e-05 | -1.7 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 9.5e-06 | 0 | 3.2e-05 | 4.4e-05 | 0 | 0.00011 | 2.2 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 2.7.7.85 | Diadenylate cyclase. | 0.71 | 0.86 | 1 | 0.071 | 1 | 1 | 0.13 | 0.82 | 1 | 0.81 | 0.93 | 1 | 0.41 | 0.9 | 1 | 0.57 | 0.9 | 1 | 0.014 | 0.014 | 0.014 | 0.015 | 0.014 | 0.0037 | 0.013 | 0.014 | 0.0038 | -0.21 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.014 | 0.015 | 0.0032 | 0.014 | 0.014 | 0.0021 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.5.3 | Glycerol-3-phosphate dehydrogenase. | 0.26 | 0.56 | 1 | 0.51 | 1 | 1 | 0.48 | 0.93 | 1 | 0.45 | 0.7 | 1 | 0.57 | 0.93 | 1 | 0.55 | 0.9 | 1 | 0.0016 | 0.0016 | 0.0012 | 0.0021 | 0.0011 | 0.002 | 0.0019 | 0.0012 | 0.0023 | -0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0013 | 0.0015 | 0.00081 | 0.0013 | 0.0011 | 0.00083 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.2.- | 0.004 | 0.49 | 1 | 0.63 | 1 | 1 | 0.43 | 0.93 | 1 | 0.0083 | 0.38 | 1 | 0.87 | 0.99 | 1 | 0.17 | 0.71 | 1 | 0.011 | 0.011 | 0.01 | 0.0087 | 0.0086 | 0.002 | 0.008 | 0.0081 | 0.0012 | -0.12 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.015 | 0.013 | 0.0075 | 0.012 | 0.013 | 0.0028 | -0.32 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 3.1.6.1 | Arylsulfatase (type I). | 0.15 | 0.5 | 1 | 0.71 | 1 | 1 | 0.27 | 0.89 | 1 | 0.04 | 0.43 | 1 | 0.7 | 0.95 | 1 | 0.34 | 0.8 | 1 | 0.012 | 0.012 | 0.0065 | 0.013 | 0.013 | 0.0066 | 0.013 | 0.0085 | 0.015 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0076 | 0.0032 | 0.011 | 0.015 | 0.0039 | 0.025 | 0.98 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.2.40 | Tagatose-bisphosphate aldolase. | 0.047 | 0.49 | 1 | 0.93 | 1 | 1 | 0.53 | 0.95 | 1 | 0.22 | 0.52 | 1 | 0.61 | 0.95 | 1 | 0.99 | 1 | 1 | 0.0076 | 0.0076 | 0.0072 | 0.0089 | 0.009 | 0.0034 | 0.0088 | 0.0084 | 0.0029 | -0.016 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0064 | 0.0059 | 0.0026 | 0.0071 | 0.0077 | 0.0024 | 0.15 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.1.180 | Beta-ketoacyl-[acyl-carrier-protein] synthase III. | 0.063 | 0.49 | 1 | 0.39 | 1 | 1 | 1 | 1 | 1 | 0.093 | 0.45 | 1 | 0.032 | 0.79 | 1 | 0.19 | 0.72 | 1 | 0.011 | 0.011 | 0.011 | 0.0095 | 0.0095 | 0.0016 | 0.01 | 0.011 | 0.0018 | 0.074 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.011 | 0.0022 | 0.012 | 0.012 | 0.003 | 0.13 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.3.3 | Phosphoserine phosphatase. | 0.38 | 0.65 | 1 | 0.46 | 1 | 1 | 0.17 | 0.85 | 1 | 0.27 | 0.56 | 1 | 0.21 | 0.88 | 1 | 0.07 | 0.66 | 1 | 0.011 | 0.011 | 0.011 | 0.011 | 0.01 | 0.0019 | 0.012 | 0.011 | 0.0036 | 0.13 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.012 | 0.011 | 0.0049 | 0.01 | 0.011 | 0.0021 | -0.26 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.2.1.48 | Carnitine--CoA ligase. | 0.08 | 0.49 | 1 | 0.27 | 1 | 1 | 0.27 | 0.89 | 1 | 0.051 | 0.43 | 1 | 0.25 | 0.88 | 1 | 0.29 | 0.78 | 1 | 0.00029 | 4.8e-05 | 0 | 0.00011 | 0 | 0.00022 | 0.00012 | 0 | 0.00031 | 0.13 | 6 / 36 (17%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1e-05 | 0 | 3.4e-05 | 1.9 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 4.4.1.24 | (2R)-sulfolactate sulfo-lyase. | 0.28 | 0.56 | 1 | 1 | 1 | 1 | 0.81 | 1 | 1 | 0.15 | 0.47 | 1 | 0.87 | 0.99 | 1 | 0.86 | 0.96 | 1 | 0.00027 | 3.8e-05 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 5 / 36 (14%) | 0 / 7 (0%) | 0 / 7 (0%) | 0 | 0 | 3.6e-05 | 0 | 7.3e-05 | 8.8e-05 | 0 | 0.00023 | 1.3 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 2.1.1.195 | Cobalt-precorrin-5B (C(1))-methyltransferase. | 0.82 | 0.92 | 1 | 0.33 | 1 | 1 | 0.75 | 0.99 | 1 | 0.64 | 0.84 | 1 | 0.017 | 0.76 | 1 | 0.11 | 0.66 | 1 | 0.011 | 0.011 | 0.01 | 0.01 | 0.0099 | 0.0013 | 0.011 | 0.012 | 0.0029 | 0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.01 | 0.01 | 0.0027 | 0.011 | 0.01 | 0.0017 | 0.14 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.97.1.- | 0.32 | 0.6 | 1 | 0.5 | 1 | 1 | 0.45 | 0.93 | 1 | 0.23 | 0.53 | 1 | 0.2 | 0.87 | 1 | 0.15 | 0.71 | 1 | 0.017 | 0.017 | 0.017 | 0.016 | 0.017 | 0.0034 | 0.017 | 0.014 | 0.0075 | 0.087 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.018 | 0.017 | 0.0031 | 0.017 | 0.016 | 0.0017 | -0.082 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 5.1.99.- | 0.014 | 0.49 | 1 | 0.0071 | 0.7 | 1 | 0.015 | 0.67 | 1 | 0.0066 | 0.37 | 1 | 0.029 | 0.76 | 1 | 0.042 | 0.63 | 1 | 0.00017 | 2.4e-05 | 0 | 9.8e-05 | 0 | 0.00022 | 1.9e-05 | 0 | 5.1e-05 | -2.4 | 5 / 36 (14%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 | |
| 2.2.1.7 | 1-deoxy-D-xylulose-5-phosphate synthase. | 0.31 | 0.59 | 1 | 0.38 | 1 | 1 | 0.23 | 0.88 | 1 | 0.27 | 0.56 | 1 | 0.87 | 0.99 | 1 | 0.83 | 0.96 | 1 | 0.053 | 0.053 | 0.054 | 0.049 | 0.046 | 0.01 | 0.047 | 0.052 | 0.014 | -0.06 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.055 | 0.056 | 0.013 | 0.057 | 0.055 | 0.0064 | 0.052 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.179 | Gellan tetrasaccharide unsaturated glucuronosyl hydrolase. | 0.36 | 0.63 | 1 | 0.84 | 1 | 1 | 0.31 | 0.89 | 1 | 0.22 | 0.52 | 1 | 0.68 | 0.95 | 1 | 0.31 | 0.79 | 1 | 0.0029 | 0.0029 | 0.0014 | 0.0029 | 0.0016 | 0.0026 | 0.0033 | 0.00096 | 0.0048 | 0.19 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0018 | 0.0014 | 0.0018 | 0.0037 | 0.0014 | 0.0056 | 1 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.5.98.1 | Methylenetetrahydromethanopterin dehydrogenase. | 0.48 | 0.72 | 1 | 0.31 | 1 | 1 | 0.27 | 0.89 | 1 | 0.68 | 0.87 | 1 | 0.41 | 0.9 | 1 | 0.43 | 0.85 | 1 | 0.00029 | 2.4e-05 | 0 | 4.5e-05 | 0 | 0.00012 | 0 | 0 | 0 | -Inf | 3 / 36 (8.3%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 9.5e-06 | 0 | 3.2e-05 | 4.1e-05 | 0 | 0.00014 | 2.1 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.5.1.1 | Dimethylallyltranstransferase. | 0.65 | 0.84 | 1 | 0.075 | 1 | 1 | 0.025 | 0.71 | 1 | 0.58 | 0.8 | 1 | 0.097 | 0.86 | 1 | 0.049 | 0.65 | 1 | 0.0013 | 0.0012 | 0.00091 | 0.0013 | 0.00096 | 0.00093 | 9e-04 | 0.00095 | 0.00087 | -0.53 | 34 / 36 (94%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.0012 | 8e-04 | 0.0011 | 0.0015 | 0.0012 | 0.0014 | 0.32 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 2.4.1.10 | Levansucrase. | 0.93 | 0.97 | 1 | 0.49 | 1 | 1 | 0.77 | 0.99 | 1 | 0.86 | 0.95 | 1 | 0.57 | 0.93 | 1 | 0.85 | 0.96 | 1 | 0.00043 | 0.00027 | 0.00012 | 0.00023 | 9.6e-05 | 0.00038 | 0.00035 | 0.00027 | 0.00038 | 0.61 | 23 / 36 (64%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.00024 | 0.00013 | 0.00035 | 0.00028 | 0.00014 | 0.00041 | 0.22 | 6 / 11 (55%) | 7 / 11 (64%) | 0.545 | 0.636 |
| 4.99.1.3 | Sirohydrochlorin cobaltochelatase. | 0.21 | 0.54 | 1 | 0.97 | 1 | 1 | 0.43 | 0.93 | 1 | 0.6 | 0.81 | 1 | 0.49 | 0.92 | 1 | 0.76 | 0.95 | 1 | 0.014 | 0.014 | 0.014 | 0.015 | 0.017 | 0.0049 | 0.016 | 0.015 | 0.0064 | 0.093 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.013 | 0.013 | 0.0042 | 0.014 | 0.013 | 0.0032 | 0.11 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.1.15 | Glutamate decarboxylase. | 0.17 | 0.51 | 1 | 0.44 | 1 | 1 | 0.36 | 0.9 | 1 | 0.11 | 0.47 | 1 | 0.32 | 0.9 | 1 | 0.29 | 0.78 | 1 | 0.003 | 0.003 | 0.002 | 0.0016 | 0.0013 | 0.0012 | 0.0022 | 0.0023 | 0.0015 | 0.46 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0041 | 0.0022 | 0.0063 | 0.0032 | 0.0019 | 0.0028 | -0.36 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
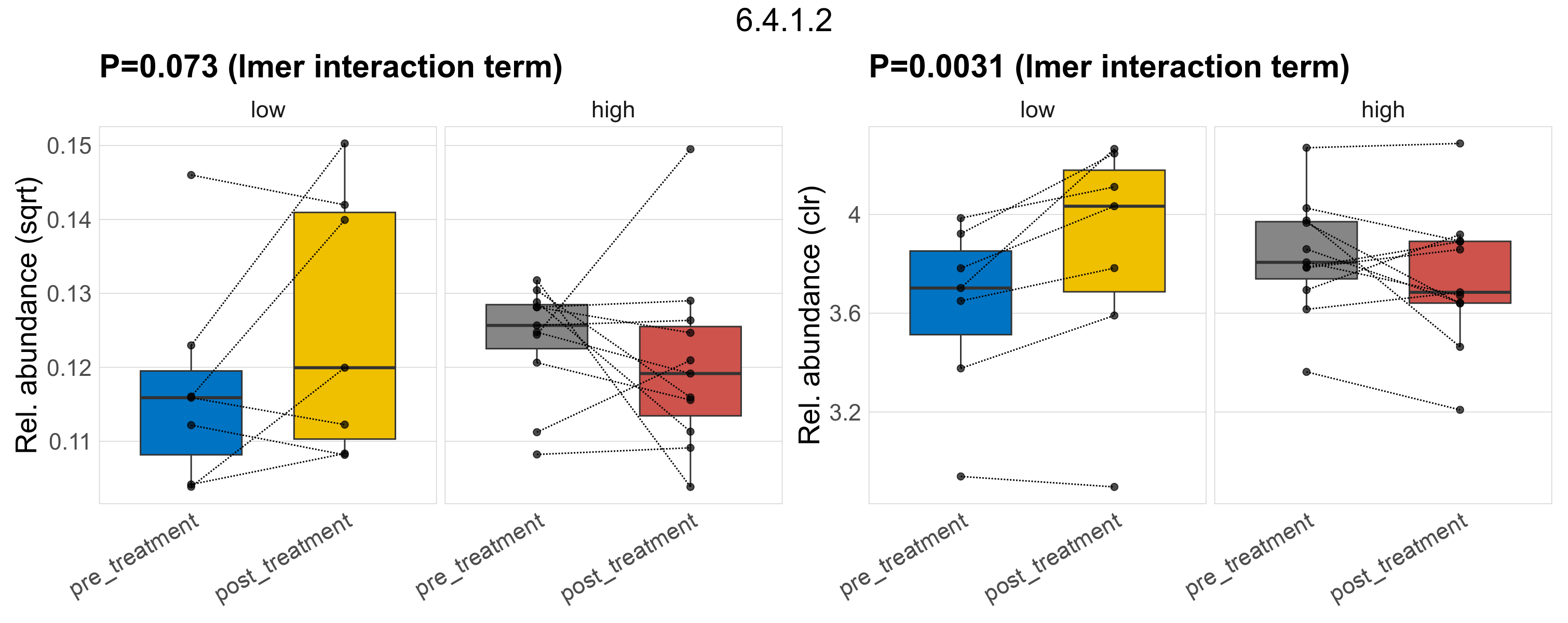
| 6.4.1.7 | 2-oxoglutarate carboxylase. | 0.58 | 0.79 | 1 | 0.29 | 1 | 1 | 0.91 | 1 | 1 | 0.75 | 0.9 | 1 | 0.31 | 0.9 | 1 | 0.72 | 0.94 | 1 | 0.0024 | 0.0024 | 0.0015 | 0.0023 | 0.0011 | 0.0024 | 0.0035 | 0.0017 | 0.004 | 0.61 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0016 | 0.002 | 0.0011 | 0.0026 | 0.0014 | 0.0029 | 0.7 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.2.12 | Xanthan lyase. | 0.2 | 0.53 | 1 | 0.12 | 1 | 1 | 0.03 | 0.71 | 1 | 0.17 | 0.49 | 1 | 0.31 | 0.9 | 1 | 0.11 | 0.66 | 1 | 0.0012 | 0.00078 | 0.00011 | 0.001 | 0.00048 | 0.0012 | 0.00032 | 0.00012 | 0.00059 | -1.6 | 24 / 36 (67%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.00047 | 2.8e-05 | 0.0011 | 0.0012 | 1e-04 | 0.0022 | 1.4 | 6 / 11 (55%) | 8 / 11 (73%) | 0.545 | 0.727 |
| 2.7.7.15 | Choline-phosphate cytidylyltransferase. | 0.95 | 0.98 | 1 | 0.28 | 1 | 1 | 0.27 | 0.89 | 1 | 0.9 | 0.97 | 1 | 0.63 | 0.95 | 1 | 0.24 | 0.76 | 1 | 0.00039 | 9.7e-05 | 0 | 4e-05 | 0 | 6e-05 | 0.00024 | 0 | 0.00058 | 2.6 | 9 / 36 (25%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 5.8e-05 | 0 | 0.00012 | 8.2e-05 | 0 | 0.00027 | 0.5 | 3 / 11 (27%) | 1 / 11 (9.1%) | 0.273 | 0.0909 |
| 3.1.3.15 | Histidinol-phosphatase. | 0.71 | 0.86 | 1 | 0.8 | 1 | 1 | 0.34 | 0.9 | 1 | 0.74 | 0.9 | 1 | 0.28 | 0.88 | 1 | 0.77 | 0.95 | 1 | 0.018 | 0.018 | 0.017 | 0.018 | 0.018 | 0.002 | 0.017 | 0.016 | 0.0019 | -0.082 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.017 | 0.017 | 0.0037 | 0.018 | 0.018 | 0.0026 | 0.082 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.7.1.4 | Nitrite reductase (NAD(P)H). | 0.27 | 0.56 | 1 | 0.54 | 1 | 1 | 0.75 | 0.99 | 1 | 0.49 | 0.74 | 1 | 0.92 | 0.99 | 1 | 0.77 | 0.95 | 1 | 0.00056 | 2e-04 | 0 | 0.00047 | 0 | 0.0011 | 0.00035 | 0 | 0.00081 | -0.43 | 13 / 36 (36%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 7e-05 | 0 | 0.00015 | 6.6e-05 | 0 | 0.00014 | -0.085 | 4 / 11 (36%) | 3 / 11 (27%) | 0.364 | 0.273 |
| 2.7.7.13 | Mannose-1-phosphate guanylyltransferase. | 0.2 | 0.53 | 1 | 0.67 | 1 | 1 | 0.26 | 0.89 | 1 | 0.23 | 0.53 | 1 | 0.91 | 0.99 | 1 | 0.34 | 0.8 | 1 | 0.004 | 0.0039 | 0.0031 | 0.0046 | 0.0047 | 0.0027 | 0.0043 | 0.0027 | 0.0033 | -0.097 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0032 | 0.0029 | 0.0021 | 0.0039 | 0.0031 | 0.0027 | 0.29 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 3.1.27.6 | Transferred entry: 4.6.1.21. | 0.019 | 0.49 | 1 | 0.023 | 0.92 | 1 | 0.12 | 0.82 | 1 | 0.013 | 0.42 | 1 | 0.16 | 0.87 | 1 | 0.49 | 0.88 | 1 | 0.00033 | 5.5e-05 | 0 | 0.00019 | 0 | 0.00039 | 8.5e-05 | 0 | 0.00018 | -1.2 | 6 / 36 (17%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 3e-06 | 0 | 9.9e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 1.14.12.1 | Anthranilate 1.2-dioxygenase (deaminating. decarboxylating). | 0.016 | 0.49 | 1 | 0.028 | 0.97 | 1 | 0.02 | 0.71 | 1 | 0.046 | 0.43 | 1 | 0.06 | 0.86 | 1 | 0.039 | 0.63 | 1 | 1e-04 | 1.1e-05 | 0 | 4.3e-05 | 0 | 7.4e-05 | 0 | 0 | 0 | -Inf | 4 / 36 (11%) | 2 / 7 (29%) | 0 / 7 (0%) | 0.286 | 0 | 0 | 0 | 0 | 9.2e-06 | 0 | 2.2e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 3.4.21.83 | Oligopeptidase B. | 0.086 | 0.49 | 1 | 0.67 | 1 | 1 | 0.5 | 0.95 | 1 | 0.048 | 0.43 | 1 | 0.37 | 0.9 | 1 | 0.18 | 0.72 | 1 | 0.0048 | 0.0044 | 0.0025 | 0.0016 | 0.0011 | 0.0017 | 0.0021 | 0.0014 | 0.0024 | 0.39 | 33 / 36 (92%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.007 | 0.0037 | 0.013 | 0.0051 | 0.003 | 0.0051 | -0.46 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 3.2.2.4 | AMP nucleosidase. | 0.19 | 0.53 | 1 | 0.54 | 1 | 1 | 0.29 | 0.89 | 1 | 0.19 | 0.5 | 1 | 0.71 | 0.95 | 1 | 0.56 | 0.9 | 1 | 0.00083 | 0.00051 | 0.00011 | 0.00071 | 0.00042 | 9e-04 | 0.00056 | 0.00024 | 0.00076 | -0.34 | 22 / 36 (61%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.00029 | 2.8e-05 | 0.00055 | 0.00056 | 5.9e-05 | 0.0011 | 0.95 | 6 / 11 (55%) | 6 / 11 (55%) | 0.545 | 0.545 |
| 1.15.1.1 | Superoxide dismutase. | 0.088 | 0.49 | 1 | 0.22 | 1 | 1 | 0.46 | 0.93 | 1 | 0.058 | 0.43 | 1 | 0.34 | 0.9 | 1 | 0.72 | 0.94 | 1 | 0.0014 | 0.0014 | 0.0012 | 0.0019 | 0.0017 | 0.0011 | 0.0015 | 0.0012 | 0.0011 | -0.34 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0012 | 0.00096 | 0.0011 | 0.0011 | 0.00057 | 0.0011 | -0.13 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.3.7.11 | 2.3-bis-O-geranylgeranyl-sn-glycero-phospholipid reductase. | 0.82 | 0.92 | 1 | 0.76 | 1 | 1 | 0.92 | 1 | 1 | 0.91 | 0.97 | 1 | 0.99 | 1 | 1 | 0.79 | 0.95 | 1 | 0.00024 | 2.7e-05 | 0 | 4.1e-05 | 0 | 0.00011 | 2.2e-05 | 0 | 5.8e-05 | -0.9 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3.8e-05 | 0 | 0.00013 | 1e-05 | 0 | 3.4e-05 | -1.9 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 1.1.1.51 | 3(or 17)-beta-hydroxysteroid dehydrogenase. | 0.71 | 0.87 | 1 | 1 | 1 | 1 | 0.25 | 0.89 | 1 | 0.44 | 0.69 | 1 | 0.65 | 0.95 | 1 | 0.4 | 0.83 | 1 | 0.00012 | 1e-05 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 3 / 36 (8.3%) | 0 / 7 (0%) | 0 / 7 (0%) | 0 | 0 | 3e-06 | 0 | 9.9e-06 | 3e-05 | 0 | 8.9e-05 | 3.3 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 1.18.1.3 | Ferredoxin--NAD(+) reductase. | 0.1 | 0.49 | 1 | 0.27 | 1 | 1 | 0.047 | 0.71 | 1 | 0.033 | 0.43 | 1 | 0.15 | 0.87 | 1 | 0.014 | 0.55 | 1 | 8.8e-05 | 9.8e-06 | 0 | 0 | 0 | 0 | 1.9e-05 | 0 | 5.1e-05 | Inf | 4 / 36 (11%) | 0 / 7 (0%) | 1 / 7 (14%) | 0 | 0.143 | 2e-05 | 0 | 4.1e-05 | 0 | 0 | 0 | -Inf | 3 / 11 (27%) | 0 / 11 (0%) | 0.273 | 0 |
| 1.1.1.373 | Sulfolactaldehyde 3-reductase. | 0.095 | 0.49 | 1 | 0.52 | 1 | 1 | 0.38 | 0.9 | 1 | 0.049 | 0.43 | 1 | 0.45 | 0.92 | 1 | 0.36 | 0.81 | 1 | 0.00025 | 2.8e-05 | 0 | 6.2e-05 | 0 | 0.00013 | 7.7e-05 | 0 | 2e-04 | 0.31 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.4.1.3 | Glutamate dehydrogenase (NAD(P)(+)). | 0.2 | 0.53 | 1 | 0.87 | 1 | 1 | 0.37 | 0.9 | 1 | 0.15 | 0.47 | 1 | 0.92 | 0.99 | 1 | 0.5 | 0.89 | 1 | 0.00077 | 0.00057 | 0.00016 | 0.00058 | 0.00042 | 6e-04 | 0.00081 | 0.00012 | 0.0012 | 0.48 | 27 / 36 (75%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 2e-04 | 8e-05 | 0.00028 | 8e-04 | 0.00021 | 0.0016 | 2 | 7 / 11 (64%) | 8 / 11 (73%) | 0.636 | 0.727 |
| 1.2.4.2 | Oxoglutarate dehydrogenase (succinyl-transferring). | 0.052 | 0.49 | 1 | 0.44 | 1 | 1 | 0.13 | 0.82 | 1 | 0.016 | 0.42 | 1 | 0.57 | 0.93 | 1 | 0.1 | 0.66 | 1 | 0.00072 | 0.00042 | 6.6e-05 | 0.00078 | 0.00042 | 0.0013 | 0.00073 | 0.00012 | 0.0017 | -0.096 | 21 / 36 (58%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.00015 | 0 | 0.00044 | 0.00026 | 7.5e-05 | 0.00039 | 0.79 | 3 / 11 (27%) | 8 / 11 (73%) | 0.273 | 0.727 |
| 6.2.1.22 | [Citrate (pro-3S)-lyase] ligase. | 0.68 | 0.85 | 1 | 0.83 | 1 | 1 | 0.6 | 0.96 | 1 | 0.96 | 0.99 | 1 | 0.71 | 0.95 | 1 | 0.34 | 0.8 | 1 | 0.0015 | 0.0015 | 0.0014 | 0.0017 | 0.0019 | 0.00074 | 0.0016 | 0.0015 | 0.00031 | -0.087 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0016 | 0.0014 | 8e-04 | 0.0013 | 0.0014 | 0.00058 | -0.3 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.1.111 | 2-oxoglutaramate amidase. | 0.85 | 0.93 | 1 | 0.071 | 1 | 1 | 0.13 | 0.82 | 1 | 0.64 | 0.85 | 1 | 0.028 | 0.76 | 1 | 0.1 | 0.66 | 1 | 0.00033 | 0.00024 | 0.00013 | 0.00024 | 8.5e-05 | 0.00047 | 0.00037 | 0.00025 | 0.00029 | 0.62 | 26 / 36 (72%) | 4 / 7 (57%) | 7 / 7 (100%) | 0.571 | 1 | 0.00023 | 9.7e-05 | 0.00037 | 0.00016 | 0.00014 | 0.00014 | -0.52 | 7 / 11 (64%) | 8 / 11 (73%) | 0.636 | 0.727 |
| 3.4.25.2 | HslU--HslV peptidase. | 0.44 | 0.69 | 1 | 0.55 | 1 | 1 | 0.15 | 0.84 | 1 | 0.73 | 0.89 | 1 | 0.25 | 0.88 | 1 | 0.051 | 0.65 | 1 | 3e-04 | 0.00024 | 0.00023 | 0.00033 | 0.00033 | 3e-04 | 0.00033 | 0.00024 | 0.00017 | 0 | 29 / 36 (81%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.00023 | 0.00024 | 2e-04 | 0.00014 | 1e-04 | 0.00016 | -0.72 | 9 / 11 (82%) | 7 / 11 (64%) | 0.818 | 0.636 |
| 3.5.3.- | 0.22 | 0.54 | 1 | 0.043 | 0.97 | 1 | 0.23 | 0.88 | 1 | 0.12 | 0.47 | 1 | 0.015 | 0.76 | 1 | 0.098 | 0.66 | 1 | 0.0016 | 0.0015 | 0.0015 | 0.001 | 0.00097 | 0.00057 | 0.0017 | 0.0018 | 0.0012 | 0.77 | 35 / 36 (97%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.0016 | 0.0015 | 0.001 | 0.0018 | 0.0017 | 0.001 | 0.17 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 3.6.3.1 | Transferred entry: 7.6.2.1. | 0.21 | 0.53 | 1 | 0.72 | 1 | 1 | 0.42 | 0.92 | 1 | 0.24 | 0.53 | 1 | 0.54 | 0.93 | 1 | 0.4 | 0.83 | 1 | 0.00076 | 0.00015 | 0 | 0.00031 | 0 | 0.00075 | 0.00037 | 0 | 0.00097 | 0.26 | 7 / 36 (19%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 8.1e-06 | 0 | 1.9e-05 | 4.9e-05 | 0 | 0.00014 | 2.6 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 3.1.1.27 | 4-pyridoxolactonase. | 0.13 | 0.5 | 1 | 0.94 | 1 | 1 | 0.16 | 0.85 | 1 | 0.12 | 0.47 | 1 | 0.95 | 0.99 | 1 | 0.24 | 0.76 | 1 | 0.00033 | 0.00017 | 2.4e-05 | 0.00013 | 0 | 0.00023 | 0.00014 | 0 | 0.00026 | 0.11 | 19 / 36 (53%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 3e-04 | 0.00019 | 0.00036 | 9.2e-05 | 3.3e-05 | 0.00011 | -1.7 | 8 / 11 (73%) | 6 / 11 (55%) | 0.727 | 0.545 |
| 1.97.1.4 | [Formate-C-acetyltransferase]-activating enzyme. | 0.22 | 0.54 | 1 | 1 | 1 | 1 | 0.79 | 1 | 1 | 0.12 | 0.47 | 1 | 0.32 | 0.9 | 1 | 0.48 | 0.87 | 1 | 0.018 | 0.018 | 0.017 | 0.016 | 0.016 | 0.0039 | 0.016 | 0.017 | 0.0055 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.019 | 0.017 | 0.0051 | 0.019 | 0.018 | 0.0025 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.35 | 3-hydroxyacyl-CoA dehydrogenase. | 0.92 | 0.96 | 1 | 0.62 | 1 | 1 | 0.25 | 0.89 | 1 | 0.69 | 0.87 | 1 | 0.82 | 0.98 | 1 | 0.68 | 0.93 | 1 | 0.0055 | 0.0055 | 0.005 | 0.0052 | 0.0051 | 0.0014 | 0.0049 | 0.0047 | 0.0013 | -0.086 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0055 | 0.005 | 0.0024 | 0.0062 | 0.0052 | 0.0031 | 0.17 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.2.7 | Butyrate kinase. | 0.93 | 0.97 | 1 | 0.91 | 1 | 1 | 0.74 | 0.99 | 1 | 0.97 | 0.99 | 1 | 0.88 | 0.99 | 1 | 0.83 | 0.96 | 1 | 0.0027 | 0.0027 | 0.0021 | 0.0025 | 0.0024 | 0.00098 | 0.0027 | 0.0027 | 0.0017 | 0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0029 | 0.0014 | 0.0024 | 0.0025 | 0.002 | 0.0018 | -0.21 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.2.10 | UDP-N-acetylmuramoylpentapeptide-lysine N(6)-alanyltransferase. | 0.055 | 0.49 | 1 | 0.43 | 1 | 1 | 0.29 | 0.89 | 1 | 0.078 | 0.44 | 1 | 0.27 | 0.88 | 1 | 0.3 | 0.78 | 1 | 0.00022 | 0.00011 | 1.5e-05 | 2.1e-05 | 0 | 3.6e-05 | 6.3e-05 | 0 | 9e-05 | 1.6 | 18 / 36 (50%) | 2 / 7 (29%) | 3 / 7 (43%) | 0.286 | 0.429 | 0.00021 | 0.00011 | 0.00033 | 1e-04 | 0.00011 | 0.00012 | -1.1 | 6 / 11 (55%) | 7 / 11 (64%) | 0.545 | 0.636 |
| 3.2.1.41 | Pullulanase. | 0.15 | 0.5 | 1 | 0.74 | 1 | 1 | 0.69 | 0.98 | 1 | 0.19 | 0.5 | 1 | 0.98 | 0.99 | 1 | 0.83 | 0.96 | 1 | 0.027 | 0.027 | 0.024 | 0.021 | 0.02 | 0.0039 | 0.021 | 0.022 | 0.01 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.031 | 0.033 | 0.017 | 0.03 | 0.031 | 0.011 | -0.047 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.199 | Protein-N(pi)-phosphohistidine--D-glucose phosphotransferase. | 0.096 | 0.49 | 1 | 0.82 | 1 | 1 | 0.88 | 1 | 1 | 0.077 | 0.44 | 1 | 0.69 | 0.95 | 1 | 0.67 | 0.93 | 1 | 0.00067 | 0.00022 | 0 | 0.00061 | 1.6e-05 | 0.0015 | 0.00048 | 5.4e-05 | 0.0011 | -0.35 | 12 / 36 (33%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 2.1e-05 | 0 | 4.6e-05 | 2.3e-05 | 0 | 6.8e-05 | 0.13 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 1.7.-.- | 0.057 | 0.49 | 1 | 0.11 | 1 | 1 | 0.14 | 0.83 | 1 | 0.051 | 0.43 | 1 | 0.0032 | 0.62 | 1 | 0.0049 | 0.48 | 1 | 0.0091 | 0.0091 | 0.0087 | 0.0073 | 0.0065 | 0.0023 | 0.0086 | 0.007 | 0.0034 | 0.24 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.01 | 0.012 | 0.0034 | 0.0097 | 0.0097 | 0.0029 | -0.044 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 1.4.99.5 | Glycine dehydrogenase (cyanide-forming). | 0.88 | 0.95 | 1 | 0.14 | 1 | 1 | 0.49 | 0.95 | 1 | 0.88 | 0.96 | 1 | 0.23 | 0.88 | 1 | 0.63 | 0.92 | 1 | 0.00022 | 8.7e-05 | 0 | 9.5e-05 | 0 | 0.00012 | 1.6e-05 | 0 | 3.1e-05 | -2.6 | 14 / 36 (39%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 0.00012 | 0 | 0.00021 | 9e-05 | 0 | 2e-04 | -0.42 | 5 / 11 (45%) | 4 / 11 (36%) | 0.455 | 0.364 |
| 1.3.1.108 | Caffeoyl-CoA reductase. | 0.099 | 0.49 | 1 | 0.016 | 0.92 | 1 | 0.28 | 0.89 | 1 | 0.19 | 0.5 | 1 | 0.071 | 0.86 | 1 | 0.83 | 0.96 | 1 | 0.00027 | 5.2e-05 | 0 | 0.00019 | 0 | 0.00044 | 4.8e-05 | 0 | 0.00013 | -2 | 7 / 36 (19%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 1.8e-05 | 0 | 3.5e-05 | 0 | 0 | 0 | -Inf | 3 / 11 (27%) | 0 / 11 (0%) | 0.273 | 0 |
| 5.1.1.4 | Proline racemase. | 0.29 | 0.57 | 1 | 0.87 | 1 | 1 | 0.24 | 0.89 | 1 | 0.24 | 0.53 | 1 | 0.79 | 0.97 | 1 | 0.39 | 0.83 | 1 | 0.00048 | 0.00045 | 0.00036 | 0.00044 | 0.00036 | 0.00044 | 0.00046 | 0.00024 | 5e-04 | 0.064 | 34 / 36 (94%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00059 | 0.00048 | 0.00042 | 0.00032 | 0.00032 | 0.00016 | -0.88 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 4.1.-.- | 0.12 | 0.5 | 1 | 0.44 | 1 | 1 | 0.48 | 0.93 | 1 | 0.074 | 0.43 | 1 | 0.32 | 0.9 | 1 | 0.44 | 0.86 | 1 | 0.00045 | 0.00019 | 0 | 0.00032 | 0.00011 | 0.00055 | 0.00036 | 3.1e-05 | 0.00083 | 0.17 | 15 / 36 (42%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 7.8e-05 | 0 | 0.00015 | 1e-04 | 0 | 0.00026 | 0.36 | 3 / 11 (27%) | 3 / 11 (27%) | 0.273 | 0.273 | |
| 6.1.1.2 | Tryptophan--tRNA ligase. | 0.23 | 0.54 | 1 | 0.89 | 1 | 1 | 0.67 | 0.98 | 1 | 0.23 | 0.53 | 1 | 0.42 | 0.91 | 1 | 0.64 | 0.92 | 1 | 0.019 | 0.019 | 0.02 | 0.018 | 0.018 | 0.0018 | 0.018 | 0.019 | 0.0043 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.02 | 0.02 | 0.0057 | 0.021 | 0.02 | 0.003 | 0.07 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.1.148 | Thymidylate synthase (FAD). | 0.83 | 0.92 | 1 | 0.79 | 1 | 1 | 0.25 | 0.89 | 1 | 0.68 | 0.87 | 1 | 0.27 | 0.88 | 1 | 0.78 | 0.95 | 1 | 0.0049 | 0.0049 | 0.0049 | 0.0045 | 0.0034 | 0.0017 | 0.0046 | 0.0049 | 0.0012 | 0.032 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0048 | 0.0047 | 0.002 | 0.0054 | 0.0056 | 0.0013 | 0.17 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.21.- | 0.39 | 0.66 | 1 | 0.91 | 1 | 1 | 0.24 | 0.88 | 1 | 0.96 | 0.99 | 1 | 0.19 | 0.87 | 1 | 0.82 | 0.96 | 1 | 0.069 | 0.069 | 0.069 | 0.069 | 0.071 | 0.0091 | 0.07 | 0.072 | 0.012 | 0.021 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.063 | 0.067 | 0.01 | 0.076 | 0.062 | 0.028 | 0.27 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 1.1.5.4 | Malate dehydrogenase (quinone). | 0.12 | 0.5 | 1 | 0.081 | 1 | 1 | 0.21 | 0.87 | 1 | 0.14 | 0.47 | 1 | 0.36 | 0.9 | 1 | 0.61 | 0.92 | 1 | 0.00032 | 4.5e-05 | 0 | 0.00014 | 0 | 0.00035 | 7.7e-05 | 0 | 2e-04 | -0.86 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 5.1e-06 | 0 | 1.7e-05 | 2.7e-06 | 0 | 8.9e-06 | -0.92 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.1.1.45 | Thymidylate synthase. | 0.061 | 0.49 | 1 | 0.9 | 1 | 1 | 0.033 | 0.71 | 1 | 0.05 | 0.43 | 1 | 0.34 | 0.9 | 1 | 0.0048 | 0.48 | 1 | 0.012 | 0.012 | 0.012 | 0.012 | 0.011 | 0.0029 | 0.012 | 0.012 | 0.0042 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.015 | 0.014 | 0.0041 | 0.011 | 0.011 | 0.0019 | -0.45 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.99.11 | Benzylsuccinate synthase. | 0.8 | 0.9 | 1 | 0.46 | 1 | 1 | 0.11 | 0.82 | 1 | 0.36 | 0.63 | 1 | 0.088 | 0.86 | 1 | 0.02 | 0.55 | 1 | 9e-04 | 8e-04 | 0.00034 | 0.00054 | 0.00043 | 0.00042 | 0.00055 | 0.00021 | 0.00073 | 0.026 | 32 / 36 (89%) | 7 / 7 (100%) | 5 / 7 (71%) | 1 | 0.714 | 0.00067 | 0.00029 | 0.0011 | 0.0012 | 0.00038 | 0.0022 | 0.84 | 9 / 11 (82%) | 11 / 11 (100%) | 0.818 | 1 |
| 3.4.23.36 | Signal peptidase II. | 0.71 | 0.86 | 1 | 0.49 | 1 | 1 | 0.9 | 1 | 1 | 0.42 | 0.68 | 1 | 0.79 | 0.97 | 1 | 0.43 | 0.85 | 1 | 0.0094 | 0.0094 | 0.0089 | 0.0094 | 0.0088 | 0.0026 | 0.0089 | 0.0091 | 0.0024 | -0.079 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0098 | 0.0089 | 0.0029 | 0.0092 | 0.0091 | 0.0013 | -0.091 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.3.1.3 | Histidine ammonia-lyase. | 0.77 | 0.89 | 1 | 0.44 | 1 | 1 | 0.53 | 0.95 | 1 | 0.54 | 0.77 | 1 | 0.42 | 0.91 | 1 | 0.63 | 0.92 | 1 | 0.0025 | 0.0025 | 0.0015 | 0.002 | 0.0019 | 9e-04 | 0.0026 | 0.0014 | 0.0019 | 0.38 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0021 | 0.0013 | 0.0025 | 0.0031 | 0.0022 | 0.0035 | 0.56 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.1.8 | Oxalyl-CoA decarboxylase. | 0.73 | 0.87 | 1 | 0.74 | 1 | 1 | 0.7 | 0.98 | 1 | 0.82 | 0.93 | 1 | 0.74 | 0.96 | 1 | 0.96 | 0.99 | 1 | 0.0011 | 0.00055 | 8e-06 | 0.00073 | 1.6e-05 | 0.00095 | 7e-04 | 0 | 0.0011 | -0.061 | 18 / 36 (50%) | 4 / 7 (57%) | 3 / 7 (43%) | 0.571 | 0.429 | 0.00061 | 0 | 0.001 | 0.00029 | 3e-05 | 0.00061 | -1.1 | 5 / 11 (45%) | 6 / 11 (55%) | 0.455 | 0.545 |
| 2.8.1.10 | Thiazole synthase. | 0.7 | 0.86 | 1 | 0.69 | 1 | 1 | 0.4 | 0.92 | 1 | 0.47 | 0.72 | 1 | 0.28 | 0.88 | 1 | 0.15 | 0.71 | 1 | 0.0093 | 0.0093 | 0.0079 | 0.0089 | 0.008 | 0.0031 | 0.0095 | 0.0096 | 0.0037 | 0.094 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.01 | 0.0079 | 0.0067 | 0.0086 | 0.0075 | 0.0028 | -0.22 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.2.19 | Nicotinate-nucleotide diphosphorylase (carboxylating). | 0.64 | 0.83 | 1 | 0.71 | 1 | 1 | 0.88 | 1 | 1 | 0.44 | 0.69 | 1 | 0.91 | 0.99 | 1 | 0.86 | 0.96 | 1 | 0.0099 | 0.0099 | 0.0098 | 0.0096 | 0.0095 | 0.0028 | 0.0095 | 0.011 | 0.0045 | -0.015 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.01 | 0.01 | 0.0026 | 0.0099 | 0.0097 | 0.0019 | -0.014 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.1.228 | tRNA (guanine(37)-N(1))-methyltransferase. | 0.0055 | 0.49 | 1 | 0.71 | 1 | 1 | 0.36 | 0.9 | 1 | 0.018 | 0.42 | 1 | 0.16 | 0.87 | 1 | 0.056 | 0.66 | 1 | 0.014 | 0.014 | 0.015 | 0.012 | 0.012 | 0.0015 | 0.012 | 0.013 | 0.0034 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.016 | 0.015 | 0.0039 | 0.015 | 0.015 | 0.0028 | -0.093 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.4.1.11 | L-erythro-3.5-diaminohexanoate dehydrogenase. | 0.3 | 0.58 | 1 | 0.94 | 1 | 1 | 0.47 | 0.93 | 1 | 0.33 | 0.6 | 1 | 0.98 | 0.99 | 1 | 0.47 | 0.87 | 1 | 0.00036 | 5e-05 | 0 | 8.1e-06 | 0 | 2.1e-05 | 4.4e-06 | 0 | 1.2e-05 | -0.88 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0.00015 | 0 | 0.00045 | 9e-06 | 0 | 3e-05 | -4.1 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 1.1.1.58 | Tagaturonate reductase. | 0.068 | 0.49 | 1 | 0.57 | 1 | 1 | 0.12 | 0.82 | 1 | 0.21 | 0.52 | 1 | 0.85 | 0.98 | 1 | 0.34 | 0.8 | 1 | 0.0071 | 0.0071 | 0.0067 | 0.0079 | 0.0082 | 0.0019 | 0.0073 | 0.0073 | 0.0028 | -0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0058 | 0.006 | 0.0021 | 0.0079 | 0.0095 | 0.003 | 0.45 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.121 | Phosphoenolpyruvate--glycerone phosphotransferase. | 0.56 | 0.78 | 1 | 0.24 | 1 | 1 | 0.58 | 0.96 | 1 | 0.75 | 0.9 | 1 | 0.41 | 0.9 | 1 | 0.67 | 0.93 | 1 | 0.003 | 0.003 | 0.0026 | 0.003 | 0.0025 | 0.0019 | 0.0042 | 0.0043 | 0.003 | 0.49 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0025 | 0.0021 | 0.0014 | 0.0028 | 0.003 | 0.0015 | 0.16 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.7.56 | tRNA nucleotidyltransferase. | 0.095 | 0.49 | 1 | 0.71 | 1 | 1 | 0.33 | 0.89 | 1 | 0.067 | 0.43 | 1 | 0.65 | 0.95 | 1 | 0.25 | 0.76 | 1 | 0.0025 | 0.0024 | 0.0021 | 0.0015 | 0.0014 | 0.00084 | 0.0018 | 0.0014 | 0.0017 | 0.26 | 34 / 36 (94%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.0032 | 0.0023 | 0.0038 | 0.0024 | 0.0023 | 0.0016 | -0.42 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.202 | Protein-N(pi)-phosphohistidine--D-fructose phosphotransferase. | 0.048 | 0.49 | 1 | 0.021 | 0.92 | 1 | 0.046 | 0.71 | 1 | 0.03 | 0.43 | 1 | 0.11 | 0.87 | 1 | 0.18 | 0.72 | 1 | 0.00073 | 0.00028 | 0 | 0.00087 | 0.00012 | 0.0021 | 0.00047 | 0 | 0.0011 | -0.89 | 14 / 36 (39%) | 5 / 7 (71%) | 3 / 7 (43%) | 0.714 | 0.429 | 3e-05 | 0 | 5.2e-05 | 4.6e-05 | 0 | 1e-04 | 0.62 | 3 / 11 (27%) | 3 / 11 (27%) | 0.273 | 0.273 |
| 2.7.1.85 | Beta-glucoside kinase. | 0.57 | 0.79 | 1 | 0.2 | 1 | 1 | 0.4 | 0.91 | 1 | 0.48 | 0.72 | 1 | 0.26 | 0.88 | 1 | 0.62 | 0.92 | 1 | 0.0043 | 0.0043 | 0.0033 | 0.0039 | 0.0036 | 0.0015 | 0.0031 | 0.0033 | 0.0013 | -0.33 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.005 | 0.0037 | 0.0039 | 0.0048 | 0.0032 | 0.0032 | -0.059 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.7.2.5 | Nitric-oxide reductase (cytochrome c). | 0.59 | 0.79 | 1 | 0.65 | 1 | 1 | 0.61 | 0.96 | 1 | 0.79 | 0.92 | 1 | 0.43 | 0.91 | 1 | 0.44 | 0.85 | 1 | 0.00015 | 2e-05 | 0 | 3.5e-05 | 0 | 9.2e-05 | 3.7e-05 | 0 | 6.3e-05 | 0.08 | 5 / 36 (14%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 1.5e-05 | 0 | 4.9e-05 | 6e-06 | 0 | 2e-05 | -1.3 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.3.1.85 | Fatty-acid synthase system. | 0.9 | 0.96 | 1 | 0.4 | 1 | 1 | 0.33 | 0.89 | 1 | 0.74 | 0.9 | 1 | 0.49 | 0.92 | 1 | 0.51 | 0.89 | 1 | 0.00015 | 2.1e-05 | 0 | 1.7e-05 | 0 | 4.5e-05 | 0 | 0 | 0 | -Inf | 5 / 36 (14%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 1.8e-05 | 0 | 4.3e-05 | 4.1e-05 | 0 | 0.00011 | 1.2 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 2.5.1.31 | Ditrans.polycis-undecaprenyl-diphosphate synthase ((2E.6E)-farnesyl- | 0.31 | 0.59 | 1 | 0.7 | 1 | 1 | 0.89 | 1 | 1 | 0.38 | 0.65 | 1 | 0.38 | 0.9 | 1 | 0.5 | 0.88 | 1 | 0.0019 | 0.0018 | 0.00093 | 0.0011 | 0.00096 | 0.00077 | 0.001 | 0.00072 | 0.001 | -0.14 | 35 / 36 (97%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.0027 | 0.00089 | 0.0049 | 0.0019 | 0.0016 | 0.0016 | -0.51 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.31 | 3-hydroxyisobutyrate dehydrogenase. | 0.018 | 0.49 | 1 | 0.81 | 1 | 1 | 0.6 | 0.96 | 1 | 0.0068 | 0.37 | 1 | 0.3 | 0.89 | 1 | 0.13 | 0.67 | 1 | 0.0011 | 0.00094 | 0.00059 | 4e-04 | 9.6e-05 | 0.00066 | 0.00035 | 0.00024 | 0.00039 | -0.19 | 30 / 36 (83%) | 4 / 7 (57%) | 5 / 7 (71%) | 0.571 | 0.714 | 0.0013 | 0.00064 | 0.0011 | 0.0013 | 0.00073 | 0.0014 | 0 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 4.2.1.90 | L-rhamnonate dehydratase. | 0.83 | 0.92 | 1 | 0.43 | 1 | 1 | 0.68 | 0.98 | 1 | 0.9 | 0.97 | 1 | 0.33 | 0.9 | 1 | 0.39 | 0.83 | 1 | 0.00035 | 0.00021 | 0.00012 | 0.00022 | 5.7e-05 | 0.00035 | 0.00028 | 0.00012 | 0.00037 | 0.35 | 22 / 36 (61%) | 4 / 7 (57%) | 5 / 7 (71%) | 0.571 | 0.714 | 0.00015 | 0.00012 | 0.00018 | 0.00023 | 7.5e-05 | 4e-04 | 0.62 | 7 / 11 (64%) | 6 / 11 (55%) | 0.636 | 0.545 |
| 2.1.1.113 | Site-specific DNA-methyltransferase (cytosine-N(4)-specific). | 0.67 | 0.84 | 1 | 0.74 | 1 | 1 | 0.83 | 1 | 1 | 0.55 | 0.78 | 1 | 0.52 | 0.93 | 1 | 0.67 | 0.93 | 1 | 6.5e-05 | 9e-06 | 0 | 2.3e-06 | 0 | 6.1e-06 | 7.7e-06 | 0 | 2e-05 | 1.7 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 1.5e-05 | 0 | 4.9e-05 | 8.4e-06 | 0 | 2e-05 | -0.84 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 3.2.1.55 | Non-reducing end alpha-L-arabinofuranosidase. | 0.044 | 0.49 | 1 | 0.26 | 1 | 1 | 0.33 | 0.89 | 1 | 0.016 | 0.42 | 1 | 0.028 | 0.76 | 1 | 0.067 | 0.66 | 1 | 0.06 | 0.06 | 0.055 | 0.043 | 0.039 | 0.024 | 0.051 | 0.052 | 0.026 | 0.25 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.07 | 0.051 | 0.041 | 0.066 | 0.063 | 0.02 | -0.085 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.1.115 | UDP-N-acetylglucosamine 4.6-dehydratase (inverting). | 0.5 | 0.73 | 1 | 0.82 | 1 | 1 | 0.45 | 0.93 | 1 | 0.91 | 0.97 | 1 | 0.75 | 0.97 | 1 | 0.91 | 0.98 | 1 | 0.0011 | 0.00091 | 0.00043 | 0.0011 | 0.00057 | 0.0013 | 0.001 | 0.00028 | 0.0013 | -0.14 | 30 / 36 (83%) | 5 / 7 (71%) | 6 / 7 (86%) | 0.714 | 0.857 | 0.00067 | 0.00035 | 0.00092 | 0.00094 | 0.00055 | 0.001 | 0.49 | 9 / 11 (82%) | 10 / 11 (91%) | 0.818 | 0.909 |
| 3.2.1.185 | Non-reducing end beta-L-arabinofuranosidase. | 0.88 | 0.95 | 1 | 0.36 | 1 | 1 | 0.092 | 0.79 | 1 | 0.92 | 0.97 | 1 | 0.5 | 0.93 | 1 | 0.5 | 0.89 | 1 | 0.0028 | 0.0027 | 0.0018 | 0.0024 | 0.0025 | 0.0018 | 0.0021 | 0.001 | 0.0025 | -0.19 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0025 | 0.0015 | 0.003 | 0.0034 | 0.0021 | 0.0037 | 0.44 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 5.3.3.2 | Isopentenyl-diphosphate Delta-isomerase. | 0.72 | 0.87 | 1 | 0.94 | 1 | 1 | 0.98 | 1 | 1 | 0.45 | 0.7 | 1 | 0.56 | 0.93 | 1 | 0.52 | 0.9 | 1 | 0.0055 | 0.0055 | 0.0054 | 0.0054 | 0.0057 | 0.0022 | 0.0054 | 0.0055 | 0.0025 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0056 | 0.0055 | 0.0018 | 0.0056 | 0.0047 | 0.0016 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.99.1.4 | Sirohydrochlorin ferrochelatase. | 0.49 | 0.73 | 1 | 0.014 | 0.89 | 1 | 0.043 | 0.71 | 1 | 0.57 | 0.79 | 1 | 0.02 | 0.76 | 1 | 0.063 | 0.66 | 1 | 0.00088 | 0.00086 | 0.00072 | 0.00099 | 0.001 | 0.00029 | 6e-04 | 0.00063 | 0.00046 | -0.72 | 35 / 36 (97%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 9e-04 | 0.00072 | 0.00068 | 9e-04 | 8e-04 | 0.00069 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 7.6.2.5 | ABC-type heme transporter. | 0.77 | 0.89 | 1 | 0.6 | 1 | 1 | 0.92 | 1 | 1 | 0.79 | 0.92 | 1 | 0.97 | 0.99 | 1 | 0.68 | 0.93 | 1 | 0.00017 | 3.3e-05 | 0 | 6.7e-05 | 0 | 0.00018 | 3.9e-05 | 0 | 1e-04 | -0.78 | 7 / 36 (19%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.2e-05 | 0 | 4.5e-05 | 1.8e-05 | 0 | 4e-05 | -0.29 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 6.3.1.1 | Aspartate--ammonia ligase. | 0.33 | 0.6 | 1 | 0.36 | 1 | 1 | 0.28 | 0.89 | 1 | 0.28 | 0.57 | 1 | 0.96 | 0.99 | 1 | 0.78 | 0.95 | 1 | 0.012 | 0.012 | 0.011 | 0.011 | 0.01 | 0.0027 | 0.0097 | 0.0096 | 0.0013 | -0.18 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.012 | 0.013 | 0.0032 | 0.013 | 0.012 | 0.0029 | 0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.6.- | 0.97 | 0.99 | 1 | 0.95 | 1 | 1 | 0.15 | 0.84 | 1 | 0.9 | 0.97 | 1 | 0.51 | 0.93 | 1 | 0.45 | 0.86 | 1 | 0.038 | 0.038 | 0.032 | 0.034 | 0.04 | 0.011 | 0.034 | 0.032 | 0.015 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.036 | 0.026 | 0.027 | 0.046 | 0.033 | 0.037 | 0.35 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 6.1.1.15 | Proline--tRNA ligase. | 0.1 | 0.49 | 1 | 0.47 | 1 | 1 | 0.62 | 0.96 | 1 | 0.14 | 0.47 | 1 | 0.43 | 0.91 | 1 | 0.061 | 0.66 | 1 | 0.028 | 0.028 | 0.029 | 0.027 | 0.024 | 0.0047 | 0.026 | 0.027 | 0.0055 | -0.054 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.031 | 0.03 | 0.0049 | 0.029 | 0.029 | 0.0034 | -0.096 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.2.20 | Acyl-CoA hydrolase. | 0.28 | 0.57 | 1 | 0.62 | 1 | 1 | 0.91 | 1 | 1 | 0.53 | 0.77 | 1 | 0.98 | 0.99 | 1 | 0.79 | 0.96 | 1 | 0.00028 | 8.6e-05 | 0 | 0.00022 | 0 | 0.00053 | 0.00014 | 0 | 0.00028 | -0.65 | 11 / 36 (31%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 3.5e-05 | 0 | 6.7e-05 | 1.9e-05 | 0 | 3.8e-05 | -0.88 | 3 / 11 (27%) | 3 / 11 (27%) | 0.273 | 0.273 |
| 2.3.1.193 | tRNA(Met) cytidine acetyltransferase. | 0.24 | 0.55 | 1 | 0.097 | 1 | 1 | 0.38 | 0.9 | 1 | 0.38 | 0.65 | 1 | 0.58 | 0.93 | 1 | 0.85 | 0.96 | 1 | 0.00039 | 6.5e-05 | 0 | 0.00017 | 0 | 0.00044 | 0.00014 | 0 | 0.00036 | -0.28 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 8.5e-06 | 0 | 2.1e-05 | 8.1e-06 | 0 | 2.7e-05 | -0.07 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 6.3.2.5 | Phosphopantothenate--cysteine ligase (CTP). | 0.23 | 0.55 | 1 | 0.47 | 1 | 1 | 0.47 | 0.93 | 1 | 0.17 | 0.49 | 1 | 0.86 | 0.99 | 1 | 0.91 | 0.98 | 1 | 0.015 | 0.015 | 0.014 | 0.014 | 0.015 | 0.0042 | 0.013 | 0.011 | 0.0029 | -0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.016 | 0.014 | 0.0062 | 0.016 | 0.015 | 0.0037 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 7.5.2.1 | ABC-type maltose transporter. | 0.59 | 0.79 | 1 | 0.59 | 1 | 1 | 0.99 | 1 | 1 | 0.81 | 0.93 | 1 | 0.65 | 0.95 | 1 | 0.94 | 0.99 | 1 | 0.00022 | 3.7e-05 | 0 | 5e-05 | 0 | 0.00013 | 8.7e-05 | 0 | 0.00023 | 0.8 | 6 / 36 (17%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 8.1e-06 | 0 | 1.9e-05 | 2.6e-05 | 0 | 6.9e-05 | 1.7 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 5.4.99.16 | Maltose alpha-D-glucosyltransferase. | 0.26 | 0.56 | 1 | 0.056 | 1 | 1 | 0.13 | 0.82 | 1 | 0.69 | 0.88 | 1 | 0.39 | 0.9 | 1 | 0.8 | 0.96 | 1 | 0.0045 | 0.0045 | 0.0042 | 0.0057 | 0.0049 | 0.0033 | 0.0042 | 0.0039 | 0.0018 | -0.44 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0042 | 0.0041 | 0.0017 | 0.0044 | 0.0043 | 0.0023 | 0.067 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.169 | 2-dehydropantoate 2-reductase. | 0.21 | 0.54 | 1 | 0.92 | 1 | 1 | 0.78 | 1 | 1 | 0.86 | 0.95 | 1 | 0.23 | 0.88 | 1 | 0.16 | 0.71 | 1 | 0.01 | 0.01 | 0.011 | 0.011 | 0.01 | 0.0028 | 0.011 | 0.011 | 0.0019 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.01 | 0.011 | 0.0021 | 0.0098 | 0.011 | 0.002 | -0.029 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 7.1.1.1 | Proton-translocating NAD(P)(+) transhydrogenase. | 0.16 | 0.5 | 1 | 0.43 | 1 | 1 | 0.51 | 0.95 | 1 | 0.16 | 0.48 | 1 | 0.17 | 0.87 | 1 | 0.19 | 0.72 | 1 | 0.0025 | 0.0022 | 0.0014 | 0.00095 | 0.00033 | 0.0011 | 0.0014 | 0.0015 | 0.0011 | 0.56 | 32 / 36 (89%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.0032 | 0.0014 | 0.0061 | 0.0026 | 0.0014 | 0.0024 | -0.3 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 1.12.98.1 | Coenzyme F420 hydrogenase. | 0.99 | 1 | 1 | 0.67 | 1 | 1 | 0.95 | 1 | 1 | 0.86 | 0.95 | 1 | 0.93 | 0.99 | 1 | 0.64 | 0.92 | 1 | 0.00078 | 0.00011 | 0 | 0.00015 | 0 | 0.00039 | 5.7e-05 | 0 | 0.00015 | -1.4 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0.00015 | 0 | 0.00047 | 7.2e-05 | 0 | 0.00024 | -1.1 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 2.6.1.33 | dTDP-4-amino-4.6-dideoxy-D-glucose transaminase. | 0.99 | 1 | 1 | 0.31 | 1 | 1 | 0.79 | 1 | 1 | 0.79 | 0.92 | 1 | 0.46 | 0.92 | 1 | 0.98 | 0.99 | 1 | 0.00011 | 2.4e-05 | 0 | 4.2e-05 | 0 | 1e-04 | 3.9e-06 | 0 | 1e-05 | -3.4 | 8 / 36 (22%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 3.6e-05 | 0 | 8.1e-05 | 1.3e-05 | 0 | 3.3e-05 | -1.5 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 3.5.2.14 | N-methylhydantoinase (ATP-hydrolyzing). | 0.2 | 0.53 | 1 | 0.031 | 0.97 | 1 | 0.029 | 0.71 | 1 | 0.14 | 0.47 | 1 | 0.026 | 0.76 | 1 | 0.018 | 0.55 | 1 | 0.00016 | 4.5e-05 | 0 | 2.3e-06 | 0 | 6.1e-06 | 5.3e-05 | 0 | 6.7e-05 | 4.5 | 10 / 36 (28%) | 1 / 7 (14%) | 3 / 7 (43%) | 0.143 | 0.429 | 5.6e-05 | 0 | 0.00012 | 5.6e-05 | 0 | 0.00016 | 0 | 4 / 11 (36%) | 2 / 11 (18%) | 0.364 | 0.182 |
| 3.1.3.45 | 3-deoxy-manno-octulosonate-8-phosphatase. | 0.37 | 0.65 | 1 | 0.93 | 1 | 1 | 0.62 | 0.96 | 1 | 0.63 | 0.84 | 1 | 0.59 | 0.93 | 1 | 0.75 | 0.95 | 1 | 0.001 | 0.00094 | 0.00056 | 0.001 | 0.00059 | 0.00081 | 0.0012 | 0.00078 | 0.0011 | 0.26 | 34 / 36 (94%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00063 | 0.00052 | 0.00044 | 0.001 | 4e-04 | 0.0013 | 0.67 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 2.6.1.76 | Diaminobutyrate--2-oxoglutarate transaminase. | 0.26 | 0.56 | 1 | 0.45 | 1 | 1 | 0.91 | 1 | 1 | 0.33 | 0.6 | 1 | 0.54 | 0.93 | 1 | 0.73 | 0.94 | 1 | 0.00067 | 0.00058 | 0.00042 | 0.00048 | 0.00043 | 0.00039 | 3e-04 | 0.00025 | 0.00033 | -0.68 | 31 / 36 (86%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 8e-04 | 0.00056 | 0.00062 | 6e-04 | 4e-04 | 7e-04 | -0.42 | 10 / 11 (91%) | 9 / 11 (82%) | 0.909 | 0.818 |
| 1.13.12.- | 0.88 | 0.95 | 1 | 0.88 | 1 | 1 | 0.37 | 0.9 | 1 | 0.69 | 0.87 | 1 | 0.77 | 0.97 | 1 | 0.29 | 0.78 | 1 | 0.00012 | 1.3e-05 | 0 | 1.7e-05 | 0 | 4.5e-05 | 2.3e-05 | 0 | 6.1e-05 | 0.44 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 1.8e-05 | 0 | 4.3e-05 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 | |
| 1.1.1.405 | Ribitol-5-phosphate 2-dehydrogenase (NADP(+)). | 0.91 | 0.96 | 1 | 0.65 | 1 | 1 | 0.68 | 0.98 | 1 | 0.55 | 0.78 | 1 | 0.32 | 0.9 | 1 | 0.25 | 0.76 | 1 | 0.00079 | 0.00075 | 0.00071 | 0.00079 | 0.00079 | 0.00046 | 0.00081 | 0.00071 | 0.00042 | 0.036 | 34 / 36 (94%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.00069 | 0.00064 | 0.00038 | 0.00075 | 0.00065 | 0.00058 | 0.12 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 1.1.1.44 | Phosphogluconate dehydrogenase (NADP(+)-dependent. decarboxylating). | 0.44 | 0.69 | 1 | 0.69 | 1 | 1 | 0.99 | 1 | 1 | 0.38 | 0.65 | 1 | 0.36 | 0.9 | 1 | 0.8 | 0.96 | 1 | 0.0051 | 0.0051 | 0.0039 | 0.0036 | 0.0035 | 0.002 | 0.004 | 0.0046 | 0.0021 | 0.15 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0062 | 0.003 | 0.0095 | 0.0058 | 0.0046 | 0.0042 | -0.096 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 7.2.1.1 | NADH:ubiquinone reductase (Na(+)-transporting). | 0.33 | 0.61 | 1 | 0.55 | 1 | 1 | 0.16 | 0.85 | 1 | 0.17 | 0.49 | 1 | 0.39 | 0.9 | 1 | 0.38 | 0.82 | 1 | 0.0053 | 0.0051 | 0.0025 | 0.0048 | 0.0042 | 0.0028 | 0.005 | 0.0034 | 0.0065 | 0.059 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0035 | 0.0019 | 0.0049 | 0.0071 | 0.0026 | 0.0095 | 1 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 3.5.1.59 | N-carbamoylsarcosine amidase. | 0.16 | 0.5 | 1 | 0.62 | 1 | 1 | 0.78 | 1 | 1 | 0.25 | 0.54 | 1 | 0.74 | 0.96 | 1 | 0.95 | 0.99 | 1 | 6.2e-05 | 1.4e-05 | 0 | 2.8e-05 | 0 | 5.1e-05 | 2.3e-05 | 0 | 5.1e-05 | -0.28 | 8 / 36 (22%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 5.8e-06 | 0 | 1.3e-05 | 6.8e-06 | 0 | 2.2e-05 | 0.23 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 1.7.1.15 | Nitrite reductase (NADH). | 0.2 | 0.53 | 1 | 0.25 | 1 | 1 | 0.24 | 0.89 | 1 | 0.39 | 0.65 | 1 | 0.76 | 0.97 | 1 | 0.66 | 0.92 | 1 | 0.00076 | 0.00015 | 0 | 0.00044 | 0 | 0.0012 | 0.00026 | 0 | 0.00069 | -0.76 | 7 / 36 (19%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 1.1e-05 | 0 | 2.9e-05 | 2.4e-05 | 0 | 5.3e-05 | 1.1 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 5.3.1.3 | D-arabinose isomerase. | 0.026 | 0.49 | 1 | 0.0063 | 0.7 | 1 | 0.02 | 0.71 | 1 | 0.014 | 0.42 | 1 | 0.036 | 0.8 | 1 | 0.075 | 0.66 | 1 | 0.00056 | 9.4e-05 | 0 | 0.00035 | 0 | 0.00079 | 0.00012 | 0 | 0.00031 | -1.5 | 6 / 36 (17%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 8.1e-06 | 0 | 2.7e-05 | 1.6 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.4.11.4 | Tripeptide aminopeptidase. | 0.55 | 0.77 | 1 | 0.63 | 1 | 1 | 0.26 | 0.89 | 1 | 0.32 | 0.59 | 1 | 0.092 | 0.86 | 1 | 0.022 | 0.56 | 1 | 0.018 | 0.018 | 0.017 | 0.018 | 0.016 | 0.0042 | 0.018 | 0.017 | 0.0052 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.019 | 0.018 | 0.0035 | 0.017 | 0.017 | 0.0023 | -0.16 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.1.3.7 | UDP-N-acetylglucosamine 4-epimerase. | 0.068 | 0.49 | 1 | 0.29 | 1 | 1 | 0.048 | 0.71 | 1 | 0.034 | 0.43 | 1 | 0.21 | 0.88 | 1 | 0.048 | 0.65 | 1 | 0.00035 | 0.00011 | 0 | 0.00018 | 8e-05 | 3e-04 | 0.00017 | 0 | 0.00033 | -0.082 | 11 / 36 (31%) | 4 / 7 (57%) | 3 / 7 (43%) | 0.571 | 0.429 | 2.1e-05 | 0 | 6.9e-05 | 0.00011 | 0 | 0.00033 | 2.4 | 1 / 11 (9.1%) | 3 / 11 (27%) | 0.0909 | 0.273 |
| 4.1.3.27 | Anthranilate synthase. | 0.064 | 0.49 | 1 | 0.38 | 1 | 1 | 0.45 | 0.93 | 1 | 0.072 | 0.43 | 1 | 0.099 | 0.86 | 1 | 0.16 | 0.71 | 1 | 0.023 | 0.023 | 0.023 | 0.019 | 0.019 | 0.0027 | 0.021 | 0.021 | 0.0061 | 0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.025 | 0.025 | 0.011 | 0.024 | 0.024 | 0.0043 | -0.059 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.11.15 | Aminopeptidase Y. | 0.46 | 0.7 | 1 | 0.21 | 1 | 1 | 0.019 | 0.71 | 1 | 0.3 | 0.58 | 1 | 0.13 | 0.87 | 1 | 0.015 | 0.55 | 1 | 0.00054 | 3e-04 | 7.3e-05 | 0.00024 | 0.00011 | 0.00034 | 9.2e-05 | 0 | 2e-04 | -1.4 | 20 / 36 (56%) | 5 / 7 (71%) | 2 / 7 (29%) | 0.714 | 0.286 | 0.00015 | 0 | 0.00025 | 0.00063 | 0.00014 | 0.0012 | 2.1 | 4 / 11 (36%) | 9 / 11 (82%) | 0.364 | 0.818 |
| 5.4.1.- | 0.51 | 0.74 | 1 | 0.3 | 1 | 1 | 0.22 | 0.88 | 1 | 0.89 | 0.97 | 1 | 0.65 | 0.95 | 1 | 0.61 | 0.91 | 1 | 0.0044 | 0.0044 | 0.0044 | 0.0047 | 0.0047 | 0.0011 | 0.0044 | 0.0046 | 0.002 | -0.095 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0043 | 0.0043 | 0.0014 | 0.0045 | 0.0038 | 0.0016 | 0.066 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 6.3.4.6 | Urea carboxylase. | 0.22 | 0.54 | 1 | 0.77 | 1 | 1 | 0.64 | 0.97 | 1 | 0.18 | 0.5 | 1 | 0.87 | 0.99 | 1 | 0.54 | 0.9 | 1 | 0.0026 | 0.0025 | 0.00068 | 0.0011 | 0.00056 | 0.0013 | 0.0016 | 0.00068 | 0.0023 | 0.54 | 34 / 36 (94%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.0033 | 0.0012 | 0.0058 | 0.0031 | 0.00067 | 0.004 | -0.09 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 7.5.2.12 | ABC-type L-arabinose transporter. | 0.29 | 0.57 | 1 | 0.1 | 1 | 1 | 0.49 | 0.95 | 1 | 0.3 | 0.58 | 1 | 0.096 | 0.86 | 1 | 0.45 | 0.86 | 1 | 3e-04 | 3.4e-05 | 0 | 5e-05 | 0 | 0.00013 | 1e-04 | 0 | 0.00023 | 1 | 4 / 36 (11%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 0 | 0 | 0 | 1.3e-05 | 0 | 4.5e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 4.2.2.2 | Pectate lyase. | 0.44 | 0.69 | 1 | 0.44 | 1 | 1 | 0.22 | 0.88 | 1 | 0.13 | 0.47 | 1 | 0.42 | 0.91 | 1 | 0.13 | 0.68 | 1 | 0.0014 | 0.0013 | 0.00058 | 0.0013 | 0.00089 | 0.0012 | 0.00093 | 0.00068 | 0.00093 | -0.48 | 32 / 36 (89%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.0011 | 0.00064 | 0.0019 | 0.0016 | 0.00047 | 0.0027 | 0.54 | 8 / 11 (73%) | 11 / 11 (100%) | 0.727 | 1 |
| 3.1.3.70 | Mannosyl-3-phosphoglycerate phosphatase. | 0.1 | 0.49 | 1 | 0.74 | 1 | 1 | 0.59 | 0.96 | 1 | 0.11 | 0.47 | 1 | 0.86 | 0.99 | 1 | 0.88 | 0.97 | 1 | 0.002 | 0.0016 | 0.00072 | 4e-04 | 4e-04 | 0.00035 | 0.00071 | 0.00048 | 0.001 | 0.83 | 28 / 36 (78%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 0.0028 | 0.00087 | 0.0061 | 0.0017 | 0.0012 | 0.0019 | -0.72 | 10 / 11 (91%) | 9 / 11 (82%) | 0.909 | 0.818 |
| 1.1.1.76 | (S.S)-butanediol dehydrogenase. | 0.2 | 0.53 | 1 | 0.38 | 1 | 1 | 0.32 | 0.89 | 1 | 0.17 | 0.49 | 1 | 0.25 | 0.88 | 1 | 0.25 | 0.76 | 1 | 0.00012 | 1.7e-05 | 0 | 0 | 0 | 0 | 1.2e-05 | 0 | 3.1e-05 | Inf | 5 / 36 (14%) | 0 / 7 (0%) | 1 / 7 (14%) | 0 | 0.143 | 3.2e-05 | 0 | 8e-05 | 1.7e-05 | 0 | 3.8e-05 | -0.91 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 3.5.1.49 | Formamidase. | 1 | 1 | 1 | 0.16 | 1 | 1 | 0.43 | 0.93 | 1 | 0.87 | 0.95 | 1 | 0.3 | 0.89 | 1 | 0.43 | 0.85 | 1 | 0.00023 | 0.00014 | 8.5e-05 | 0.00015 | 0.00011 | 0.00019 | 8.5e-05 | 6.8e-05 | 0.00013 | -0.82 | 22 / 36 (61%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 0.00018 | 9.7e-05 | 0.00024 | 0.00013 | 8.9e-05 | 0.00016 | -0.47 | 6 / 11 (55%) | 7 / 11 (64%) | 0.545 | 0.636 |
| 2.8.3.11 | Citramalate CoA-transferase. | 0.92 | 0.96 | 1 | 0.99 | 1 | 1 | 0.74 | 0.99 | 1 | 0.85 | 0.95 | 1 | 0.64 | 0.95 | 1 | 0.92 | 0.98 | 1 | 0.00036 | 0.00018 | 4.4e-05 | 0.00016 | 0 | 0.00025 | 0.00012 | 0.00013 | 0.00013 | -0.42 | 18 / 36 (50%) | 3 / 7 (43%) | 4 / 7 (57%) | 0.429 | 0.571 | 0.00019 | 0 | 0.00031 | 0.00022 | 8.9e-05 | 0.00031 | 0.21 | 5 / 11 (45%) | 6 / 11 (55%) | 0.455 | 0.545 |
| 4.2.1.147 | 5.6.7.8-tetrahydromethanopterin hydro-lyase. | 0.76 | 0.89 | 1 | 0.37 | 1 | 1 | 0.79 | 1 | 1 | 0.69 | 0.87 | 1 | 0.75 | 0.97 | 1 | 0.44 | 0.85 | 1 | 0.00025 | 4.1e-05 | 0 | 4.9e-05 | 0 | 0.00013 | 1.8e-05 | 0 | 4.7e-05 | -1.4 | 6 / 36 (17%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 7e-05 | 0 | 0.00017 | 2.3e-05 | 0 | 5.2e-05 | -1.6 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 1.7.2.3 | Trimethylamine-N-oxide reductase. | 0.088 | 0.49 | 1 | 0.22 | 1 | 1 | 0.14 | 0.83 | 1 | 0.037 | 0.43 | 1 | 0.076 | 0.86 | 1 | 0.063 | 0.66 | 1 | 0.00071 | 0.00027 | 0 | 0.00057 | 8.1e-05 | 0.0013 | 0.00069 | 0 | 0.0018 | 0.28 | 14 / 36 (39%) | 5 / 7 (71%) | 2 / 7 (29%) | 0.714 | 0.286 | 3e-05 | 0 | 6.2e-05 | 6.8e-05 | 0 | 0.00012 | 1.2 | 3 / 11 (27%) | 4 / 11 (36%) | 0.273 | 0.364 |
| 3.4.22.40 | Bleomycin hydrolase. | 0.2 | 0.53 | 1 | 0.63 | 1 | 1 | 0.85 | 1 | 1 | 0.15 | 0.47 | 1 | 0.2 | 0.87 | 1 | 0.52 | 0.9 | 1 | 0.0076 | 0.0076 | 0.0052 | 0.0044 | 0.0035 | 0.0035 | 0.005 | 0.0043 | 0.003 | 0.18 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0098 | 0.0063 | 0.014 | 0.0091 | 0.0068 | 0.0071 | -0.11 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.1.104 | Peptidoglycan-N-acetylglucosamine deacetylase. | 0.56 | 0.77 | 1 | 0.97 | 1 | 1 | 0.75 | 0.99 | 1 | 0.5 | 0.74 | 1 | 0.92 | 0.99 | 1 | 0.9 | 0.98 | 1 | 7e-04 | 7e-04 | 0.00026 | 0.00068 | 0.00056 | 0.00057 | 0.00075 | 0.00024 | 0.00084 | 0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00053 | 0.00029 | 0.00067 | 0.00084 | 0.00023 | 0.0014 | 0.66 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.1.80 | Protein-glutamate O-methyltransferase. | 0.15 | 0.5 | 1 | 0.68 | 1 | 1 | 0.17 | 0.85 | 1 | 0.091 | 0.45 | 1 | 0.63 | 0.95 | 1 | 0.077 | 0.66 | 1 | 0.0013 | 0.0013 | 9e-04 | 0.0014 | 0.0013 | 0.00062 | 0.0014 | 0.001 | 0.0011 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00097 | 0.00058 | 0.0013 | 0.0014 | 0.00082 | 0.0012 | 0.53 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.1.96 | Succinylglutamate desuccinylase. | 0.084 | 0.49 | 1 | 0.11 | 1 | 1 | 0.39 | 0.91 | 1 | 0.078 | 0.44 | 1 | 0.29 | 0.89 | 1 | 0.74 | 0.95 | 1 | 0.00014 | 1.5e-05 | 0 | 4.5e-05 | 0 | 8.9e-05 | 2.9e-05 | 0 | 7.7e-05 | -0.63 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.3.1.5 | Arylamine N-acetyltransferase. | 0.66 | 0.84 | 1 | 0.95 | 1 | 1 | 0.36 | 0.9 | 1 | 0.9 | 0.97 | 1 | 0.68 | 0.95 | 1 | 0.4 | 0.83 | 1 | 0.00076 | 0.00074 | 0.00057 | 0.00075 | 0.00026 | 7e-04 | 0.00064 | 0.00061 | 0.00034 | -0.23 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.001 | 0.00063 | 0.0011 | 0.00054 | 0.00028 | 0.00054 | -0.89 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 2.7.7.43 | N-acylneuraminate cytidylyltransferase. | 0.062 | 0.49 | 1 | 0.32 | 1 | 1 | 0.096 | 0.81 | 1 | 0.11 | 0.47 | 1 | 0.44 | 0.91 | 1 | 0.25 | 0.76 | 1 | 0.00081 | 0.00072 | 0.00038 | 0.0011 | 0.00056 | 0.0011 | 0.00074 | 0.00053 | 0.00078 | -0.57 | 32 / 36 (89%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00036 | 0.00029 | 0.00036 | 0.00083 | 0.00038 | 0.001 | 1.2 | 9 / 11 (82%) | 10 / 11 (91%) | 0.818 | 0.909 |
| 1.6.99.- | 0.34 | 0.62 | 1 | 0.55 | 1 | 1 | 0.23 | 0.88 | 1 | 0.14 | 0.47 | 1 | 0.83 | 0.98 | 1 | 0.33 | 0.8 | 1 | 0.0044 | 0.0044 | 0.0042 | 0.0037 | 0.0036 | 0.0012 | 0.0034 | 0.0033 | 0.0015 | -0.12 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0047 | 0.0036 | 0.0029 | 0.0051 | 0.0046 | 0.0015 | 0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 2.4.1.230 | Kojibiose phosphorylase. | 0.014 | 0.49 | 1 | 0.12 | 1 | 1 | 0.0074 | 0.61 | 1 | 0.058 | 0.43 | 1 | 0.17 | 0.87 | 1 | 0.0039 | 0.48 | 1 | 0.0017 | 0.0017 | 0.0013 | 0.0024 | 0.0024 | 0.00096 | 0.003 | 0.0029 | 0.002 | 0.32 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0013 | 0.0013 | 0.00055 | 9e-04 | 0.00091 | 0.00043 | -0.53 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.1.3.10 | CDP-paratose 2-epimerase. | 0.053 | 0.49 | 1 | 1 | 1 | 1 | 0.4 | 0.91 | 1 | 0.041 | 0.43 | 1 | 0.85 | 0.98 | 1 | 0.35 | 0.81 | 1 | 0.00013 | 1.8e-05 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 5 / 36 (14%) | 0 / 7 (0%) | 0 / 7 (0%) | 0 | 0 | 4.8e-05 | 0 | 8.9e-05 | 1.2e-05 | 0 | 3e-05 | -2 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 1.1.1.381 | 3-hydroxy acid dehydrogenase. | 0.27 | 0.56 | 1 | 0.22 | 1 | 1 | 0.12 | 0.82 | 1 | 0.35 | 0.62 | 1 | 0.13 | 0.87 | 1 | 0.065 | 0.66 | 1 | 0.00031 | 3.5e-05 | 0 | 8.4e-05 | 0 | 0.00022 | 9.1e-05 | 0 | 0.00023 | 0.12 | 4 / 36 (11%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 3e-06 | 0 | 9.9e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.5.1.7 | UDP-N-acetylglucosamine 1-carboxyvinyltransferase. | 0.025 | 0.49 | 1 | 0.55 | 1 | 1 | 0.53 | 0.95 | 1 | 0.08 | 0.44 | 1 | 0.53 | 0.93 | 1 | 0.072 | 0.66 | 1 | 0.032 | 0.032 | 0.033 | 0.029 | 0.028 | 0.0055 | 0.029 | 0.027 | 0.0088 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.036 | 0.037 | 0.0049 | 0.033 | 0.032 | 0.0045 | -0.13 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.7.76 | Molybdenum cofactor cytidylyltransferase. | 0.13 | 0.5 | 1 | 0.77 | 1 | 1 | 0.14 | 0.83 | 1 | 0.15 | 0.47 | 1 | 0.79 | 0.97 | 1 | 0.29 | 0.78 | 1 | 0.00022 | 4.3e-05 | 0 | 1e-04 | 0 | 0.00027 | 7.2e-05 | 0 | 0.00018 | -0.47 | 7 / 36 (19%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0 | 0 | 0 | 3e-05 | 0 | 6.9e-05 | Inf | 0 / 11 (0%) | 3 / 11 (27%) | 0 | 0.273 |
| 6.2.1.5 | Succinate--CoA ligase (ADP-forming). | 0.25 | 0.55 | 1 | 0.93 | 1 | 1 | 0.74 | 0.99 | 1 | 0.55 | 0.78 | 1 | 0.56 | 0.93 | 1 | 0.59 | 0.91 | 1 | 0.004 | 0.0039 | 0.0026 | 0.0023 | 0.0026 | 0.0021 | 0.0025 | 0.0012 | 0.0028 | 0.12 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0056 | 0.0031 | 0.0092 | 0.0041 | 0.0026 | 0.0035 | -0.45 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 3.6.3.43 | Transferred entry: 7.4.2.5. | 0.79 | 0.9 | 1 | 0.59 | 1 | 1 | 0.42 | 0.93 | 1 | 0.9 | 0.97 | 1 | 0.91 | 0.99 | 1 | 0.49 | 0.87 | 1 | 0.00039 | 0.00021 | 4.2e-05 | 0.00027 | 0 | 0.00047 | 0.00011 | 5.4e-05 | 0.00013 | -1.3 | 19 / 36 (53%) | 3 / 7 (43%) | 4 / 7 (57%) | 0.429 | 0.571 | 0.00022 | 0 | 0.00036 | 0.00022 | 0.00014 | 0.00036 | 0 | 4 / 11 (36%) | 8 / 11 (73%) | 0.364 | 0.727 |
| 3.4.24.3 | Microbial collagenase. | 0.014 | 0.49 | 1 | 0.46 | 1 | 1 | 0.027 | 0.71 | 1 | 0.024 | 0.42 | 1 | 0.46 | 0.92 | 1 | 0.044 | 0.65 | 1 | 2e-04 | 8.9e-05 | 0 | 2e-05 | 0 | 4.2e-05 | 6.7e-05 | 0 | 0.00014 | 1.7 | 16 / 36 (44%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0.00018 | 2e-04 | 0.00019 | 5.1e-05 | 0 | 6.8e-05 | -1.8 | 7 / 11 (64%) | 5 / 11 (45%) | 0.636 | 0.455 |
| 2.3.1.- | 0.77 | 0.89 | 1 | 0.44 | 1 | 1 | 0.42 | 0.92 | 1 | 0.29 | 0.58 | 1 | 0.85 | 0.98 | 1 | 1 | 1 | 1 | 0.046 | 0.046 | 0.043 | 0.045 | 0.04 | 0.012 | 0.042 | 0.04 | 0.011 | -0.1 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.048 | 0.042 | 0.022 | 0.048 | 0.048 | 0.012 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 6.2.1.9 | Malate--CoA ligase. | 0.29 | 0.57 | 1 | 0.22 | 1 | 1 | 0.69 | 0.98 | 1 | 0.57 | 0.8 | 1 | 0.65 | 0.95 | 1 | 0.66 | 0.92 | 1 | 0.00021 | 1.7e-05 | 0 | 6.1e-05 | 0 | 0.00016 | 1.3e-05 | 0 | 3.5e-05 | -2.2 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 9.5e-06 | 0 | 3.2e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.4.1.25 | 4-alpha-glucanotransferase. | 0.19 | 0.53 | 1 | 0.71 | 1 | 1 | 0.85 | 1 | 1 | 0.14 | 0.47 | 1 | 0.62 | 0.95 | 1 | 0.6 | 0.91 | 1 | 0.044 | 0.044 | 0.043 | 0.04 | 0.04 | 0.011 | 0.038 | 0.036 | 0.0096 | -0.074 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.049 | 0.043 | 0.024 | 0.047 | 0.045 | 0.0087 | -0.06 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.4.25 | GTP cyclohydrolase II. | 0.62 | 0.82 | 1 | 0.55 | 1 | 1 | 0.8 | 1 | 1 | 0.47 | 0.72 | 1 | 0.2 | 0.87 | 1 | 0.64 | 0.92 | 1 | 0.01 | 0.01 | 0.011 | 0.0095 | 0.009 | 0.002 | 0.01 | 0.0089 | 0.0027 | 0.074 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.01 | 0.011 | 0.0025 | 0.011 | 0.012 | 0.0026 | 0.14 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.3.37 | Sedoheptulose-bisphosphatase. | 0.26 | 0.56 | 1 | 0.69 | 1 | 1 | 0.96 | 1 | 1 | 0.2 | 0.5 | 1 | 0.47 | 0.92 | 1 | 0.93 | 0.98 | 1 | 0.0017 | 0.0014 | 0.00054 | 0.00071 | 0.00016 | 0.0011 | 9e-04 | 0.00036 | 0.0015 | 0.34 | 30 / 36 (83%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.002 | 0.00053 | 0.0039 | 0.0017 | 0.00075 | 0.0016 | -0.23 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 2.5.1.72 | Quinolinate synthase. | 0.26 | 0.56 | 1 | 0.33 | 1 | 1 | 0.95 | 1 | 1 | 0.26 | 0.55 | 1 | 0.02 | 0.76 | 1 | 0.15 | 0.7 | 1 | 0.011 | 0.011 | 0.012 | 0.01 | 0.01 | 0.0028 | 0.011 | 0.012 | 0.0034 | 0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.011 | 0.0015 | 0.012 | 0.012 | 0.0016 | 0.13 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.202 | 1.3-propanediol dehydrogenase. | 0.75 | 0.88 | 1 | 0.75 | 1 | 1 | 1 | 1 | 1 | 0.45 | 0.7 | 1 | 0.78 | 0.97 | 1 | 0.84 | 0.96 | 1 | 0.0066 | 0.0066 | 0.0066 | 0.0062 | 0.0059 | 0.0022 | 0.007 | 0.0059 | 0.0041 | 0.18 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0064 | 0.0066 | 0.00081 | 0.0067 | 0.0069 | 0.0014 | 0.066 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.1.9 | Dihydroxy-acid dehydratase. | 0.31 | 0.59 | 1 | 0.6 | 1 | 1 | 0.64 | 0.97 | 1 | 0.24 | 0.54 | 1 | 0.58 | 0.93 | 1 | 0.51 | 0.89 | 1 | 0.03 | 0.03 | 0.03 | 0.028 | 0.028 | 0.0047 | 0.027 | 0.026 | 0.0075 | -0.052 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.031 | 0.032 | 0.007 | 0.031 | 0.032 | 0.0052 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.7.5.1 | Nitrate reductase (quinone). | 0.24 | 0.55 | 1 | 0.83 | 1 | 1 | 0.94 | 1 | 1 | 0.36 | 0.63 | 1 | 0.91 | 0.99 | 1 | 0.95 | 0.99 | 1 | 0.00064 | 0.00021 | 0 | 0.00045 | 0 | 0.0011 | 0.00054 | 0 | 0.0013 | 0.26 | 12 / 36 (33%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 3.2e-05 | 0 | 5.7e-05 | 3.9e-05 | 0 | 6.9e-05 | 0.29 | 3 / 11 (27%) | 3 / 11 (27%) | 0.273 | 0.273 |
| 3.1.1.47 | 1-alkyl-2-acetylglycerophosphocholine esterase. | 0.071 | 0.49 | 1 | 0.97 | 1 | 1 | 0.88 | 1 | 1 | 0.096 | 0.45 | 1 | 0.78 | 0.97 | 1 | 0.8 | 0.96 | 1 | 0.0015 | 0.0011 | 0.00025 | 0.00015 | 0.00022 | 0.00013 | 0.00021 | 6.8e-05 | 0.00034 | 0.49 | 27 / 36 (75%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 0.0022 | 0.00052 | 0.0053 | 0.0013 | 0.00091 | 0.0014 | -0.76 | 9 / 11 (82%) | 9 / 11 (82%) | 0.818 | 0.818 |
| 3.1.3.5 | 5'-nucleotidase. | 0.37 | 0.65 | 1 | 0.71 | 1 | 1 | 0.38 | 0.9 | 1 | 0.13 | 0.47 | 1 | 0.6 | 0.94 | 1 | 0.96 | 0.99 | 1 | 0.015 | 0.015 | 0.015 | 0.014 | 0.015 | 0.0025 | 0.014 | 0.012 | 0.0041 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.016 | 0.015 | 0.0046 | 0.017 | 0.016 | 0.003 | 0.087 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.1.1.6 | Lysine--tRNA ligase. | 0.083 | 0.49 | 1 | 0.72 | 1 | 1 | 0.66 | 0.97 | 1 | 0.058 | 0.43 | 1 | 0.7 | 0.95 | 1 | 0.23 | 0.75 | 1 | 0.032 | 0.032 | 0.032 | 0.029 | 0.027 | 0.0063 | 0.028 | 0.025 | 0.0088 | -0.051 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.037 | 0.034 | 0.016 | 0.033 | 0.033 | 0.0057 | -0.17 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.53 | 3-alpha(or 20-beta)-hydroxysteroid dehydrogenase. | 0.19 | 0.53 | 1 | 0.25 | 1 | 1 | 0.21 | 0.87 | 1 | 0.2 | 0.5 | 1 | 0.55 | 0.93 | 1 | 0.47 | 0.87 | 1 | 0.00029 | 9.6e-05 | 0 | 2e-04 | 1.6e-05 | 0.00043 | 6e-05 | 0 | 1e-04 | -1.7 | 12 / 36 (33%) | 4 / 7 (57%) | 3 / 7 (43%) | 0.571 | 0.429 | 5.3e-05 | 0 | 0.00012 | 9.6e-05 | 0 | 0.00021 | 0.86 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 1.3.99.4 | 3-oxosteroid 1-dehydrogenase. | 0.076 | 0.49 | 1 | 0.18 | 1 | 1 | 0.03 | 0.71 | 1 | 0.15 | 0.47 | 1 | 0.25 | 0.88 | 1 | 0.073 | 0.66 | 1 | 0.0037 | 0.0037 | 0.0032 | 0.0046 | 0.0037 | 0.0025 | 0.0034 | 0.003 | 0.0028 | -0.44 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0027 | 0.002 | 0.0017 | 0.0044 | 0.0043 | 0.0026 | 0.7 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.1.293 | GalNAc(5)-diNAcBac-PP-undecaprenol beta-1.3-glucosyltransferase. | 0.19 | 0.53 | 1 | 0.89 | 1 | 1 | 0.89 | 1 | 1 | 0.29 | 0.58 | 1 | 0.48 | 0.92 | 1 | 0.72 | 0.94 | 1 | 0.00024 | 7.4e-05 | 0 | 0.00014 | 0 | 0.00027 | 0.00011 | 2.7e-05 | 0.00019 | -0.35 | 11 / 36 (31%) | 3 / 7 (43%) | 4 / 7 (57%) | 0.429 | 0.571 | 2.9e-05 | 0 | 8.8e-05 | 5.2e-05 | 0 | 0.00013 | 0.84 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 1.14.14.- | 0.044 | 0.49 | 1 | 0.13 | 1 | 1 | 0.043 | 0.71 | 1 | 0.028 | 0.43 | 1 | 0.18 | 0.87 | 1 | 0.054 | 0.66 | 1 | 0.00021 | 3.4e-05 | 0 | 8.7e-05 | 0 | 0.00018 | 6.8e-05 | 0 | 0.00018 | -0.36 | 6 / 36 (17%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 0 | 0 | 0 | 1.5e-05 | 0 | 3.3e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 | |
| 1.8.98.- | 0.43 | 0.69 | 1 | 0.55 | 1 | 1 | 0.71 | 0.98 | 1 | 0.22 | 0.52 | 1 | 0.52 | 0.93 | 1 | 0.54 | 0.9 | 1 | 0.0056 | 0.0056 | 0.0049 | 0.0047 | 0.0045 | 0.003 | 0.0056 | 0.0047 | 0.0038 | 0.25 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0057 | 0.005 | 0.0027 | 0.0062 | 0.0051 | 0.0046 | 0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 2.4.1.13 | Sucrose synthase. | 0.12 | 0.5 | 1 | 0.12 | 1 | 1 | 0.1 | 0.82 | 1 | 0.052 | 0.43 | 1 | 0.048 | 0.81 | 1 | 0.033 | 0.62 | 1 | 0.00069 | 0.00053 | 0.00029 | 0.00027 | 0.00012 | 0.00037 | 6e-04 | 0.00071 | 0.00064 | 1.2 | 28 / 36 (78%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.00067 | 0.00042 | 0.00092 | 0.00053 | 0.00028 | 0.00052 | -0.34 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 2.7.1.190 | Aminoglycoside 2''-phosphotransferase. | 0.12 | 0.5 | 1 | 0.82 | 1 | 1 | 0.91 | 1 | 1 | 0.1 | 0.46 | 1 | 0.71 | 0.95 | 1 | 0.8 | 0.96 | 1 | 0.00035 | 0.00015 | 0 | 2e-04 | 0.00011 | 0.00029 | 0.00028 | 0.00013 | 0.00044 | 0.49 | 15 / 36 (42%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 5.9e-05 | 0 | 0.00011 | 0.00011 | 0 | 3e-04 | 0.9 | 3 / 11 (27%) | 3 / 11 (27%) | 0.273 | 0.273 |
| 4.2.2.- | 0.81 | 0.91 | 1 | 0.27 | 1 | 1 | 0.029 | 0.71 | 1 | 0.8 | 0.93 | 1 | 0.88 | 0.99 | 1 | 0.21 | 0.74 | 1 | 0.018 | 0.018 | 0.017 | 0.018 | 0.017 | 0.003 | 0.016 | 0.017 | 0.0027 | -0.17 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.018 | 0.018 | 0.0058 | 0.021 | 0.017 | 0.0086 | 0.22 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 3.6.4.6 | Vesicle-fusing ATPase. | 0.43 | 0.69 | 1 | 0.73 | 1 | 1 | 0.96 | 1 | 1 | 0.32 | 0.59 | 1 | 0.52 | 0.93 | 1 | 0.76 | 0.95 | 1 | 0.00017 | 1.9e-05 | 0 | 0 | 0 | 0 | 3.9e-06 | 0 | 1e-05 | Inf | 4 / 36 (11%) | 0 / 7 (0%) | 1 / 7 (14%) | 0 | 0.143 | 2.6e-05 | 0 | 8.8e-05 | 3.3e-05 | 0 | 9.9e-05 | 0.34 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 1.2.1.46 | Formaldehyde dehydrogenase. | 0.097 | 0.49 | 1 | 0.13 | 1 | 1 | 0.073 | 0.73 | 1 | 0.22 | 0.52 | 1 | 0.19 | 0.87 | 1 | 0.12 | 0.66 | 1 | 9.2e-05 | 7.7e-06 | 0 | 1.8e-05 | 0 | 4.2e-05 | 0 | 0 | 0 | -Inf | 3 / 36 (8.3%) | 2 / 7 (29%) | 0 / 7 (0%) | 0.286 | 0 | 0 | 0 | 0 | 1.4e-05 | 0 | 4.5e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 3.1.30.2 | Serratia marcescens nuclease. | 0.87 | 0.94 | 1 | 0.36 | 1 | 1 | 0.024 | 0.71 | 1 | 0.89 | 0.97 | 1 | 0.59 | 0.93 | 1 | 0.044 | 0.65 | 1 | 0.00015 | 3.7e-05 | 0 | 3.7e-05 | 0 | 9.7e-05 | 3.9e-06 | 0 | 1e-05 | -3.2 | 9 / 36 (25%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2e-05 | 0 | 4.5e-05 | 7.6e-05 | 0 | 0.00012 | 1.9 | 2 / 11 (18%) | 5 / 11 (45%) | 0.182 | 0.455 |
| 6.1.1.5 | Isoleucine--tRNA ligase. | 0.12 | 0.5 | 1 | 0.41 | 1 | 1 | 0.53 | 0.95 | 1 | 0.16 | 0.48 | 1 | 0.64 | 0.95 | 1 | 0.074 | 0.66 | 1 | 0.058 | 0.058 | 0.059 | 0.055 | 0.053 | 0.0078 | 0.053 | 0.054 | 0.012 | -0.053 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.063 | 0.062 | 0.011 | 0.057 | 0.058 | 0.01 | -0.14 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.3.1.27 | 6-phospho-3-hexuloisomerase. | 0.25 | 0.56 | 1 | 0.36 | 1 | 1 | 0.59 | 0.96 | 1 | 0.16 | 0.49 | 1 | 0.34 | 0.9 | 1 | 0.65 | 0.92 | 1 | 0.0011 | 0.0011 | 9e-04 | 0.00074 | 0.00069 | 0.00049 | 0.0012 | 0.00068 | 0.0011 | 0.7 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0013 | 0.00072 | 0.0014 | 0.0012 | 0.001 | 0.00055 | -0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.3.8.4 | Isovaleryl-CoA dehydrogenase. | 0.085 | 0.49 | 1 | 0.058 | 1 | 1 | 0.21 | 0.87 | 1 | 0.072 | 0.43 | 1 | 0.14 | 0.87 | 1 | 0.39 | 0.83 | 1 | 0.00037 | 7.2e-05 | 0 | 2e-04 | 0 | 0.00043 | 0.00014 | 0 | 0.00036 | -0.51 | 7 / 36 (19%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 1.4e-05 | 0 | 3.7e-05 | 8.1e-06 | 0 | 2.7e-05 | -0.79 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 2.7.1.165 | Glycerate 2-kinase. | 0.064 | 0.49 | 1 | 0.23 | 1 | 1 | 0.22 | 0.88 | 1 | 0.027 | 0.43 | 1 | 0.37 | 0.9 | 1 | 0.41 | 0.84 | 1 | 0.00041 | 0.00025 | 7.2e-05 | 0.00041 | 0.00024 | 0.00039 | 0.00022 | 0.00012 | 0.00033 | -0.9 | 22 / 36 (61%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00016 | 0 | 0.00038 | 0.00026 | 0 | 0.00057 | 0.7 | 5 / 11 (45%) | 5 / 11 (45%) | 0.455 | 0.455 |
| 1.1.99.- | 0.66 | 0.84 | 1 | 0.48 | 1 | 1 | 0.34 | 0.89 | 1 | 0.99 | 1 | 1 | 0.97 | 0.99 | 1 | 0.76 | 0.95 | 1 | 0.014 | 0.014 | 0.014 | 0.014 | 0.013 | 0.0046 | 0.013 | 0.014 | 0.0033 | -0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.013 | 0.014 | 0.0049 | 0.014 | 0.014 | 0.0049 | 0.11 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 4.1.99.12 | 3.4-dihydroxy-2-butanone-4-phosphate synthase. | 0.79 | 0.9 | 1 | 0.67 | 1 | 1 | 0.68 | 0.98 | 1 | 0.56 | 0.79 | 1 | 0.29 | 0.89 | 1 | 0.8 | 0.96 | 1 | 0.01 | 0.01 | 0.011 | 0.0096 | 0.0092 | 0.002 | 0.01 | 0.0091 | 0.0028 | 0.059 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.01 | 0.01 | 0.0025 | 0.011 | 0.012 | 0.0028 | 0.14 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.99.3 | Transferred entry: 3.5.4.43. | 0.75 | 0.88 | 1 | 0.51 | 1 | 1 | 0.5 | 0.95 | 1 | 0.82 | 0.93 | 1 | 0.94 | 0.99 | 1 | 0.91 | 0.98 | 1 | 0.00035 | 0.00017 | 0 | 0.00017 | 3.2e-05 | 0.00035 | 0.00037 | 0 | 0.00068 | 1.1 | 17 / 36 (47%) | 4 / 7 (57%) | 3 / 7 (43%) | 0.571 | 0.429 | 0.00012 | 0 | 0.00022 | 7.9e-05 | 0 | 0.00011 | -0.6 | 5 / 11 (45%) | 5 / 11 (45%) | 0.455 | 0.455 |
| 2.1.2.5 | Glutamate formimidoyltransferase. | 0.21 | 0.54 | 1 | 0.63 | 1 | 1 | 0.7 | 0.98 | 1 | 0.14 | 0.47 | 1 | 0.67 | 0.95 | 1 | 0.98 | 1 | 1 | 0.00087 | 0.00078 | 0.00053 | 9e-04 | 0.00096 | 0.00035 | 0.00082 | 0.00093 | 0.00067 | -0.13 | 32 / 36 (89%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00065 | 0.00038 | 0.00094 | 0.00079 | 0.00021 | 0.0011 | 0.28 | 10 / 11 (91%) | 8 / 11 (73%) | 0.909 | 0.727 |
| 2.1.4.2 | Scyllo-inosamine-4-phosphate amidinotransferase. | 0.43 | 0.69 | 1 | 0.15 | 1 | 1 | 1 | 1 | 1 | 0.6 | 0.81 | 1 | 0.2 | 0.87 | 1 | 0.76 | 0.95 | 1 | 0.00015 | 9.3e-05 | 8e-05 | 0.00015 | 0.00012 | 0.00011 | 8.1e-05 | 6.8e-05 | 8.7e-05 | -0.89 | 22 / 36 (61%) | 6 / 7 (86%) | 4 / 7 (57%) | 0.857 | 0.571 | 1e-04 | 8.7e-05 | 9.7e-05 | 5.5e-05 | 0 | 8.3e-05 | -0.86 | 8 / 11 (73%) | 4 / 11 (36%) | 0.727 | 0.364 |
| 2.4.2.53 | Undecaprenyl-phosphate 4-deoxy-4-formamido-L-arabinose transferase. | 0.78 | 0.9 | 1 | 0.18 | 1 | 1 | 0.14 | 0.83 | 1 | 0.47 | 0.72 | 1 | 0.023 | 0.76 | 1 | 0.014 | 0.55 | 1 | 0.0081 | 0.0081 | 0.0077 | 0.0079 | 0.0084 | 0.0026 | 0.0091 | 0.011 | 0.0029 | 0.2 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0082 | 0.0072 | 0.0029 | 0.0077 | 0.0074 | 0.0025 | -0.091 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.8.7 | Holo-[acyl-carrier-protein] synthase. | 0.22 | 0.54 | 1 | 0.54 | 1 | 1 | 0.34 | 0.89 | 1 | 0.13 | 0.47 | 1 | 0.38 | 0.9 | 1 | 0.24 | 0.76 | 1 | 0.0035 | 0.0035 | 0.0031 | 0.0027 | 0.0022 | 0.0012 | 0.0032 | 0.0032 | 0.0016 | 0.25 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0041 | 0.0031 | 0.0033 | 0.0034 | 0.0032 | 0.0016 | -0.27 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.6.-.- | 0.8 | 0.9 | 1 | 0.22 | 1 | 1 | 0.67 | 0.98 | 1 | 0.6 | 0.81 | 1 | 0.076 | 0.86 | 1 | 0.38 | 0.82 | 1 | 0.004 | 0.004 | 0.0033 | 0.0033 | 0.0034 | 0.0017 | 0.0043 | 0.0044 | 0.0018 | 0.38 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0039 | 0.003 | 0.0041 | 0.0043 | 0.0033 | 0.0029 | 0.14 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 3.1.3.74 | Pyridoxal phosphatase. | 0.42 | 0.68 | 1 | 0.14 | 1 | 1 | 0.18 | 0.85 | 1 | 0.14 | 0.47 | 1 | 0.26 | 0.88 | 1 | 0.21 | 0.74 | 1 | 0.00067 | 0.00052 | 0.00035 | 4e-04 | 0.00024 | 0.00059 | 0.00069 | 6e-04 | 0.00089 | 0.79 | 28 / 36 (78%) | 4 / 7 (57%) | 5 / 7 (71%) | 0.571 | 0.714 | 0.00052 | 0.00032 | 0.00076 | 0.00049 | 0.00045 | 0.00046 | -0.086 | 10 / 11 (91%) | 9 / 11 (82%) | 0.909 | 0.818 |
| 2.7.7.65 | Diguanylate cyclase. | 0.26 | 0.56 | 1 | 0.99 | 1 | 1 | 0.8 | 1 | 1 | 0.21 | 0.51 | 1 | 0.54 | 0.93 | 1 | 0.72 | 0.94 | 1 | 0.046 | 0.046 | 0.046 | 0.041 | 0.04 | 0.0077 | 0.042 | 0.048 | 0.015 | 0.035 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.048 | 0.045 | 0.016 | 0.049 | 0.051 | 0.011 | 0.03 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.8.4.2 | Protein-disulfide reductase (glutathione). | 0.63 | 0.82 | 1 | 0.4 | 1 | 1 | 0.78 | 1 | 1 | 0.83 | 0.94 | 1 | 0.19 | 0.87 | 1 | 0.46 | 0.86 | 1 | 0.0017 | 0.0016 | 0.0016 | 0.0018 | 0.0018 | 0.00046 | 0.0017 | 0.0016 | 0.00089 | -0.082 | 35 / 36 (97%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.0016 | 0.0017 | 0.00077 | 0.0015 | 0.0012 | 0.00099 | -0.093 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.4.1.2 | Acetyl-CoA carboxylase. | 0.27 | 0.56 | 1 | 0.098 | 1 | 1 | 0.073 | 0.73 | 1 | 0.18 | 0.49 | 1 | 0.0059 | 0.7 | 1 | 0.0031 | 0.48 | 1 | 0.015 | 0.015 | 0.015 | 0.014 | 0.013 | 0.0036 | 0.016 | 0.014 | 0.0045 | 0.19 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.015 | 0.016 | 0.0018 | 0.015 | 0.014 | 0.0031 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.8.3.10 | Citrate CoA-transferase. | 0.63 | 0.83 | 1 | 0.41 | 1 | 1 | 0.95 | 1 | 1 | 0.8 | 0.92 | 1 | 0.67 | 0.95 | 1 | 0.67 | 0.93 | 1 | 0.0025 | 0.0025 | 0.0024 | 0.0027 | 0.0026 | 0.00055 | 0.0023 | 0.0026 | 0.00072 | -0.23 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0027 | 0.0023 | 0.0019 | 0.0021 | 0.0022 | 0.001 | -0.36 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.10.1.1 | Molybdopterin molybdotransferase. | 0.38 | 0.65 | 1 | 0.99 | 1 | 1 | 0.54 | 0.95 | 1 | 0.29 | 0.58 | 1 | 0.58 | 0.93 | 1 | 0.21 | 0.74 | 1 | 0.014 | 0.014 | 0.014 | 0.013 | 0.014 | 0.0039 | 0.013 | 0.013 | 0.0054 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.015 | 0.015 | 0.0042 | 0.013 | 0.013 | 0.0042 | -0.21 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.21.87 | Transferred entry: 3.4.23.49. | 0.068 | 0.49 | 1 | 0.11 | 1 | 1 | 0.076 | 0.74 | 1 | 0.045 | 0.43 | 1 | 0.28 | 0.88 | 1 | 0.18 | 0.72 | 1 | 0.00048 | 5.4e-05 | 0 | 0.00015 | 0 | 0.00035 | 0.00011 | 0 | 0.00028 | -0.45 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 1e-05 | 0 | 3.4e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 5.4.3.9 | Glutamate 2.3-aminomutase. | 0.26 | 0.56 | 1 | 0.81 | 1 | 1 | 0.94 | 1 | 1 | 0.29 | 0.58 | 1 | 0.83 | 0.98 | 1 | 0.76 | 0.95 | 1 | 0.00051 | 0.00027 | 8.4e-05 | 0.00034 | 0.00014 | 0.00051 | 0.00039 | 0.00011 | 0.00051 | 0.2 | 19 / 36 (53%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.00012 | 0 | 0.00017 | 0.00029 | 0 | 0.00064 | 1.3 | 5 / 11 (45%) | 4 / 11 (36%) | 0.455 | 0.364 |
| 2.3.1.57 | Diamine N-acetyltransferase. | 0.14 | 0.5 | 1 | 0.72 | 1 | 1 | 0.89 | 1 | 1 | 0.085 | 0.44 | 1 | 0.19 | 0.87 | 1 | 0.53 | 0.9 | 1 | 0.0034 | 0.0034 | 0.0027 | 0.0021 | 0.0016 | 0.0014 | 0.0022 | 0.0015 | 0.0015 | 0.067 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0044 | 0.0027 | 0.0056 | 0.004 | 0.0038 | 0.0024 | -0.14 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.4.3.- | 0.55 | 0.77 | 1 | 0.41 | 1 | 1 | 0.25 | 0.89 | 1 | 0.53 | 0.77 | 1 | 0.57 | 0.93 | 1 | 0.31 | 0.79 | 1 | 0.00042 | 0.00041 | 3e-04 | 0.00045 | 0.00031 | 0.00038 | 0.00039 | 0.00025 | 0.00048 | -0.21 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00053 | 0.00059 | 3e-04 | 0.00028 | 0.00023 | 0.00017 | -0.92 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 | |
| 3.2.1.35 | Hyaluronoglucosaminidase. | 0.93 | 0.97 | 1 | 0.52 | 1 | 1 | 0.76 | 0.99 | 1 | 0.63 | 0.84 | 1 | 0.26 | 0.88 | 1 | 0.44 | 0.85 | 1 | 0.011 | 0.011 | 0.0088 | 0.012 | 0.0086 | 0.011 | 0.011 | 0.014 | 0.0079 | -0.13 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.0098 | 0.009 | 0.0097 | 0.0086 | 0.0053 | -0.18 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.1.23 | Ceramidase. | 0.26 | 0.56 | 1 | 0.55 | 1 | 1 | 0.07 | 0.73 | 1 | 0.25 | 0.55 | 1 | 0.37 | 0.9 | 1 | 0.05 | 0.65 | 1 | 0.00017 | 4.7e-05 | 0 | 3.4e-05 | 0 | 8.4e-05 | 4.8e-05 | 0 | 6.2e-05 | 0.5 | 10 / 36 (28%) | 2 / 7 (29%) | 3 / 7 (43%) | 0.286 | 0.429 | 9.7e-05 | 0 | 0.00015 | 3e-06 | 0 | 1e-05 | -5 | 4 / 11 (36%) | 1 / 11 (9.1%) | 0.364 | 0.0909 |
| 4.4.1.8 | Cystathionine beta-lyase. | 0.17 | 0.5 | 1 | 0.91 | 1 | 1 | 0.9 | 1 | 1 | 0.079 | 0.44 | 1 | 0.57 | 0.93 | 1 | 0.76 | 0.95 | 1 | 0.024 | 0.024 | 0.022 | 0.018 | 0.018 | 0.0075 | 0.019 | 0.018 | 0.0095 | 0.078 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.029 | 0.022 | 0.027 | 0.026 | 0.026 | 0.01 | -0.16 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.291 | 2-hydroxymethylglutarate dehydrogenase. | 0.22 | 0.54 | 1 | 0.36 | 1 | 1 | 0.55 | 0.95 | 1 | 0.35 | 0.62 | 1 | 0.77 | 0.97 | 1 | 0.9 | 0.98 | 1 | 0.00022 | 4.9e-05 | 0 | 0.00013 | 0 | 0.00031 | 5.6e-05 | 0 | 0.00013 | -1.2 | 8 / 36 (22%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 2.5e-05 | 0 | 7.2e-05 | 1.4e-05 | 0 | 3.2e-05 | -0.84 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 6.2.1.1 | Acetate--CoA ligase. | 0.75 | 0.88 | 1 | 0.59 | 1 | 1 | 0.19 | 0.86 | 1 | 0.5 | 0.74 | 1 | 0.73 | 0.96 | 1 | 0.21 | 0.74 | 1 | 0.0037 | 0.0036 | 0.0039 | 0.0034 | 0.0037 | 0.0015 | 0.0033 | 0.0048 | 0.0021 | -0.043 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0035 | 0.0029 | 0.0024 | 0.0042 | 0.0048 | 0.0023 | 0.26 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 3.5.2.9 | 5-oxoprolinase (ATP-hydrolyzing). | 0.72 | 0.87 | 1 | 0.75 | 1 | 1 | 0.24 | 0.89 | 1 | 0.95 | 0.98 | 1 | 0.57 | 0.93 | 1 | 0.13 | 0.68 | 1 | 0.004 | 0.004 | 0.0037 | 0.0043 | 0.0053 | 0.0015 | 0.0047 | 0.004 | 0.0027 | 0.13 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.004 | 0.0037 | 0.0015 | 0.0032 | 0.0032 | 0.0011 | -0.32 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.2.1.54 | D-alanine--[D-alanyl-carrier protein] ligase. | 0.37 | 0.64 | 1 | 0.18 | 1 | 1 | 0.1 | 0.82 | 1 | 0.59 | 0.81 | 1 | 0.09 | 0.86 | 1 | 0.085 | 0.66 | 1 | 0.00046 | 0.00037 | 0.00021 | 0.00024 | 0.00011 | 0.00033 | 4e-04 | 0.00032 | 0.00038 | 0.74 | 29 / 36 (81%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.00048 | 0.00025 | 0.00052 | 0.00033 | 2e-04 | 0.00041 | -0.54 | 8 / 11 (73%) | 8 / 11 (73%) | 0.727 | 0.727 |
| 1.3.99.1 | Deleted entry. | 0.94 | 0.97 | 1 | 0.95 | 1 | 1 | 0.77 | 0.99 | 1 | 0.43 | 0.69 | 1 | 0.3 | 0.89 | 1 | 0.45 | 0.86 | 1 | 0.018 | 0.018 | 0.018 | 0.017 | 0.015 | 0.0056 | 0.017 | 0.017 | 0.0053 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.017 | 0.018 | 0.0033 | 0.018 | 0.018 | 0.0053 | 0.082 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.2.22 | Pectate trisaccharide-lyase. | 0.12 | 0.5 | 1 | 0.49 | 1 | 1 | 0.28 | 0.89 | 1 | 0.21 | 0.51 | 1 | 0.95 | 0.99 | 1 | 0.66 | 0.92 | 1 | 0.00038 | 0.00011 | 0 | 0.00026 | 0 | 0.00063 | 7.4e-05 | 0 | 0.00011 | -1.8 | 10 / 36 (28%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 1.2e-05 | 0 | 3e-05 | 0.00012 | 0 | 0.00033 | 3.3 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 3.5.4.2 | Adenine deaminase. | 0.91 | 0.96 | 1 | 0.19 | 1 | 1 | 0.87 | 1 | 1 | 0.59 | 0.81 | 1 | 0.93 | 0.99 | 1 | 0.17 | 0.71 | 1 | 0.016 | 0.016 | 0.017 | 0.017 | 0.018 | 0.0025 | 0.016 | 0.015 | 0.004 | -0.087 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.017 | 0.017 | 0.0036 | 0.016 | 0.015 | 0.004 | -0.087 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.1.94 | Gamma-glutamyl-gamma-aminobutyrate hydrolase. | 0.29 | 0.57 | 1 | 0.34 | 1 | 1 | 0.33 | 0.89 | 1 | 0.23 | 0.53 | 1 | 0.15 | 0.87 | 1 | 0.26 | 0.76 | 1 | 0.0016 | 0.0015 | 0.0011 | 0.0011 | 0.00082 | 0.00079 | 0.00099 | 0.00053 | 0.00087 | -0.15 | 35 / 36 (97%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.0017 | 0.0011 | 0.0014 | 0.0019 | 0.0012 | 0.0018 | 0.16 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.14.14.5 | Alkanesulfonate monooxygenase. | 0.14 | 0.5 | 1 | 0.084 | 1 | 1 | 0.042 | 0.71 | 1 | 0.17 | 0.49 | 1 | 0.46 | 0.92 | 1 | 0.21 | 0.74 | 1 | 0.00038 | 6.4e-05 | 0 | 0.00019 | 0 | 0.00049 | 0.00012 | 0 | 0.00031 | -0.66 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 3e-06 | 0 | 9.9e-06 | 1.2e-05 | 0 | 2.7e-05 | 2 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 1.15.1.2 | Superoxide reductase. | 0.62 | 0.82 | 1 | 0.31 | 1 | 1 | 0.46 | 0.93 | 1 | 0.33 | 0.6 | 1 | 0.93 | 0.99 | 1 | 0.64 | 0.92 | 1 | 0.0043 | 0.0043 | 0.0039 | 0.0042 | 0.0039 | 0.0015 | 0.0038 | 0.0031 | 0.0013 | -0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0045 | 0.0044 | 0.001 | 0.0046 | 0.0038 | 0.0018 | 0.032 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.12.1.4 | Hydrogenase (NAD(+). ferredoxin). | 0.56 | 0.78 | 1 | 0.96 | 1 | 1 | 0.86 | 1 | 1 | 0.61 | 0.82 | 1 | 0.81 | 0.98 | 1 | 0.71 | 0.94 | 1 | 0.00011 | 1.9e-05 | 0 | 2.5e-05 | 0 | 5.3e-05 | 2.1e-05 | 0 | 3.7e-05 | -0.25 | 6 / 36 (17%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 2.2e-05 | 0 | 7.3e-05 | 9.9e-06 | 0 | 3.3e-05 | -1.2 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.5.4.27 | Methenyltetrahydromethanopterin cyclohydrolase. | 0.48 | 0.72 | 1 | 0.32 | 1 | 1 | 0.92 | 1 | 1 | 0.75 | 0.9 | 1 | 0.63 | 0.95 | 1 | 0.68 | 0.93 | 1 | 1e-04 | 8.5e-06 | 0 | 2.4e-05 | 0 | 6.4e-05 | 4.4e-06 | 0 | 1.2e-05 | -2.4 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 9.5e-06 | 0 | 3.2e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 1.1.1.149 | 20-alpha-hydroxysteroid dehydrogenase. | 0.16 | 0.5 | 1 | 0.69 | 1 | 1 | 0.62 | 0.96 | 1 | 0.28 | 0.57 | 1 | 0.47 | 0.92 | 1 | 0.49 | 0.87 | 1 | 0.002 | 0.002 | 0.0017 | 0.0025 | 0.002 | 0.0014 | 0.0027 | 0.0027 | 0.0017 | 0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0018 | 0.0016 | 0.0011 | 0.0016 | 0.0016 | 0.00052 | -0.17 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.1.53 | Oleate hydratase. | 0.94 | 0.97 | 1 | 0.15 | 1 | 1 | 0.19 | 0.87 | 1 | 0.75 | 0.9 | 1 | 0.4 | 0.9 | 1 | 0.4 | 0.83 | 1 | 0.019 | 0.019 | 0.018 | 0.02 | 0.024 | 0.0085 | 0.016 | 0.016 | 0.0065 | -0.32 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.02 | 0.018 | 0.011 | 0.02 | 0.019 | 0.0073 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.3.44 | tRNA 4-demethylwyosine synthase (AdoMet-dependent). | 0.47 | 0.72 | 1 | 0.37 | 1 | 1 | 0.57 | 0.96 | 1 | 0.73 | 0.89 | 1 | 0.7 | 0.95 | 1 | 0.88 | 0.97 | 1 | 2e-04 | 2.2e-05 | 0 | 5.7e-05 | 0 | 0.00015 | 8.8e-06 | 0 | 2.3e-05 | -2.7 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 1.9e-05 | 0 | 6.3e-05 | 1e-05 | 0 | 3.4e-05 | -0.93 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.7.4.3 | Adenylate kinase. | 0.15 | 0.5 | 1 | 0.12 | 1 | 1 | 0.96 | 1 | 1 | 0.14 | 0.47 | 1 | 0.88 | 0.99 | 1 | 0.24 | 0.75 | 1 | 0.013 | 0.013 | 0.013 | 0.012 | 0.011 | 0.0028 | 0.011 | 0.011 | 0.0029 | -0.13 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.014 | 0.014 | 0.0025 | 0.013 | 0.013 | 0.0019 | -0.11 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.5.1.57 | N-acylneuraminate-9-phosphate synthase. | 0.023 | 0.49 | 1 | 0.38 | 1 | 1 | 0.64 | 0.97 | 1 | 0.016 | 0.42 | 1 | 0.98 | 0.99 | 1 | 0.73 | 0.94 | 1 | 0.00021 | 4.1e-05 | 0 | 0.00014 | 0 | 0.00031 | 6.3e-05 | 0 | 1e-04 | -1.2 | 7 / 36 (19%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 2.8e-06 | 0 | 9.4e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.3.1.16 | Acetyl-CoA C-acyltransferase. | 0.84 | 0.93 | 1 | 0.86 | 1 | 1 | 0.62 | 0.96 | 1 | 0.8 | 0.93 | 1 | 0.78 | 0.97 | 1 | 0.32 | 0.8 | 1 | 0.00085 | 0.00083 | 0.00062 | 0.001 | 0.00048 | 0.0014 | 0.00086 | 0.00064 | 0.00079 | -0.22 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00083 | 0.00067 | 0.00071 | 0.00066 | 7e-04 | 0.00047 | -0.33 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 2.6.1.82 | Putrescine--2-oxoglutarate transaminase. | 0.69 | 0.85 | 1 | 0.079 | 1 | 1 | 0.19 | 0.87 | 1 | 0.61 | 0.82 | 1 | 0.017 | 0.76 | 1 | 0.099 | 0.66 | 1 | 0.00078 | 0.00069 | 0.00036 | 0.00078 | 0.00062 | 7e-04 | 0.00039 | 0.00024 | 0.00043 | -1 | 32 / 36 (89%) | 7 / 7 (100%) | 5 / 7 (71%) | 1 | 0.714 | 0.00073 | 0.00029 | 0.00089 | 0.00078 | 0.00036 | 0.0011 | 0.096 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 1.17.5.2 | Caffeine dehydrogenase. | 0.58 | 0.79 | 1 | 0.22 | 1 | 1 | 0.59 | 0.96 | 1 | 0.9 | 0.97 | 1 | 0.26 | 0.88 | 1 | 0.59 | 0.91 | 1 | 0.00018 | 6e-05 | 0 | 8.6e-05 | 0 | 0.00022 | 0.00014 | 0 | 3e-04 | 0.7 | 12 / 36 (33%) | 2 / 7 (29%) | 3 / 7 (43%) | 0.286 | 0.429 | 2e-05 | 0 | 4.1e-05 | 3.3e-05 | 0 | 5.2e-05 | 0.72 | 3 / 11 (27%) | 4 / 11 (36%) | 0.273 | 0.364 |
| 3.1.25.- | 0.46 | 0.71 | 1 | 0.12 | 1 | 1 | 0.99 | 1 | 1 | 0.65 | 0.85 | 1 | 0.33 | 0.9 | 1 | 0.65 | 0.92 | 1 | 0.0026 | 0.0025 | 0.0016 | 0.0026 | 0.0019 | 0.0028 | 0.0037 | 0.0023 | 0.0041 | 0.51 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0017 | 0.00097 | 0.0022 | 0.0025 | 0.0021 | 0.0024 | 0.56 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 | |
| 4.2.1.135 | UDP-N-acetylglucosamine 4.6-dehydratase (configuration-retaining). | 0.79 | 0.9 | 1 | 1 | 1 | 1 | 0.85 | 1 | 1 | 0.82 | 0.93 | 1 | 0.37 | 0.9 | 1 | 0.61 | 0.92 | 1 | 0.0021 | 0.0019 | 0.0011 | 0.0018 | 0.00069 | 0.0017 | 0.0021 | 0.0019 | 0.002 | 0.22 | 33 / 36 (92%) | 7 / 7 (100%) | 5 / 7 (71%) | 1 | 0.714 | 0.002 | 0.0013 | 0.0019 | 0.0019 | 0.001 | 0.0018 | -0.074 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 1.12.7.2 | Ferredoxin hydrogenase. | 0.67 | 0.84 | 1 | 0.75 | 1 | 1 | 0.7 | 0.98 | 1 | 0.89 | 0.96 | 1 | 0.54 | 0.93 | 1 | 0.56 | 0.9 | 1 | 0.034 | 0.034 | 0.035 | 0.035 | 0.035 | 0.0076 | 0.035 | 0.037 | 0.012 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.033 | 0.035 | 0.0091 | 0.034 | 0.033 | 0.0087 | 0.043 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.3.73 | Adenosylcobalamin/alpha-ribazole phosphatase. | 0.37 | 0.64 | 1 | 0.097 | 1 | 1 | 0.24 | 0.88 | 1 | 0.24 | 0.53 | 1 | 0.25 | 0.88 | 1 | 0.51 | 0.89 | 1 | 0.01 | 0.01 | 0.0099 | 0.0098 | 0.0093 | 0.002 | 0.0082 | 0.0066 | 0.0025 | -0.26 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.011 | 0.0037 | 0.011 | 0.011 | 0.0024 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.11.1.23 | (S)-2-hydroxypropylphosphonic acid epoxidase. | 0.75 | 0.88 | 1 | 0.83 | 1 | 1 | 0.65 | 0.97 | 1 | 0.68 | 0.87 | 1 | 0.88 | 0.99 | 1 | 0.95 | 0.99 | 1 | 0.00047 | 0.00017 | 0 | 0.00011 | 0 | 0.00023 | 5e-05 | 0 | 8.3e-05 | -1.1 | 13 / 36 (36%) | 2 / 7 (29%) | 3 / 7 (43%) | 0.286 | 0.429 | 0.00017 | 0 | 0.00033 | 0.00029 | 0 | 0.00064 | 0.77 | 4 / 11 (36%) | 4 / 11 (36%) | 0.364 | 0.364 |
| 2.3.1.48 | Histone acetyltransferase. | 0.0046 | 0.49 | 1 | 0.94 | 1 | 1 | 0.038 | 0.71 | 1 | 0.0064 | 0.37 | 1 | 0.89 | 0.99 | 1 | 0.03 | 0.61 | 1 | 0.00085 | 0.00068 | 0.00053 | 4e-04 | 0.00024 | 0.00048 | 0.00037 | 0.00019 | 0.00043 | -0.11 | 29 / 36 (81%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.0013 | 0.00084 | 0.0011 | 0.00046 | 0.00034 | 0.00039 | -1.5 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 3.6.5.n1 | Elongation factor 4. | 0.017 | 0.49 | 1 | 0.63 | 1 | 1 | 0.79 | 1 | 1 | 0.064 | 0.43 | 1 | 0.39 | 0.9 | 1 | 0.14 | 0.69 | 1 | 0.029 | 0.029 | 0.029 | 0.027 | 0.027 | 0.0028 | 0.026 | 0.027 | 0.0045 | -0.054 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.032 | 0.031 | 0.005 | 0.03 | 0.03 | 0.0036 | -0.093 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.5.1.39 | 4-hydroxybenzoate polyprenyltransferase. | 0.034 | 0.49 | 1 | 0.76 | 1 | 1 | 0.81 | 1 | 1 | 0.011 | 0.42 | 1 | 0.38 | 0.9 | 1 | 0.36 | 0.81 | 1 | 0.00029 | 4.8e-05 | 0 | 0.00012 | 0 | 0.00026 | 0.00013 | 0 | 0.00025 | 0.12 | 6 / 36 (17%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 2.1.1.196 | Cobalt-precorrin-6B (C(15))-methyltransferase (decarboxylating). | 0.91 | 0.96 | 1 | 0.58 | 1 | 1 | 0.23 | 0.88 | 1 | 0.92 | 0.97 | 1 | 0.42 | 0.91 | 1 | 0.13 | 0.68 | 1 | 9.5e-05 | 7.9e-06 | 0 | 8.1e-06 | 0 | 2.1e-05 | 1.8e-05 | 0 | 4.7e-05 | 1.2 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 9.5e-06 | 0 | 3.2e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 6.3.5.5 | Carbamoyl-phosphate synthase (glutamine-hydrolyzing). | 0.097 | 0.49 | 1 | 1 | 1 | 1 | 0.81 | 1 | 1 | 0.17 | 0.49 | 1 | 0.13 | 0.87 | 1 | 0.17 | 0.71 | 1 | 0.083 | 0.083 | 0.083 | 0.076 | 0.075 | 0.015 | 0.076 | 0.074 | 0.017 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.086 | 0.086 | 0.012 | 0.088 | 0.085 | 0.011 | 0.033 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.3.18 | Phosphoglycolate phosphatase. | 0.22 | 0.54 | 1 | 0.4 | 1 | 1 | 0.52 | 0.95 | 1 | 0.13 | 0.47 | 1 | 0.52 | 0.93 | 1 | 0.5 | 0.88 | 1 | 0.023 | 0.023 | 0.021 | 0.02 | 0.017 | 0.0088 | 0.017 | 0.015 | 0.0086 | -0.23 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.027 | 0.025 | 0.02 | 0.025 | 0.023 | 0.006 | -0.11 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.3.-.- | 0.58 | 0.79 | 1 | 0.022 | 0.92 | 1 | 0.17 | 0.85 | 1 | 0.79 | 0.92 | 1 | 0.065 | 0.86 | 1 | 0.48 | 0.87 | 1 | 0.00017 | 6.4e-05 | 0 | 1e-04 | 1.6e-05 | 0.00014 | 2.8e-05 | 0 | 5.3e-05 | -1.8 | 14 / 36 (39%) | 4 / 7 (57%) | 2 / 7 (29%) | 0.571 | 0.286 | 6.3e-05 | 0 | 7.7e-05 | 6.5e-05 | 0 | 0.00011 | 0.045 | 5 / 11 (45%) | 3 / 11 (27%) | 0.455 | 0.273 | |
| 2.7.1.15 | Ribokinase. | 0.7 | 0.86 | 1 | 0.95 | 1 | 1 | 0.99 | 1 | 1 | 0.44 | 0.69 | 1 | 0.79 | 0.97 | 1 | 0.86 | 0.96 | 1 | 0.012 | 0.012 | 0.011 | 0.011 | 0.012 | 0.0044 | 0.012 | 0.015 | 0.0056 | 0.13 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.013 | 0.0099 | 0.0097 | 0.013 | 0.011 | 0.0056 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.8.1.2 | 3-mercaptopyruvate sulfurtransferase. | 0.24 | 0.55 | 1 | 0.1 | 1 | 1 | 0.25 | 0.89 | 1 | 0.33 | 0.6 | 1 | 0.14 | 0.87 | 1 | 0.088 | 0.66 | 1 | 0.00041 | 4.5e-05 | 0 | 0.00012 | 0 | 0.00031 | 0.00011 | 0 | 0.00028 | -0.13 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3e-06 | 0 | 9.9e-06 | 2.7e-06 | 0 | 8.9e-06 | -0.15 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 4.4.1.16 | Selenocysteine lyase. | 0.75 | 0.88 | 1 | 0.53 | 1 | 1 | 0.33 | 0.89 | 1 | 0.61 | 0.82 | 1 | 0.16 | 0.87 | 1 | 0.16 | 0.71 | 1 | 0.00065 | 0.00052 | 0.00028 | 0.00055 | 0.00022 | 0.00088 | 0.00052 | 5e-04 | 0.00045 | -0.081 | 29 / 36 (81%) | 5 / 7 (71%) | 6 / 7 (86%) | 0.714 | 0.857 | 0.00061 | 0.00032 | 0.00088 | 0.00041 | 2e-04 | 5e-04 | -0.57 | 9 / 11 (82%) | 9 / 11 (82%) | 0.818 | 0.818 |
| 1.9.3.- | 0.11 | 0.5 | 1 | 0.78 | 1 | 1 | 0.5 | 0.95 | 1 | 0.13 | 0.47 | 1 | 0.49 | 0.92 | 1 | 0.94 | 0.99 | 1 | 0.00065 | 9e-05 | 0 | 0.00027 | 0 | 0.00071 | 0.00017 | 0 | 4e-04 | -0.67 | 5 / 36 (14%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0 | 0 | 0 | 1.1e-05 | 0 | 3.6e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 | |
| 4.2.1.70 | Pseudouridylate synthase. | 0.58 | 0.79 | 1 | 0.4 | 1 | 1 | 0.84 | 1 | 1 | 0.56 | 0.78 | 1 | 0.67 | 0.95 | 1 | 0.89 | 0.98 | 1 | 0.0012 | 0.0011 | 0.00053 | 0.0016 | 0.00042 | 0.0024 | 0.001 | 0.00063 | 0.0012 | -0.68 | 33 / 36 (92%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.0012 | 0.00053 | 0.0015 | 0.00085 | 0.00045 | 0.0013 | -0.5 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 1.1.1.136 | UDP-N-acetylglucosamine 6-dehydrogenase. | 0.026 | 0.49 | 1 | 0.31 | 1 | 1 | 0.63 | 0.96 | 1 | 0.0062 | 0.37 | 1 | 0.13 | 0.87 | 1 | 0.21 | 0.74 | 1 | 0.00088 | 0.00056 | 0.00012 | 0.001 | 0.00071 | 0.00097 | 0.0011 | 0.00019 | 0.0022 | 0.14 | 23 / 36 (64%) | 7 / 7 (100%) | 5 / 7 (71%) | 1 | 0.714 | 0.00036 | 0 | 0.00087 | 0.00012 | 5.9e-05 | 0.00014 | -1.6 | 5 / 11 (45%) | 6 / 11 (55%) | 0.455 | 0.545 |
| 3.6.3.24 | Transferred entry: 7.2.2.11. | 0.15 | 0.5 | 1 | 0.97 | 1 | 1 | 0.8 | 1 | 1 | 0.18 | 0.5 | 1 | 0.46 | 0.92 | 1 | 0.56 | 0.9 | 1 | 0.0072 | 0.007 | 0.0036 | 0.0031 | 0.0014 | 0.0033 | 0.0035 | 0.0023 | 0.0041 | 0.18 | 35 / 36 (97%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.011 | 0.0049 | 0.02 | 0.0078 | 0.0065 | 0.0075 | -0.5 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.5.1.120 | Aminodeoxyfutalosine synthase. | 0.16 | 0.5 | 1 | 0.15 | 1 | 1 | 0.03 | 0.71 | 1 | 0.22 | 0.52 | 1 | 0.22 | 0.88 | 1 | 0.06 | 0.66 | 1 | 0.00011 | 3.1e-05 | 0 | 5.1e-05 | 0 | 8.4e-05 | 1.8e-05 | 0 | 4.7e-05 | -1.5 | 10 / 36 (28%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 8.5e-06 | 0 | 2.1e-05 | 4.9e-05 | 0 | 9.1e-05 | 2.5 | 2 / 11 (18%) | 4 / 11 (36%) | 0.182 | 0.364 |
| 2.1.3.9 | N-acetylornithine carbamoyltransferase. | 0.21 | 0.54 | 1 | 0.63 | 1 | 1 | 0.12 | 0.82 | 1 | 0.075 | 0.43 | 1 | 0.25 | 0.88 | 1 | 0.096 | 0.66 | 1 | 0.00098 | 0.00074 | 0.00051 | 0.00065 | 0.00057 | 0.00029 | 0.00077 | 0.00068 | 0.00087 | 0.24 | 27 / 36 (75%) | 7 / 7 (100%) | 5 / 7 (71%) | 1 | 0.714 | 0.00045 | 0.00019 | 0.00062 | 0.0011 | 0.00055 | 0.0015 | 1.3 | 7 / 11 (64%) | 8 / 11 (73%) | 0.636 | 0.727 |
| 3.4.24.84 | Ste24 endopeptidase. | 0.076 | 0.49 | 1 | 0.34 | 1 | 1 | 0.62 | 0.96 | 1 | 0.053 | 0.43 | 1 | 0.39 | 0.9 | 1 | 0.58 | 0.91 | 1 | 9.9e-05 | 3e-05 | 0 | 1.9e-05 | 0 | 4.4e-05 | 0 | 0 | 0 | -Inf | 11 / 36 (31%) | 2 / 7 (29%) | 0 / 7 (0%) | 0.286 | 0 | 6.8e-05 | 3.1e-05 | 8.8e-05 | 1.9e-05 | 0 | 3.3e-05 | -1.8 | 6 / 11 (55%) | 3 / 11 (27%) | 0.545 | 0.273 |
| 6.3.5.10 | Adenosylcobyric acid synthase (glutamine-hydrolyzing). | 0.15 | 0.5 | 1 | 0.68 | 1 | 1 | 0.3 | 0.89 | 1 | 0.13 | 0.47 | 1 | 0.52 | 0.93 | 1 | 0.14 | 0.69 | 1 | 0.00012 | 3.6e-05 | 0 | 6.3e-06 | 0 | 1.1e-05 | 2e-05 | 0 | 4e-05 | 1.7 | 11 / 36 (31%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 5.6e-05 | 0 | 8e-05 | 4.7e-05 | 0 | 0.00012 | -0.25 | 5 / 11 (45%) | 2 / 11 (18%) | 0.455 | 0.182 |
| 3.2.1.15 | Endo-polygalacturonase. | 0.78 | 0.9 | 1 | 0.36 | 1 | 1 | 0.13 | 0.82 | 1 | 0.66 | 0.85 | 1 | 0.23 | 0.88 | 1 | 0.18 | 0.72 | 1 | 0.0064 | 0.0064 | 0.0027 | 0.0051 | 0.0039 | 0.0044 | 0.0035 | 0.0016 | 0.0047 | -0.54 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0048 | 0.003 | 0.0064 | 0.011 | 0.0019 | 0.019 | 1.2 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.4.99.5 | Chorismate mutase. | 0.63 | 0.82 | 1 | 0.26 | 1 | 1 | 0.1 | 0.82 | 1 | 0.68 | 0.87 | 1 | 0.38 | 0.9 | 1 | 0.15 | 0.71 | 1 | 0.0039 | 0.0039 | 0.0034 | 0.004 | 0.0042 | 0.0019 | 0.0036 | 0.0034 | 0.0024 | -0.15 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0037 | 0.0025 | 0.0031 | 0.0041 | 0.0035 | 0.0024 | 0.15 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.122 | Maltose-6'-phosphate glucosidase. | 0.23 | 0.54 | 1 | 0.78 | 1 | 1 | 0.67 | 0.98 | 1 | 0.13 | 0.47 | 1 | 0.73 | 0.96 | 1 | 0.69 | 0.94 | 1 | 0.0025 | 0.0025 | 0.0024 | 0.002 | 0.0013 | 0.0017 | 0.0019 | 0.0018 | 0.0015 | -0.074 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0027 | 0.0028 | 0.0013 | 0.0029 | 0.0026 | 0.0014 | 0.1 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.1.3.29 | L-fucose mutarotase. | 0.99 | 1 | 1 | 0.69 | 1 | 1 | 0.45 | 0.93 | 1 | 0.73 | 0.89 | 1 | 0.69 | 0.95 | 1 | 0.36 | 0.81 | 1 | 0.00036 | 0.00026 | 0.00017 | 0.00028 | 0.00016 | 3e-04 | 0.00033 | 0.00036 | 0.00028 | 0.24 | 26 / 36 (72%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.00024 | 0.00017 | 0.00022 | 0.00022 | 0.00015 | 0.00028 | -0.13 | 9 / 11 (82%) | 7 / 11 (64%) | 0.818 | 0.636 |
| 3.1.2.- | 0.68 | 0.85 | 1 | 0.49 | 1 | 1 | 0.8 | 1 | 1 | 0.31 | 0.58 | 1 | 0.96 | 0.99 | 1 | 0.75 | 0.95 | 1 | 0.006 | 0.006 | 0.0053 | 0.0059 | 0.0053 | 0.0031 | 0.0051 | 0.0044 | 0.0021 | -0.21 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0066 | 0.005 | 0.0049 | 0.0059 | 0.0059 | 0.0021 | -0.16 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 3.4.25.1 | Proteasome endopeptidase complex. | 0.13 | 0.5 | 1 | 0.041 | 0.97 | 1 | 0.047 | 0.71 | 1 | 0.23 | 0.53 | 1 | 0.13 | 0.87 | 1 | 0.15 | 0.71 | 1 | 0.00025 | 6.3e-05 | 0 | 0.00015 | 0 | 0.00025 | 9e-05 | 0 | 0.00018 | -0.74 | 9 / 36 (25%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 2.1e-05 | 0 | 4.6e-05 | 3.4e-05 | 0 | 7.7e-05 | 0.7 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 4.2.1.1 | Carbonic anhydrase. | 0.31 | 0.59 | 1 | 0.71 | 1 | 1 | 0.7 | 0.98 | 1 | 0.19 | 0.5 | 1 | 0.37 | 0.9 | 1 | 0.52 | 0.89 | 1 | 0.0036 | 0.0036 | 0.003 | 0.0027 | 0.003 | 0.001 | 0.0031 | 0.0026 | 0.002 | 0.2 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0044 | 0.003 | 0.0051 | 0.0038 | 0.0032 | 0.0022 | -0.21 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.10.99.2 | Transferred entry: 1.10.5.1. | 0.25 | 0.56 | 1 | 0.047 | 0.97 | 1 | 0.12 | 0.82 | 1 | 0.24 | 0.53 | 1 | 0.028 | 0.76 | 1 | 0.11 | 0.66 | 1 | 0.00018 | 6.1e-05 | 0 | 4.6e-06 | 0 | 1.2e-05 | 9e-05 | 8.1e-05 | 9.9e-05 | 4.3 | 12 / 36 (33%) | 1 / 7 (14%) | 4 / 7 (57%) | 0.143 | 0.571 | 8.1e-05 | 0 | 0.00018 | 5.9e-05 | 0 | 9.1e-05 | -0.46 | 3 / 11 (27%) | 4 / 11 (36%) | 0.273 | 0.364 |
| 1.1.99.38 | 2-deoxy-scyllo-inosamine dehydrogenase (AdoMet-dependent). | 0.05 | 0.49 | 1 | 0.019 | 0.92 | 1 | 0.16 | 0.85 | 1 | 0.11 | 0.47 | 1 | 0.026 | 0.76 | 1 | 0.28 | 0.78 | 1 | 0.00017 | 1.9e-05 | 0 | 8.3e-05 | 0 | 0.00015 | 0 | 0 | 0 | -Inf | 4 / 36 (11%) | 2 / 7 (29%) | 0 / 7 (0%) | 0.286 | 0 | 8.5e-06 | 0 | 2.1e-05 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 4.2.2.1 | Hyaluronate lyase. | 0.022 | 0.49 | 1 | 0.13 | 1 | 1 | 0.28 | 0.89 | 1 | 0.03 | 0.43 | 1 | 0.45 | 0.92 | 1 | 0.62 | 0.92 | 1 | 0.00042 | 0.00018 | 0 | 0.00044 | 0.00014 | 0.00063 | 0.00031 | 0.00012 | 0.00054 | -0.51 | 15 / 36 (42%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 5.4e-05 | 0 | 0.00011 | 4.3e-05 | 0 | 9.3e-05 | -0.33 | 3 / 11 (27%) | 3 / 11 (27%) | 0.273 | 0.273 |
| 3.4.17.13 | Muramoyltetrapeptide carboxypeptidase. | 0.017 | 0.49 | 1 | 0.81 | 1 | 1 | 0.77 | 0.99 | 1 | 0.068 | 0.43 | 1 | 0.68 | 0.95 | 1 | 0.97 | 0.99 | 1 | 0.0015 | 0.0015 | 0.0015 | 0.002 | 0.002 | 0.00084 | 0.0021 | 0.0021 | 0.001 | 0.07 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.001 | 0.00097 | 0.00058 | 0.0013 | 0.0015 | 0.00087 | 0.38 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.3.9 | Phosphoenolpyruvate--protein phosphotransferase. | 0.78 | 0.9 | 1 | 0.45 | 1 | 1 | 0.66 | 0.97 | 1 | 0.71 | 0.88 | 1 | 0.9 | 0.99 | 1 | 0.62 | 0.92 | 1 | 0.034 | 0.034 | 0.034 | 0.035 | 0.033 | 0.0084 | 0.033 | 0.032 | 0.0092 | -0.085 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.034 | 0.035 | 0.0074 | 0.033 | 0.035 | 0.006 | -0.043 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.1.131 | Precorrin-3B C(17)-methyltransferase. | 0.69 | 0.86 | 1 | 0.91 | 1 | 1 | 1 | 1 | 1 | 0.92 | 0.97 | 1 | 0.52 | 0.93 | 1 | 0.51 | 0.89 | 1 | 0.0069 | 0.0069 | 0.0066 | 0.0072 | 0.0064 | 0.0026 | 0.0071 | 0.0073 | 0.0018 | -0.02 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0069 | 0.0066 | 0.0028 | 0.0067 | 0.0058 | 0.002 | -0.042 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.1.3.20 | ADP-glyceromanno-heptose 6-epimerase. | 0.53 | 0.76 | 1 | 0.29 | 1 | 1 | 0.88 | 1 | 1 | 0.79 | 0.92 | 1 | 0.5 | 0.93 | 1 | 0.65 | 0.92 | 1 | 0.00045 | 3e-04 | 9.1e-05 | 0.00051 | 0.00033 | 0.00068 | 0.00034 | 9.3e-05 | 0.00059 | -0.58 | 24 / 36 (67%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.00034 | 8e-05 | 0.00051 | 0.00011 | 8.9e-05 | 1e-04 | -1.6 | 7 / 11 (64%) | 9 / 11 (82%) | 0.636 | 0.818 |
| 2.1.3.1 | Methylmalonyl-CoA carboxytransferase. | 0.43 | 0.69 | 1 | 0.79 | 1 | 1 | 0.32 | 0.89 | 1 | 0.86 | 0.95 | 1 | 0.37 | 0.9 | 1 | 0.83 | 0.96 | 1 | 0.027 | 0.027 | 0.027 | 0.027 | 0.027 | 0.005 | 0.027 | 0.029 | 0.006 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.025 | 0.024 | 0.0089 | 0.027 | 0.028 | 0.0061 | 0.11 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.6.1.22 | NAD(+) diphosphatase. | 0.37 | 0.65 | 1 | 0.24 | 1 | 1 | 0.49 | 0.95 | 1 | 0.22 | 0.52 | 1 | 0.081 | 0.86 | 1 | 0.24 | 0.76 | 1 | 0.0095 | 0.0095 | 0.0091 | 0.0083 | 0.0076 | 0.0023 | 0.0097 | 0.0086 | 0.0036 | 0.22 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0098 | 0.0077 | 0.0047 | 0.0098 | 0.0095 | 0.0019 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.12.99.6 | Hydrogenase (acceptor). | 0.075 | 0.49 | 1 | 0.037 | 0.97 | 1 | 0.059 | 0.71 | 1 | 0.023 | 0.42 | 1 | 0.073 | 0.86 | 1 | 0.12 | 0.66 | 1 | 0.00099 | 0.00066 | 0.00012 | 0.0014 | 0.00026 | 0.0029 | 0.00087 | 0.00011 | 0.002 | -0.69 | 24 / 36 (67%) | 7 / 7 (100%) | 5 / 7 (71%) | 1 | 0.714 | 0.00031 | 3.1e-05 | 0.00061 | 0.00039 | 5.9e-05 | 7e-04 | 0.33 | 6 / 11 (55%) | 6 / 11 (55%) | 0.545 | 0.545 |
| 3.6.3.12 | Transferred entry: 7.2.2.6. | 0.66 | 0.84 | 1 | 0.5 | 1 | 1 | 0.83 | 1 | 1 | 0.98 | 0.99 | 1 | 0.9 | 0.99 | 1 | 0.7 | 0.94 | 1 | 0.0022 | 0.0015 | 0.00017 | 0.0023 | 0.00017 | 0.0034 | 0.0014 | 0.00043 | 0.0019 | -0.72 | 25 / 36 (69%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.0016 | 0.00016 | 0.0027 | 0.0011 | 0.00011 | 0.002 | -0.54 | 8 / 11 (73%) | 7 / 11 (64%) | 0.727 | 0.636 |
| 3.5.1.2 | Glutaminase. | 0.055 | 0.49 | 1 | 0.33 | 1 | 1 | 0.98 | 1 | 1 | 0.04 | 0.43 | 1 | 0.011 | 0.75 | 1 | 0.13 | 0.68 | 1 | 0.016 | 0.016 | 0.016 | 0.014 | 0.014 | 0.0023 | 0.015 | 0.015 | 0.0038 | 0.1 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.017 | 0.016 | 0.0035 | 0.018 | 0.019 | 0.0032 | 0.082 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.8.42 | Kdo(2)-lipid A phosphoethanolamine 7''-transferase. | 0.08 | 0.49 | 1 | 0.02 | 0.92 | 1 | 0.11 | 0.82 | 1 | 0.06 | 0.43 | 1 | 0.15 | 0.87 | 1 | 0.5 | 0.88 | 1 | 0.00078 | 8.7e-05 | 0 | 0.00029 | 0 | 0.00071 | 0.00015 | 0 | 0.00041 | -0.95 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 1.3.99.33 | Urocanate reductase. | 0.0014 | 0.49 | 1 | 0.063 | 1 | 1 | 0.34 | 0.89 | 1 | 0.0017 | 0.31 | 1 | 0.079 | 0.86 | 1 | 0.4 | 0.83 | 1 | 0.00015 | 7.6e-05 | 1.4e-05 | 0.00019 | 0.00011 | 0.00019 | 1e-04 | 9.3e-05 | 0.00013 | -0.93 | 18 / 36 (50%) | 7 / 7 (100%) | 4 / 7 (57%) | 1 | 0.571 | 4.7e-05 | 0 | 8.3e-05 | 1.5e-05 | 0 | 2.7e-05 | -1.6 | 4 / 11 (36%) | 3 / 11 (27%) | 0.364 | 0.273 |
| 1.2.1.50 | Long-chain-fatty-acyl-CoA reductase. | 0.2 | 0.53 | 1 | 0.79 | 1 | 1 | 0.85 | 1 | 1 | 0.084 | 0.44 | 1 | 0.29 | 0.89 | 1 | 0.2 | 0.73 | 1 | 0.0024 | 0.0019 | 0.0014 | 0.0012 | 0.00091 | 0.0013 | 0.0012 | 0.00084 | 0.0013 | 0 | 28 / 36 (78%) | 4 / 7 (57%) | 5 / 7 (71%) | 0.571 | 0.714 | 0.0023 | 0.0014 | 0.0032 | 0.0024 | 0.0016 | 0.0024 | 0.061 | 10 / 11 (91%) | 9 / 11 (82%) | 0.909 | 0.818 |
| 2.6.1.29 | Diamine transaminase. | 0.69 | 0.85 | 1 | 0.16 | 1 | 1 | 0.23 | 0.88 | 1 | 0.88 | 0.96 | 1 | 0.067 | 0.86 | 1 | 0.11 | 0.66 | 1 | 0.00061 | 0.00042 | 0.00011 | 0.00032 | 0.00012 | 0.00047 | 0.00012 | 0 | 0.00025 | -1.4 | 25 / 36 (69%) | 6 / 7 (86%) | 3 / 7 (43%) | 0.857 | 0.429 | 0.00056 | 9.7e-05 | 0.00092 | 0.00054 | 0.00017 | 0.00082 | -0.052 | 8 / 11 (73%) | 8 / 11 (73%) | 0.727 | 0.727 |
| 3.1.1.61 | Protein-glutamate methylesterase. | 0.21 | 0.53 | 1 | 0.91 | 1 | 1 | 0.89 | 1 | 1 | 0.24 | 0.54 | 1 | 0.86 | 0.98 | 1 | 0.82 | 0.96 | 1 | 0.0089 | 0.0089 | 0.0052 | 0.0047 | 0.0028 | 0.0037 | 0.0053 | 0.0034 | 0.0052 | 0.17 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.013 | 0.0061 | 0.021 | 0.0099 | 0.007 | 0.0083 | -0.39 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.1.216 | tRNA (guanine(26)-N(2))-dimethyltransferase. | 0.96 | 0.99 | 1 | 0.1 | 1 | 1 | 0.24 | 0.89 | 1 | 0.93 | 0.98 | 1 | 0.53 | 0.93 | 1 | 0.82 | 0.96 | 1 | 0.00019 | 2.1e-05 | 0 | 1.6e-05 | 0 | 4.3e-05 | 4.4e-06 | 0 | 1.2e-05 | -1.9 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.9e-05 | 0 | 9.5e-05 | 2.6e-05 | 0 | 8.6e-05 | -0.16 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 4.2.1.49 | Urocanate hydratase. | 0.46 | 0.71 | 1 | 0.86 | 1 | 1 | 0.8 | 1 | 1 | 0.21 | 0.52 | 1 | 0.71 | 0.95 | 1 | 0.65 | 0.92 | 1 | 0.0019 | 0.0019 | 0.0015 | 0.0019 | 0.0015 | 0.00074 | 0.0021 | 0.0019 | 0.002 | 0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0019 | 0.00072 | 0.0031 | 0.0018 | 0.0011 | 0.0021 | -0.078 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.4.41 | 5'-deoxyadenosine deaminase. | 0.66 | 0.84 | 1 | 0.65 | 1 | 1 | 0.53 | 0.95 | 1 | 0.8 | 0.93 | 1 | 0.87 | 0.99 | 1 | 0.82 | 0.96 | 1 | 0.00045 | 5e-05 | 0 | 6.5e-05 | 0 | 0.00017 | 1.8e-05 | 0 | 4.7e-05 | -1.9 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.9e-05 | 0 | 9.5e-05 | 8.3e-05 | 0 | 0.00027 | 1.5 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 4.2.1.40 | Glucarate dehydratase. | 0.24 | 0.55 | 1 | 0.3 | 1 | 1 | 0.86 | 1 | 1 | 0.26 | 0.56 | 1 | 0.42 | 0.91 | 1 | 0.95 | 0.99 | 1 | 0.00067 | 0.00058 | 0.00032 | 0.00086 | 9e-04 | 0.00057 | 0.00069 | 0.00046 | 7e-04 | -0.32 | 31 / 36 (86%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00062 | 0.00029 | 0.00066 | 0.00028 | 3e-04 | 0.00024 | -1.1 | 9 / 11 (82%) | 9 / 11 (82%) | 0.818 | 0.818 |
| 1.97.1.9 | Selenate reductase. | 0.19 | 0.53 | 1 | 0.15 | 1 | 1 | 0.95 | 1 | 1 | 0.28 | 0.57 | 1 | 0.35 | 0.9 | 1 | 0.76 | 0.95 | 1 | 0.0013 | 0.0012 | 0.00061 | 0.0021 | 0.0019 | 0.0023 | 0.0015 | 0.00062 | 0.0017 | -0.49 | 33 / 36 (92%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.0011 | 0.001 | 0.001 | 0.00054 | 0.00044 | 0.00046 | -1 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 1.5.3.1 | Sarcosine oxidase. | 0.45 | 0.7 | 1 | 0.94 | 1 | 1 | 0.79 | 1 | 1 | 0.36 | 0.63 | 1 | 0.96 | 0.99 | 1 | 0.83 | 0.96 | 1 | 0.0036 | 0.0036 | 0.0033 | 0.0029 | 0.0031 | 0.0013 | 0.0033 | 0.002 | 0.0027 | 0.19 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0037 | 0.0032 | 0.0021 | 0.004 | 0.0035 | 0.0029 | 0.11 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.2.2.- | 0.28 | 0.57 | 1 | 0.3 | 1 | 1 | 0.8 | 1 | 1 | 0.44 | 0.69 | 1 | 0.68 | 0.95 | 1 | 0.74 | 0.94 | 1 | 0.00019 | 3.2e-05 | 0 | 8.6e-05 | 0 | 0.00022 | 5.8e-05 | 0 | 0.00015 | -0.57 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 9.9e-06 | 0 | 2.5e-05 | 2.7e-06 | 0 | 8.9e-06 | -1.9 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 | |
| 1.8.1.7 | Glutathione-disulfide reductase. | 1 | 1 | 1 | 0.7 | 1 | 1 | 0.38 | 0.9 | 1 | 0.73 | 0.89 | 1 | 0.34 | 0.9 | 1 | 0.62 | 0.92 | 1 | 0.001 | 0.00089 | 0.00041 | 0.00067 | 0.00042 | 0.00074 | 7e-04 | 0.00024 | 0.0011 | 0.063 | 32 / 36 (89%) | 7 / 7 (100%) | 5 / 7 (71%) | 1 | 0.714 | 0.00059 | 4e-04 | 0.00048 | 0.0014 | 0.00052 | 0.0021 | 1.2 | 11 / 11 (100%) | 9 / 11 (82%) | 1 | 0.818 |
| 4.3.2.7 | Glutathione-specific gamma-glutamylcyclotransferase. | 0.081 | 0.49 | 1 | 0.072 | 1 | 1 | 0.05 | 0.71 | 1 | 0.066 | 0.43 | 1 | 0.38 | 0.9 | 1 | 0.23 | 0.75 | 1 | 0.00015 | 1.7e-05 | 0 | 5.3e-05 | 0 | 0.00013 | 2.9e-05 | 0 | 7.7e-05 | -0.87 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.7.7.38 | 3-deoxy-manno-octulosonate cytidylyltransferase. | 0.079 | 0.49 | 1 | 0.2 | 1 | 1 | 0.076 | 0.74 | 1 | 0.063 | 0.43 | 1 | 0.36 | 0.9 | 1 | 0.11 | 0.66 | 1 | 0.0013 | 0.0012 | 8e-04 | 0.0016 | 0.0016 | 0.00098 | 0.0012 | 0.00081 | 0.0011 | -0.42 | 33 / 36 (92%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00088 | 0.00049 | 0.0012 | 0.0011 | 0.0011 | 0.0011 | 0.32 | 9 / 11 (82%) | 10 / 11 (91%) | 0.818 | 0.909 |
| 3.2.1.145 | Galactan 1.3-beta-galactosidase. | 0.41 | 0.67 | 1 | 0.57 | 1 | 1 | 0.91 | 1 | 1 | 0.11 | 0.47 | 1 | 0.13 | 0.87 | 1 | 0.26 | 0.76 | 1 | 0.0035 | 0.0028 | 0.0022 | 0.0023 | 0.0024 | 0.0023 | 0.0027 | 0.0022 | 0.003 | 0.23 | 29 / 36 (81%) | 4 / 7 (57%) | 5 / 7 (71%) | 0.571 | 0.714 | 0.0029 | 0.0021 | 0.0037 | 0.0032 | 0.0023 | 0.0025 | 0.14 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 4.3.99.- | 0.094 | 0.49 | 1 | 0.17 | 1 | 1 | 0.028 | 0.71 | 1 | 0.12 | 0.47 | 1 | 0.54 | 0.93 | 1 | 0.079 | 0.66 | 1 | 2e-04 | 3.4e-05 | 0 | 1e-04 | 0 | 0.00027 | 3.9e-05 | 0 | 1e-04 | -1.4 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2e-05 | 0 | 3.9e-05 | Inf | 0 / 11 (0%) | 3 / 11 (27%) | 0 | 0.273 | |
| 3.6.3.30 | Transferred entry: 7.2.2.7. | 0.23 | 0.54 | 1 | 0.87 | 1 | 1 | 0.57 | 0.96 | 1 | 0.18 | 0.5 | 1 | 0.65 | 0.95 | 1 | 0.5 | 0.88 | 1 | 0.022 | 0.022 | 0.019 | 0.017 | 0.014 | 0.007 | 0.018 | 0.014 | 0.011 | 0.082 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.027 | 0.019 | 0.028 | 0.022 | 0.02 | 0.0092 | -0.3 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.3.13 | Deleted entry. | 0.52 | 0.75 | 1 | 0.44 | 1 | 1 | 0.77 | 0.99 | 1 | 0.53 | 0.77 | 1 | 0.27 | 0.88 | 1 | 0.8 | 0.96 | 1 | 0.00058 | 4e-04 | 0.00013 | 0.00029 | 8.1e-05 | 0.00043 | 0.00035 | 0.00014 | 0.00044 | 0.27 | 25 / 36 (69%) | 4 / 7 (57%) | 5 / 7 (71%) | 0.571 | 0.714 | 0.00047 | 0.00012 | 0.00064 | 0.00044 | 0.00034 | 0.00052 | -0.095 | 7 / 11 (64%) | 9 / 11 (82%) | 0.636 | 0.818 |
| 6.-.-.- | 0.9 | 0.96 | 1 | 0.29 | 1 | 1 | 0.2 | 0.87 | 1 | 0.86 | 0.95 | 1 | 0.33 | 0.9 | 1 | 0.28 | 0.78 | 1 | 0.001 | 0.00095 | 0.00074 | 0.00085 | 0.0011 | 0.00051 | 0.0012 | 0.00088 | 0.00065 | 0.5 | 34 / 36 (94%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.001 | 0.00064 | 0.00081 | 0.00079 | 5e-04 | 0.00076 | -0.34 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 | |
| 2.3.1.54 | Formate C-acetyltransferase. | 0.39 | 0.66 | 1 | 0.83 | 1 | 1 | 0.72 | 0.98 | 1 | 0.24 | 0.53 | 1 | 0.52 | 0.93 | 1 | 0.59 | 0.91 | 1 | 0.043 | 0.043 | 0.045 | 0.04 | 0.04 | 0.0099 | 0.04 | 0.04 | 0.014 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.044 | 0.047 | 0.0093 | 0.045 | 0.045 | 0.0054 | 0.032 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.7.1.6 | Azobenzene reductase. | 0.5 | 0.73 | 1 | 0.97 | 1 | 1 | 1 | 1 | 1 | 0.28 | 0.57 | 1 | 0.63 | 0.95 | 1 | 0.74 | 0.94 | 1 | 0.00051 | 0.00048 | 0.00045 | 0.00044 | 0.00047 | 0.00035 | 0.00043 | 0.00049 | 0.00029 | -0.033 | 34 / 36 (94%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00051 | 0.00042 | 0.00034 | 0.00051 | 0.00037 | 0.00035 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.6.1.42 | Branched-chain-amino-acid transaminase. | 0.06 | 0.49 | 1 | 0.25 | 1 | 1 | 0.26 | 0.89 | 1 | 0.052 | 0.43 | 1 | 0.97 | 0.99 | 1 | 0.96 | 0.99 | 1 | 0.018 | 0.018 | 0.018 | 0.016 | 0.014 | 0.0037 | 0.014 | 0.014 | 0.0033 | -0.19 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.02 | 0.019 | 0.0064 | 0.02 | 0.02 | 0.0033 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.1.52 | Poly(glycerol-phosphate) alpha-glucosyltransferase. | 0.73 | 0.87 | 1 | 0.062 | 1 | 1 | 0.0011 | 0.51 | 1 | 0.47 | 0.72 | 1 | 0.026 | 0.76 | 1 | 0.0015 | 0.48 | 1 | 0.0049 | 0.0049 | 0.0035 | 0.0039 | 0.0031 | 0.0017 | 0.0026 | 0.0023 | 0.0018 | -0.58 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0044 | 0.0035 | 0.0024 | 0.0077 | 0.0064 | 0.0047 | 0.81 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.3.4.18 | 5-(carboxyamino)imidazole ribonucleotide synthase. | 0.07 | 0.49 | 1 | 0.73 | 1 | 1 | 0.52 | 0.95 | 1 | 0.055 | 0.43 | 1 | 0.91 | 0.99 | 1 | 0.86 | 0.96 | 1 | 0.0027 | 0.0026 | 0.0015 | 0.0012 | 0.00082 | 0.0011 | 0.0015 | 0.0014 | 0.0011 | 0.32 | 35 / 36 (97%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.0041 | 0.002 | 0.0067 | 0.0029 | 0.0022 | 0.0023 | -0.5 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.4.1.15 | D-cysteine desulfhydrase. | 0.28 | 0.57 | 1 | 0.42 | 1 | 1 | 0.35 | 0.9 | 1 | 0.5 | 0.74 | 1 | 0.35 | 0.9 | 1 | 0.27 | 0.77 | 1 | 0.0012 | 0.0012 | 0.00092 | 0.0015 | 0.0012 | 0.0011 | 0.0017 | 0.0015 | 0.0012 | 0.18 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.001 | 0.00087 | 0.00069 | 0.00091 | 0.00082 | 0.00058 | -0.14 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.2.24 | ADP-ribosyl-[dinitrogen reductase] hydrolase. | 0.027 | 0.49 | 1 | 0.46 | 1 | 1 | 0.58 | 0.96 | 1 | 0.0053 | 0.36 | 1 | 0.4 | 0.9 | 1 | 0.57 | 0.9 | 1 | 0.0024 | 0.0024 | 0.0015 | 0.0012 | 0.00097 | 0.00096 | 0.00091 | 0.00072 | 0.00056 | -0.4 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0032 | 0.0021 | 0.0028 | 0.0032 | 0.0022 | 0.0033 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.3.3.8 | Delta(3)-Delta(2)-enoyl-CoA isomerase. | 0.27 | 0.56 | 1 | 0.49 | 1 | 1 | 0.42 | 0.92 | 1 | 0.45 | 0.7 | 1 | 0.33 | 0.9 | 1 | 0.24 | 0.76 | 1 | 0.00048 | 9.4e-05 | 0 | 0.00022 | 0 | 0.00058 | 0.00023 | 0 | 0.00056 | 0.064 | 7 / 36 (19%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 1.1e-05 | 0 | 2.5e-05 | 1.1e-05 | 0 | 3.6e-05 | 0 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 2.3.-.- | 0.17 | 0.5 | 1 | 1 | 1 | 1 | 0.6 | 0.96 | 1 | 0.13 | 0.47 | 1 | 0.77 | 0.97 | 1 | 0.68 | 0.93 | 1 | 7e-04 | 0.00064 | 0.00052 | 8e-04 | 0.00049 | 0.00065 | 0.00074 | 0.00059 | 0.00039 | -0.11 | 33 / 36 (92%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00054 | 0.00031 | 0.00056 | 0.00058 | 0.00055 | 0.00039 | 0.1 | 9 / 11 (82%) | 10 / 11 (91%) | 0.818 | 0.909 | |
| 2.4.2.10 | Orotate phosphoribosyltransferase. | 0.13 | 0.5 | 1 | 0.69 | 1 | 1 | 0.9 | 1 | 1 | 0.13 | 0.47 | 1 | 0.16 | 0.87 | 1 | 0.41 | 0.84 | 1 | 0.018 | 0.018 | 0.017 | 0.016 | 0.016 | 0.0015 | 0.016 | 0.017 | 0.0037 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.019 | 0.017 | 0.0064 | 0.02 | 0.018 | 0.0037 | 0.074 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.1.46 | dTDP-glucose 4.6-dehydratase. | 0.63 | 0.83 | 1 | 0.79 | 1 | 1 | 0.29 | 0.89 | 1 | 0.28 | 0.57 | 1 | 0.41 | 0.9 | 1 | 0.91 | 0.98 | 1 | 0.025 | 0.025 | 0.024 | 0.024 | 0.024 | 0.0027 | 0.023 | 0.023 | 0.0042 | -0.061 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.025 | 0.024 | 0.0059 | 0.027 | 0.028 | 0.0041 | 0.11 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.1.2 | Arylesterase. | 0.7 | 0.86 | 1 | 0.87 | 1 | 1 | 0.9 | 1 | 1 | 0.95 | 0.98 | 1 | 0.99 | 1 | 1 | 0.72 | 0.94 | 1 | 0.001 | 0.001 | 0.00095 | 0.0011 | 0.00094 | 0.00054 | 0.0011 | 0.00083 | 0.00073 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00098 | 0.00079 | 0.00054 | 0.00092 | 0.00098 | 0.00055 | -0.091 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.1.96 | D-aminoacyl-tRNA deacylase. | 0.86 | 0.94 | 1 | 0.59 | 1 | 1 | 0.76 | 0.99 | 1 | 0.48 | 0.72 | 1 | 0.98 | 0.99 | 1 | 0.78 | 0.95 | 1 | 0.0082 | 0.0082 | 0.0079 | 0.0082 | 0.0081 | 0.0018 | 0.0078 | 0.0077 | 0.0026 | -0.072 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0084 | 0.0075 | 0.0024 | 0.0081 | 0.008 | 0.001 | -0.052 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.1.25 | Tyrosine decarboxylase. | 0.26 | 0.56 | 1 | 0.3 | 1 | 1 | 0.078 | 0.74 | 1 | 0.36 | 0.63 | 1 | 0.4 | 0.9 | 1 | 0.12 | 0.66 | 1 | 0.00038 | 3.2e-05 | 0 | 5.7e-05 | 0 | 0.00015 | 0 | 0 | 0 | -Inf | 3 / 36 (8.3%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 0 | 0 | 0 | 6.7e-05 | 0 | 0.00015 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 1.21.-.- | 0.78 | 0.9 | 1 | 0.35 | 1 | 1 | 0.34 | 0.9 | 1 | 0.65 | 0.85 | 1 | 0.23 | 0.88 | 1 | 0.25 | 0.76 | 1 | 9.1e-05 | 2e-05 | 0 | 1.6e-05 | 0 | 4.2e-05 | 3.1e-05 | 0 | 4.8e-05 | 0.95 | 8 / 36 (22%) | 1 / 7 (14%) | 3 / 7 (43%) | 0.143 | 0.429 | 2.3e-05 | 0 | 5.4e-05 | 1.3e-05 | 0 | 3.5e-05 | -0.82 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 | |
| 3.2.2.21 | DNA-3-methyladenine glycosylase II. | 0.43 | 0.69 | 1 | 0.91 | 1 | 1 | 0.35 | 0.9 | 1 | 0.64 | 0.84 | 1 | 0.64 | 0.95 | 1 | 0.24 | 0.76 | 1 | 0.0042 | 0.0042 | 0.0039 | 0.0047 | 0.0039 | 0.0017 | 0.0049 | 0.0043 | 0.0024 | 0.06 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0042 | 0.0046 | 0.002 | 0.0033 | 0.0034 | 0.0012 | -0.35 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.3.153 | (5-formylfuran-3-yl)methyl phosphate synthase. | 0.56 | 0.78 | 1 | 0.1 | 1 | 1 | 0.14 | 0.83 | 1 | 0.79 | 0.92 | 1 | 0.39 | 0.9 | 1 | 0.52 | 0.89 | 1 | 0.00011 | 1.2e-05 | 0 | 2.4e-05 | 0 | 6.4e-05 | 4.4e-06 | 0 | 1.2e-05 | -2.4 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 9.5e-06 | 0 | 3.2e-05 | 1.3e-05 | 0 | 4.3e-05 | 0.45 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.4.1.- | 0.13 | 0.5 | 1 | 0.85 | 1 | 1 | 0.42 | 0.93 | 1 | 0.083 | 0.44 | 1 | 0.45 | 0.92 | 1 | 0.99 | 1 | 1 | 0.068 | 0.068 | 0.065 | 0.058 | 0.056 | 0.011 | 0.057 | 0.049 | 0.017 | -0.025 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.072 | 0.073 | 0.026 | 0.077 | 0.075 | 0.018 | 0.097 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 3.6.1.34 | Transferred entry: 7.1.2.2. | 0.41 | 0.67 | 1 | 0.12 | 1 | 1 | 0.13 | 0.82 | 1 | 0.4 | 0.67 | 1 | 0.17 | 0.87 | 1 | 0.073 | 0.66 | 1 | 6e-04 | 5e-05 | 0 | 6.7e-05 | 0 | 0.00018 | 0.00018 | 0 | 0.00049 | 1.4 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 1.1.1.61 | 4-hydroxybutyrate dehydrogenase. | 0.54 | 0.77 | 1 | 0.98 | 1 | 1 | 0.81 | 1 | 1 | 0.4 | 0.66 | 1 | 0.83 | 0.98 | 1 | 0.57 | 0.9 | 1 | 1e-04 | 2.3e-05 | 0 | 1.7e-05 | 0 | 4.5e-05 | 1.8e-05 | 0 | 4.8e-05 | 0.082 | 8 / 36 (22%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.4e-05 | 0 | 4.1e-05 | 2.9e-05 | 0 | 6.5e-05 | 0.27 | 4 / 11 (36%) | 2 / 11 (18%) | 0.364 | 0.182 |
| 2.1.1.52 | Transferred entry: 2.1.1.171. 2.1.1.172. 2.1.1.173 and 2.1.1.174. | 0.46 | 0.7 | 1 | 0.84 | 1 | 1 | 0.49 | 0.94 | 1 | 0.1 | 0.46 | 1 | 0.65 | 0.95 | 1 | 0.37 | 0.82 | 1 | 0.00055 | 0.00042 | 3e-04 | 0.00037 | 5.7e-05 | 0.00056 | 0.00042 | 0 | 0.00079 | 0.18 | 28 / 36 (78%) | 4 / 7 (57%) | 3 / 7 (43%) | 0.571 | 0.429 | 0.00042 | 0.00024 | 6e-04 | 0.00046 | 0.00057 | 0.00029 | 0.13 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 2.7.8.30 | Transferred entry: 2.4.2.53. | 0.19 | 0.53 | 1 | 0.98 | 1 | 1 | 0.96 | 1 | 1 | 0.22 | 0.52 | 1 | 0.97 | 0.99 | 1 | 0.73 | 0.94 | 1 | 0.00079 | 0.00077 | 0.00045 | 0.001 | 0.0011 | 0.00086 | 0.0012 | 0.00064 | 0.0018 | 0.26 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00059 | 0.00039 | 0.00071 | 0.00051 | 0.00038 | 0.00041 | -0.21 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 2.1.1.220 | tRNA (adenine(58)-N(1))-methyltransferase. | 0.19 | 0.53 | 1 | 0.41 | 1 | 1 | 0.58 | 0.96 | 1 | 0.28 | 0.57 | 1 | 0.22 | 0.88 | 1 | 0.52 | 0.9 | 1 | 0.0022 | 0.0019 | 0.00088 | 0.00063 | 0.00062 | 0.00061 | 0.0011 | 0.0014 | 0.0011 | 0.8 | 30 / 36 (83%) | 5 / 7 (71%) | 6 / 7 (86%) | 0.714 | 0.857 | 0.0027 | 0.0012 | 0.0053 | 0.0022 | 0.00068 | 0.0023 | -0.3 | 9 / 11 (82%) | 10 / 11 (91%) | 0.818 | 0.909 |
| 2.7.8.5 | CDP-diacylglycerol--glycerol-3-phosphate 1-phosphatidyltransferase. | 0.076 | 0.49 | 1 | 0.89 | 1 | 1 | 0.55 | 0.95 | 1 | 0.041 | 0.43 | 1 | 0.48 | 0.92 | 1 | 0.17 | 0.71 | 1 | 0.015 | 0.015 | 0.015 | 0.013 | 0.014 | 0.003 | 0.013 | 0.013 | 0.0044 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.017 | 0.016 | 0.0071 | 0.015 | 0.016 | 0.003 | -0.18 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.23.48 | Plasminogen activator Pla. | 0.068 | 0.49 | 1 | 0.11 | 1 | 1 | 0.076 | 0.74 | 1 | 0.045 | 0.43 | 1 | 0.28 | 0.88 | 1 | 0.18 | 0.72 | 1 | 0.00048 | 5.4e-05 | 0 | 0.00015 | 0 | 0.00035 | 0.00011 | 0 | 0.00028 | -0.45 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 1e-05 | 0 | 3.4e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.1.1.301 | D-arabitol-phosphate dehydrogenase. | 0.57 | 0.78 | 1 | 0.54 | 1 | 1 | 0.79 | 1 | 1 | 0.42 | 0.68 | 1 | 0.2 | 0.87 | 1 | 0.87 | 0.97 | 1 | 0.0015 | 0.0011 | 0.00044 | 0.00093 | 0.00031 | 0.0012 | 0.00091 | 0 | 0.0013 | -0.031 | 25 / 36 (69%) | 5 / 7 (71%) | 3 / 7 (43%) | 0.714 | 0.429 | 0.0013 | 0.00048 | 0.0019 | 0.001 | 0.00056 | 0.0015 | -0.38 | 10 / 11 (91%) | 7 / 11 (64%) | 0.909 | 0.636 |
| 3.6.3.54 | Transferred entry: 7.2.2.8. | 0.14 | 0.5 | 1 | 0.57 | 1 | 1 | 0.83 | 1 | 1 | 0.17 | 0.49 | 1 | 0.57 | 0.93 | 1 | 0.99 | 1 | 1 | 0.0062 | 0.0062 | 0.0062 | 0.0042 | 0.0043 | 0.0025 | 0.0053 | 0.0039 | 0.0045 | 0.34 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0071 | 0.007 | 0.0047 | 0.007 | 0.0071 | 0.0027 | -0.02 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.74 | Glucan 1.4-beta-glucosidase. | 0.74 | 0.88 | 1 | 0.8 | 1 | 1 | 0.72 | 0.98 | 1 | 0.7 | 0.88 | 1 | 0.91 | 0.99 | 1 | 0.57 | 0.9 | 1 | 0.00032 | 9.7e-05 | 0 | 1e-04 | 0 | 0.00022 | 6.9e-05 | 0 | 0.00014 | -0.54 | 11 / 36 (31%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 0.00013 | 0 | 0.00021 | 8e-05 | 0 | 2e-04 | -0.7 | 4 / 11 (36%) | 2 / 11 (18%) | 0.364 | 0.182 |
| 1.17.1.9 | Formate dehydrogenase. | 0.057 | 0.49 | 1 | 0.99 | 1 | 1 | 0.065 | 0.72 | 1 | 0.0047 | 0.36 | 1 | 0.36 | 0.9 | 1 | 0.025 | 0.58 | 1 | 0.0011 | 0.00093 | 0.00067 | 0.00086 | 0.00029 | 0.0013 | 0.00076 | 0.00021 | 0.0012 | -0.18 | 31 / 36 (86%) | 4 / 7 (57%) | 5 / 7 (71%) | 0.571 | 0.714 | 0.0014 | 0.0011 | 0.0011 | 0.00062 | 0.00043 | 0.00048 | -1.2 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.3.1.95 | Acryloyl-CoA reductase (NADH). | 0.55 | 0.77 | 1 | 0.52 | 1 | 1 | 0.23 | 0.88 | 1 | 0.49 | 0.73 | 1 | 0.4 | 0.9 | 1 | 0.26 | 0.76 | 1 | 0.00027 | 0.00014 | 5.2e-05 | 0.00014 | 0 | 0.00026 | 0.00017 | 0.00024 | 0.00016 | 0.28 | 19 / 36 (53%) | 3 / 7 (43%) | 4 / 7 (57%) | 0.429 | 0.571 | 2e-04 | 0.00011 | 0.00033 | 6.9e-05 | 3e-05 | 8.8e-05 | -1.5 | 6 / 11 (55%) | 6 / 11 (55%) | 0.545 | 0.545 |
| 3.5.1.25 | N-acetylglucosamine-6-phosphate deacetylase. | 0.91 | 0.96 | 1 | 0.63 | 1 | 1 | 0.38 | 0.9 | 1 | 0.49 | 0.73 | 1 | 0.52 | 0.93 | 1 | 0.8 | 0.96 | 1 | 0.018 | 0.018 | 0.018 | 0.018 | 0.019 | 0.0041 | 0.017 | 0.017 | 0.0053 | -0.082 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.018 | 0.018 | 0.0043 | 0.019 | 0.017 | 0.0047 | 0.078 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.3.99.31 | Phytoene desaturase (lycopene-forming). | 0.094 | 0.49 | 1 | 0.042 | 0.97 | 1 | 0.1 | 0.82 | 1 | 0.12 | 0.47 | 1 | 0.00094 | 0.62 | 1 | 0.0022 | 0.48 | 1 | 0.00017 | 1.9e-05 | 0 | 3.8e-05 | 0 | 7.5e-05 | 6e-05 | 0 | 0.00013 | 0.66 | 4 / 36 (11%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 2.7.8.28 | Phosphoenolpyruvate transferase. | 0.4 | 0.66 | 1 | 0.51 | 1 | 1 | 0.8 | 1 | 1 | 0.66 | 0.86 | 1 | 0.98 | 0.99 | 1 | 0.37 | 0.82 | 1 | 0.00018 | 1.5e-05 | 0 | 4.1e-05 | 0 | 0.00011 | 2.2e-05 | 0 | 5.8e-05 | -0.9 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 9.5e-06 | 0 | 3.2e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.4.1.289 | N-acetylglucosaminyl-diphospho-decaprenol L-rhamnosyltransferase. | 0.91 | 0.96 | 1 | 0.49 | 1 | 1 | 0.21 | 0.87 | 1 | 0.98 | 0.99 | 1 | 0.5 | 0.93 | 1 | 0.28 | 0.78 | 1 | 0.00053 | 0.00039 | 0.00012 | 0.00036 | 0.00012 | 0.00059 | 0.00025 | 0.00011 | 0.00046 | -0.53 | 26 / 36 (72%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 0.00032 | 9.7e-05 | 0.00042 | 0.00056 | 0.00023 | 0.00089 | 0.81 | 8 / 11 (73%) | 9 / 11 (82%) | 0.727 | 0.818 |
| 3.1.1.11 | Pectinesterase. | 0.42 | 0.68 | 1 | 0.071 | 1 | 1 | 0.012 | 0.61 | 1 | 0.54 | 0.78 | 1 | 0.068 | 0.86 | 1 | 0.039 | 0.63 | 1 | 0.0047 | 0.0047 | 0.0035 | 0.0049 | 0.0042 | 0.0019 | 0.0034 | 0.0026 | 0.002 | -0.53 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0039 | 0.0032 | 0.0019 | 0.0061 | 0.0054 | 0.0052 | 0.65 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.1.297 | Peptide chain release factor N(5)-glutamine methyltransferase. | 0.098 | 0.49 | 1 | 0.73 | 1 | 1 | 0.88 | 1 | 1 | 0.076 | 0.44 | 1 | 0.65 | 0.95 | 1 | 0.36 | 0.82 | 1 | 0.012 | 0.012 | 0.012 | 0.011 | 0.0099 | 0.0032 | 0.011 | 0.011 | 0.0033 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.014 | 0.013 | 0.0047 | 0.013 | 0.013 | 0.0019 | -0.11 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.4.-.- | 0.28 | 0.57 | 1 | 0.99 | 1 | 1 | 0.48 | 0.93 | 1 | 0.085 | 0.44 | 1 | 0.68 | 0.95 | 1 | 0.55 | 0.9 | 1 | 0.0013 | 0.001 | 0.00024 | 0.00095 | 0.00056 | 0.00095 | 0.0012 | 0.00053 | 0.0013 | 0.34 | 29 / 36 (81%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 6e-04 | 9.7e-05 | 0.0012 | 0.0014 | 2e-04 | 0.0027 | 1.2 | 8 / 11 (73%) | 8 / 11 (73%) | 0.727 | 0.727 | |
| 1.5.1.43 | Carboxynorspermidine synthase. | 0.19 | 0.53 | 1 | 0.27 | 1 | 1 | 0.13 | 0.82 | 1 | 0.065 | 0.43 | 1 | 0.23 | 0.88 | 1 | 0.13 | 0.68 | 1 | 0.00089 | 0.00072 | 0.00045 | 0.00081 | 0.00072 | 0.00049 | 0.00058 | 0.00075 | 0.00047 | -0.48 | 29 / 36 (81%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00056 | 0.00038 | 0.00084 | 0.00091 | 0.00028 | 0.0014 | 0.7 | 7 / 11 (64%) | 9 / 11 (82%) | 0.636 | 0.818 |
| 1.20.4.- | 0.76 | 0.88 | 1 | 0.71 | 1 | 1 | 0.16 | 0.85 | 1 | 0.53 | 0.77 | 1 | 0.52 | 0.93 | 1 | 0.088 | 0.66 | 1 | 0.0015 | 0.0015 | 0.0013 | 0.0015 | 0.0013 | 0.00084 | 0.0017 | 0.0014 | 0.0013 | 0.18 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0016 | 0.0014 | 0.00075 | 0.0011 | 0.0011 | 0.00052 | -0.54 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 2.7.1.21 | Thymidine kinase. | 0.77 | 0.89 | 1 | 0.82 | 1 | 1 | 0.22 | 0.88 | 1 | 0.93 | 0.98 | 1 | 0.79 | 0.97 | 1 | 0.4 | 0.83 | 1 | 0.0042 | 0.0042 | 0.004 | 0.0039 | 0.0038 | 0.00087 | 0.0038 | 0.0039 | 0.00057 | -0.037 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.004 | 0.0035 | 0.0021 | 0.0048 | 0.0048 | 0.0013 | 0.26 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.66 | Undecaprenol kinase. | 0.003 | 0.49 | 1 | 0.11 | 1 | 1 | 0.066 | 0.72 | 1 | 0.018 | 0.42 | 1 | 0.018 | 0.76 | 1 | 0.017 | 0.55 | 1 | 0.0025 | 0.0025 | 0.0023 | 0.0016 | 0.0018 | 0.00054 | 0.0022 | 0.0023 | 0.00084 | 0.46 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0031 | 0.0035 | 0.0015 | 0.0026 | 0.0024 | 0.00065 | -0.25 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.3.2.8 | 4-oxalomesaconate tautomerase. | 0.17 | 0.5 | 1 | 0.21 | 1 | 1 | 0.22 | 0.88 | 1 | 0.3 | 0.58 | 1 | 0.78 | 0.97 | 1 | 0.67 | 0.93 | 1 | 0.00034 | 3.8e-05 | 0 | 0.00013 | 0 | 0.00036 | 3.9e-05 | 0 | 1e-04 | -1.7 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1e-05 | 0 | 3.4e-05 | 1.9 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 1.3.1.106 | Cobalt-precorrin-6A reductase. | 0.88 | 0.95 | 1 | 0.27 | 1 | 1 | 0.57 | 0.96 | 1 | 0.67 | 0.86 | 1 | 0.39 | 0.9 | 1 | 0.84 | 0.96 | 1 | 8.8e-05 | 9.8e-06 | 0 | 1.2e-05 | 0 | 3.1e-05 | 0 | 0 | 0 | -Inf | 4 / 36 (11%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 1.2e-05 | 0 | 2.8e-05 | 1.2e-05 | 0 | 4e-05 | 0 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 3.1.3.6 | 3'-nucleotidase. | 0.24 | 0.55 | 1 | 0.21 | 1 | 1 | 0.31 | 0.89 | 1 | 0.28 | 0.57 | 1 | 0.29 | 0.89 | 1 | 0.22 | 0.74 | 1 | 0.00067 | 0.00056 | 0.00025 | 0.00095 | 0.00028 | 0.0014 | 0.00062 | 0.00025 | 0.00089 | -0.62 | 30 / 36 (83%) | 7 / 7 (100%) | 5 / 7 (71%) | 1 | 0.714 | 0.00046 | 0.00013 | 0.00064 | 0.00036 | 0.00023 | 0.00045 | -0.35 | 8 / 11 (73%) | 10 / 11 (91%) | 0.727 | 0.909 |
| 4.1.1.5 | Acetolactate decarboxylase. | 0.29 | 0.57 | 1 | 0.71 | 1 | 1 | 0.13 | 0.82 | 1 | 0.55 | 0.78 | 1 | 0.78 | 0.97 | 1 | 0.18 | 0.71 | 1 | 3e-04 | 0.00023 | 0.00012 | 0.00015 | 0.00014 | 9.5e-05 | 0.00023 | 0.00016 | 0.00025 | 0.62 | 27 / 36 (75%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00038 | 0.00014 | 0.00047 | 0.00013 | 7.5e-05 | 0.00018 | -1.5 | 9 / 11 (82%) | 6 / 11 (55%) | 0.818 | 0.545 |
| 7.2.2.12 | P-type Zn(2+) transporter. | 0.094 | 0.49 | 1 | 0.14 | 1 | 1 | 0.13 | 0.82 | 1 | 0.047 | 0.43 | 1 | 0.22 | 0.88 | 1 | 0.19 | 0.72 | 1 | 0.00075 | 8.3e-05 | 0 | 0.00022 | 0 | 0.00053 | 2e-04 | 0 | 0.00054 | -0.14 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.4.2.21 | Nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase. | 0.2 | 0.53 | 1 | 0.14 | 1 | 1 | 0.23 | 0.88 | 1 | 0.74 | 0.9 | 1 | 0.44 | 0.91 | 1 | 0.69 | 0.93 | 1 | 0.0088 | 0.0088 | 0.009 | 0.0098 | 0.01 | 0.0018 | 0.0084 | 0.0089 | 0.0023 | -0.22 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0086 | 0.0091 | 0.002 | 0.0086 | 0.0082 | 0.0025 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.14.5 | Dipeptidyl-peptidase IV. | 0.098 | 0.49 | 1 | 0.22 | 1 | 1 | 0.23 | 0.88 | 1 | 0.12 | 0.47 | 1 | 0.19 | 0.87 | 1 | 0.32 | 0.79 | 1 | 0.004 | 0.004 | 0.0018 | 0.0011 | 0.0013 | 0.00068 | 0.0028 | 0.0018 | 0.0029 | 1.3 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0063 | 0.0028 | 0.012 | 0.0043 | 0.0033 | 0.0046 | -0.55 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.132 | GDP-mannose 6-dehydrogenase. | 0.021 | 0.49 | 1 | 0.71 | 1 | 1 | 0.21 | 0.87 | 1 | 0.091 | 0.45 | 1 | 0.83 | 0.98 | 1 | 0.21 | 0.74 | 1 | 0.00054 | 4e-04 | 0.00021 | 8e-04 | 0.00055 | 7e-04 | 0.00077 | 0.00048 | 0.00077 | -0.055 | 27 / 36 (75%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00024 | 0.00019 | 0.00021 | 8.2e-05 | 2.9e-05 | 0.00013 | -1.5 | 8 / 11 (73%) | 6 / 11 (55%) | 0.727 | 0.545 |
| 2.9.1.- | 0.39 | 0.65 | 1 | 0.54 | 1 | 1 | 0.96 | 1 | 1 | 0.47 | 0.72 | 1 | 0.54 | 0.93 | 1 | 0.94 | 0.99 | 1 | 0.00027 | 4.5e-05 | 0 | 8.7e-05 | 0 | 0.00018 | 8.7e-05 | 0 | 0.00023 | 0 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.9e-05 | 0 | 7.4e-05 | 8.1e-06 | 0 | 2.7e-05 | -1.8 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 | |
| 3.2.1.82 | Exo-poly-alpha-digalacturonosidase. | 0.32 | 0.6 | 1 | 0.23 | 1 | 1 | 0.065 | 0.72 | 1 | 0.3 | 0.58 | 1 | 0.27 | 0.88 | 1 | 0.16 | 0.71 | 1 | 0.0083 | 0.0083 | 0.0059 | 0.0089 | 0.0072 | 0.0036 | 0.0065 | 0.0052 | 0.0047 | -0.45 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0067 | 0.0054 | 0.0052 | 0.011 | 0.0066 | 0.01 | 0.72 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.1.113 | o-succinylbenzoate synthase. | 0.74 | 0.88 | 1 | 0.34 | 1 | 1 | 0.43 | 0.93 | 1 | 0.74 | 0.9 | 1 | 0.41 | 0.9 | 1 | 0.58 | 0.91 | 1 | 0.00048 | 0.00048 | 0.00036 | 0.00052 | 0.00042 | 0.00035 | 0.00037 | 0.00024 | 0.00027 | -0.49 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 5e-04 | 0.00039 | 0.00051 | 0.00052 | 0.00028 | 0.00051 | 0.057 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.141 | 4-alpha-D-((1->4)-alpha-D-glucano)trehalose trehalohydrolase. | 0.7 | 0.86 | 1 | 0.29 | 1 | 1 | 0.34 | 0.89 | 1 | 0.67 | 0.86 | 1 | 0.37 | 0.9 | 1 | 0.26 | 0.76 | 1 | 9.9e-05 | 2.8e-05 | 0 | 4e-05 | 0 | 5.1e-05 | 1.3e-05 | 0 | 2.4e-05 | -1.6 | 10 / 36 (28%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 5.7e-05 | 0 | 8.2e-05 | 0 | 0 | 0 | -Inf | 5 / 11 (45%) | 0 / 11 (0%) | 0.455 | 0 |
| 1.1.1.391 | 3-beta-hydroxycholanate 3-dehydrogenase (NAD(+)). | 0.74 | 0.88 | 1 | 0.94 | 1 | 1 | 0.71 | 0.98 | 1 | 0.95 | 0.98 | 1 | 0.9 | 0.99 | 1 | 0.55 | 0.9 | 1 | 0.00015 | 2e-05 | 0 | 4e-05 | 0 | 1e-04 | 3.4e-05 | 0 | 9.1e-05 | -0.23 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 1.7e-05 | 0 | 3.9e-05 | 2.7e-06 | 0 | 8.9e-06 | -2.7 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 1.6.5.11 | NADH dehydrogenase (quinone). | 0.029 | 0.49 | 1 | 0.48 | 1 | 1 | 0.68 | 0.98 | 1 | 0.19 | 0.5 | 1 | 0.95 | 0.99 | 1 | 0.7 | 0.94 | 1 | 0.018 | 0.018 | 0.016 | 0.022 | 0.022 | 0.0074 | 0.02 | 0.019 | 0.0077 | -0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.016 | 0.015 | 0.0045 | 0.015 | 0.015 | 0.0045 | -0.093 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.1.83 | 4-hydroxyphenylacetate decarboxylase. | 0.51 | 0.74 | 1 | 0.69 | 1 | 1 | 0.65 | 0.97 | 1 | 0.94 | 0.98 | 1 | 0.77 | 0.97 | 1 | 0.78 | 0.95 | 1 | 0.015 | 0.015 | 0.016 | 0.016 | 0.017 | 0.0064 | 0.016 | 0.012 | 0.012 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.014 | 0.016 | 0.0041 | 0.014 | 0.013 | 0.0034 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.3.1.43 | Arogenate dehydrogenase. | 0.29 | 0.57 | 1 | 0.24 | 1 | 1 | 0.44 | 0.93 | 1 | 0.26 | 0.55 | 1 | 0.47 | 0.92 | 1 | 0.73 | 0.94 | 1 | 0.012 | 0.012 | 0.012 | 0.011 | 0.0098 | 0.0037 | 0.0092 | 0.0093 | 0.0037 | -0.26 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.013 | 0.013 | 0.005 | 0.012 | 0.012 | 0.0031 | -0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.7.12 | UDP-glucose--hexose-1-phosphate uridylyltransferase. | 0.078 | 0.49 | 1 | 0.83 | 1 | 1 | 0.8 | 1 | 1 | 0.086 | 0.44 | 1 | 0.32 | 0.9 | 1 | 0.29 | 0.78 | 1 | 0.022 | 0.022 | 0.022 | 0.019 | 0.017 | 0.0045 | 0.02 | 0.021 | 0.0058 | 0.074 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.024 | 0.023 | 0.0068 | 0.024 | 0.025 | 0.0053 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.6.1.48 | 5-aminovalerate transaminase. | 0.096 | 0.49 | 1 | 0.084 | 1 | 1 | 0.21 | 0.87 | 1 | 0.063 | 0.43 | 1 | 0.2 | 0.87 | 1 | 0.41 | 0.84 | 1 | 0.00033 | 5.6e-05 | 0 | 0.00015 | 0 | 0.00035 | 0.00013 | 0 | 0.00033 | -0.21 | 6 / 36 (17%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 5.1e-06 | 0 | 1.7e-05 | 2.7e-06 | 0 | 8.9e-06 | -0.92 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 6.1.-.- | 0.19 | 0.53 | 1 | 0.06 | 1 | 1 | 0.038 | 0.71 | 1 | 0.28 | 0.57 | 1 | 0.12 | 0.87 | 1 | 0.22 | 0.74 | 1 | 0.00054 | 0.00039 | 2e-04 | 0.00054 | 0.00035 | 0.00043 | 0.00026 | 0.00025 | 3e-04 | -1.1 | 26 / 36 (72%) | 6 / 7 (86%) | 4 / 7 (57%) | 0.857 | 0.571 | 0.00025 | 0.00011 | 0.00037 | 0.00052 | 0.00011 | 0.00074 | 1.1 | 9 / 11 (82%) | 7 / 11 (64%) | 0.818 | 0.636 | |
| 2.4.2.22 | Xanthine phosphoribosyltransferase. | 0.48 | 0.72 | 1 | 0.54 | 1 | 1 | 0.53 | 0.95 | 1 | 0.3 | 0.58 | 1 | 0.93 | 0.99 | 1 | 1 | 1 | 1 | 0.01 | 0.01 | 0.01 | 0.0097 | 0.0097 | 0.0014 | 0.0092 | 0.0088 | 0.003 | -0.076 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.0092 | 0.0031 | 0.011 | 0.01 | 0.0035 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.3.- | 0.3 | 0.58 | 1 | 0.49 | 1 | 1 | 0.79 | 1 | 1 | 0.3 | 0.58 | 1 | 0.39 | 0.9 | 1 | 0.6 | 0.91 | 1 | 0.0021 | 0.0021 | 0.0018 | 0.0015 | 0.0012 | 0.0012 | 0.0018 | 0.0014 | 0.0014 | 0.26 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0023 | 0.0024 | 0.0017 | 0.0025 | 0.0022 | 0.002 | 0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 4.2.1.80 | 2-oxopent-4-enoate hydratase. | 0.028 | 0.49 | 1 | 0.045 | 0.97 | 1 | 0.047 | 0.71 | 1 | 0.079 | 0.44 | 1 | 0.1 | 0.86 | 1 | 0.1 | 0.66 | 1 | 8.3e-05 | 6.9e-06 | 0 | 2.7e-05 | 0 | 6.1e-05 | 0 | 0 | 0 | -Inf | 3 / 36 (8.3%) | 2 / 7 (29%) | 0 / 7 (0%) | 0.286 | 0 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.2.-.- | 0.22 | 0.54 | 1 | 0.59 | 1 | 1 | 0.11 | 0.82 | 1 | 0.23 | 0.53 | 1 | 0.58 | 0.93 | 1 | 0.2 | 0.73 | 1 | 0.005 | 0.005 | 0.0031 | 0.0054 | 0.0057 | 0.0026 | 0.0053 | 0.0031 | 0.0049 | -0.027 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0036 | 0.0026 | 0.0028 | 0.0058 | 0.0031 | 0.0059 | 0.69 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 3.2.1.18 | Exo-alpha-sialidase. | 0.77 | 0.89 | 1 | 0.81 | 1 | 1 | 0.7 | 0.98 | 1 | 0.95 | 0.98 | 1 | 0.88 | 0.99 | 1 | 0.95 | 0.99 | 1 | 0.005 | 0.005 | 0.0042 | 0.0052 | 0.0042 | 0.0041 | 0.0049 | 0.0061 | 0.0031 | -0.086 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0046 | 0.0043 | 0.0035 | 0.0053 | 0.0035 | 0.0057 | 0.2 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.210 | 3-beta-(or 20-alpha)-hydroxysteroid dehydrogenase. | 0.95 | 0.98 | 1 | 0.03 | 0.97 | 1 | 0.31 | 0.89 | 1 | 0.84 | 0.94 | 1 | 0.029 | 0.76 | 1 | 0.26 | 0.76 | 1 | 9.3e-05 | 2.3e-05 | 0 | 4.6e-06 | 0 | 1.2e-05 | 5.3e-05 | 5.4e-05 | 5.5e-05 | 3.5 | 9 / 36 (25%) | 1 / 7 (14%) | 4 / 7 (57%) | 0.143 | 0.571 | 9.5e-06 | 0 | 3.2e-05 | 3e-05 | 0 | 5.4e-05 | 1.7 | 1 / 11 (9.1%) | 3 / 11 (27%) | 0.0909 | 0.273 |
| 1.4.99.- | 0.28 | 0.57 | 1 | 0.16 | 1 | 1 | 0.91 | 1 | 1 | 0.51 | 0.75 | 1 | 0.58 | 0.93 | 1 | 0.48 | 0.87 | 1 | 0.00024 | 3.4e-05 | 0 | 1e-04 | 0 | 0.00027 | 4.8e-05 | 0 | 0.00013 | -1.1 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 1.4e-05 | 0 | 3.2e-05 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 | |
| 2.1.1.163 | Demethylmenaquinone methyltransferase. | 0.13 | 0.5 | 1 | 0.57 | 1 | 1 | 0.52 | 0.95 | 1 | 0.14 | 0.47 | 1 | 0.94 | 0.99 | 1 | 0.88 | 0.97 | 1 | 0.0066 | 0.0066 | 0.0068 | 0.0057 | 0.006 | 0.0019 | 0.0054 | 0.0049 | 0.0023 | -0.078 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0072 | 0.0074 | 0.0019 | 0.0074 | 0.0069 | 0.0019 | 0.04 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.5.1.147 | 5-amino-6-(D-ribitylamino)uracil--L-tyrosine 4-hydroxyphenyl transferase. | 0.89 | 0.95 | 1 | 0.42 | 1 | 1 | 0.89 | 1 | 1 | 0.72 | 0.89 | 1 | 0.65 | 0.95 | 1 | 0.77 | 0.95 | 1 | 0.00016 | 2.2e-05 | 0 | 2.8e-05 | 0 | 7.5e-05 | 4.4e-06 | 0 | 1.2e-05 | -2.7 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3e-05 | 0 | 7e-05 | 2.1e-05 | 0 | 6.8e-05 | -0.51 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 1.6.1.2 | NAD(P)(+) transhydrogenase (Re/Si-specific). | 0.18 | 0.52 | 1 | 0.32 | 1 | 1 | 0.55 | 0.95 | 1 | 0.073 | 0.43 | 1 | 0.4 | 0.9 | 1 | 0.51 | 0.89 | 1 | 0.0016 | 0.0013 | 0.00084 | 6e-04 | 0.00028 | 0.00069 | 0.0011 | 0.00084 | 0.0011 | 0.87 | 28 / 36 (78%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.0016 | 7e-04 | 0.0029 | 0.0015 | 0.0014 | 0.0014 | -0.093 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 2.7.7.27 | Glucose-1-phosphate adenylyltransferase. | 0.3 | 0.58 | 1 | 0.4 | 1 | 1 | 0.61 | 0.96 | 1 | 0.28 | 0.57 | 1 | 0.92 | 0.99 | 1 | 0.71 | 0.94 | 1 | 0.052 | 0.052 | 0.053 | 0.049 | 0.048 | 0.011 | 0.046 | 0.051 | 0.018 | -0.091 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.056 | 0.057 | 0.013 | 0.055 | 0.054 | 0.015 | -0.026 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.2.3.3 | Pyruvate oxidase. | 0.64 | 0.83 | 1 | 0.45 | 1 | 1 | 0.51 | 0.95 | 1 | 0.4 | 0.67 | 1 | 0.45 | 0.92 | 1 | 0.46 | 0.86 | 1 | 0.00028 | 0.00016 | 5.8e-05 | 0.00013 | 0 | 0.00026 | 0.00022 | 0 | 3e-04 | 0.76 | 21 / 36 (58%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 0.00015 | 0.00012 | 0.00019 | 0.00016 | 5.9e-05 | 0.00036 | 0.093 | 7 / 11 (64%) | 8 / 11 (73%) | 0.636 | 0.727 |
| 4.1.3.30 | Methylisocitrate lyase. | 0.6 | 0.8 | 1 | 0.63 | 1 | 1 | 0.13 | 0.82 | 1 | 0.83 | 0.94 | 1 | 0.82 | 0.98 | 1 | 0.21 | 0.74 | 1 | 7.3e-05 | 6e-06 | 0 | 2.3e-06 | 0 | 6.1e-06 | 0 | 0 | 0 | -Inf | 3 / 36 (8.3%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 0 | 0 | 0 | 1.8e-05 | 0 | 4.5e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 1.2.98.1 | Formaldehyde dismutase. | 0.66 | 0.84 | 1 | 0.74 | 1 | 1 | 0.43 | 0.93 | 1 | 0.65 | 0.85 | 1 | 0.71 | 0.95 | 1 | 0.37 | 0.82 | 1 | 0.00022 | 0.00011 | 1.6e-05 | 0.00011 | 8e-05 | 0.00017 | 1e-04 | 0 | 0.00016 | -0.14 | 18 / 36 (50%) | 4 / 7 (57%) | 3 / 7 (43%) | 0.571 | 0.429 | 0.00016 | 9.7e-05 | 0.00027 | 5.8e-05 | 0 | 0.00011 | -1.5 | 7 / 11 (64%) | 4 / 11 (36%) | 0.636 | 0.364 |
| 3.2.1.10 | Oligo-1.6-glucosidase. | 0.5 | 0.73 | 1 | 0.74 | 1 | 1 | 0.38 | 0.9 | 1 | 0.42 | 0.68 | 1 | 0.6 | 0.94 | 1 | 0.34 | 0.8 | 1 | 0.037 | 0.037 | 0.034 | 0.034 | 0.026 | 0.015 | 0.038 | 0.041 | 0.022 | 0.16 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.042 | 0.034 | 0.028 | 0.035 | 0.04 | 0.01 | -0.26 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.165 | Exo-1.4-beta-D-glucosaminidase. | 0.99 | 1 | 1 | 0.44 | 1 | 1 | 0.88 | 1 | 1 | 0.83 | 0.94 | 1 | 0.81 | 0.98 | 1 | 0.56 | 0.9 | 1 | 0.01 | 0.01 | 0.0089 | 0.01 | 0.011 | 0.0044 | 0.0088 | 0.0094 | 0.0027 | -0.18 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.0079 | 0.0095 | 0.0098 | 0.0087 | 0.0063 | -0.17 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.1.188 | 23S rRNA (guanine(748)-N(1))-methyltransferase. | 0.3 | 0.58 | 1 | 0.024 | 0.92 | 1 | 0.13 | 0.82 | 1 | 0.24 | 0.54 | 1 | 0.025 | 0.76 | 1 | 0.096 | 0.66 | 1 | 0.00015 | 6.8e-05 | 0 | 1.1e-05 | 0 | 2e-05 | 0.00011 | 8.1e-05 | 0.00014 | 3.3 | 16 / 36 (44%) | 2 / 7 (29%) | 4 / 7 (57%) | 0.286 | 0.571 | 5.7e-05 | 0 | 8.1e-05 | 8.8e-05 | 0 | 0.00014 | 0.63 | 5 / 11 (45%) | 5 / 11 (45%) | 0.455 | 0.455 |
| 3.1.-.- | 0.42 | 0.68 | 1 | 0.79 | 1 | 1 | 0.67 | 0.98 | 1 | 0.33 | 0.6 | 1 | 0.13 | 0.87 | 1 | 0.17 | 0.71 | 1 | 0.32 | 0.32 | 0.33 | 0.31 | 0.31 | 0.05 | 0.3 | 0.34 | 0.068 | -0.047 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.33 | 0.35 | 0.055 | 0.33 | 0.33 | 0.04 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 1.9.3.1 | Cytochrome-c oxidase. | 0.2 | 0.53 | 1 | 0.73 | 1 | 1 | 0.73 | 0.99 | 1 | 0.42 | 0.68 | 1 | 0.19 | 0.87 | 1 | 0.27 | 0.77 | 1 | 4e-04 | 0.00012 | 0 | 0.00033 | 0 | 0.00079 | 0.00023 | 0 | 0.00045 | -0.52 | 11 / 36 (31%) | 2 / 7 (29%) | 3 / 7 (43%) | 0.286 | 0.429 | 2.7e-05 | 0 | 7.2e-05 | 1.9e-05 | 0 | 3.8e-05 | -0.51 | 3 / 11 (27%) | 3 / 11 (27%) | 0.273 | 0.273 |
| 1.6.5.5 | NADPH:quinone reductase. | 0.37 | 0.64 | 1 | 0.71 | 1 | 1 | 0.86 | 1 | 1 | 0.58 | 0.8 | 1 | 0.83 | 0.98 | 1 | 0.91 | 0.98 | 1 | 0.00049 | 0.00041 | 0.00025 | 0.00063 | 0.00013 | 0.00075 | 0.00045 | 0.00053 | 0.00036 | -0.49 | 30 / 36 (83%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00036 | 0.00019 | 0.00042 | 3e-04 | 3e-04 | 0.00032 | -0.26 | 9 / 11 (82%) | 9 / 11 (82%) | 0.818 | 0.818 |
| 2.4.2.7 | Adenine phosphoribosyltransferase. | 0.44 | 0.69 | 1 | 0.035 | 0.97 | 1 | 0.1 | 0.82 | 1 | 0.3 | 0.58 | 1 | 0.3 | 0.89 | 1 | 0.57 | 0.91 | 1 | 0.0081 | 0.0081 | 0.0079 | 0.0079 | 0.0073 | 0.0015 | 0.0067 | 0.0072 | 0.0017 | -0.24 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0087 | 0.008 | 0.0024 | 0.0085 | 0.0085 | 0.0016 | -0.034 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.14.99.- | 0.076 | 0.49 | 1 | 0.04 | 0.97 | 1 | 0.2 | 0.87 | 1 | 0.18 | 0.5 | 1 | 0.0076 | 0.71 | 1 | 0.067 | 0.66 | 1 | 0.00016 | 4.3e-05 | 0 | 9.7e-05 | 0 | 0.00021 | 0.00011 | 0.00011 | 0.00012 | 0.18 | 10 / 36 (28%) | 2 / 7 (29%) | 5 / 7 (71%) | 0.286 | 0.714 | 2.8e-06 | 0 | 9.4e-06 | 9.1e-06 | 0 | 2.2e-05 | 1.7 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 | |
| 3.5.3.12 | Agmatine deiminase. | 0.71 | 0.87 | 1 | 0.23 | 1 | 1 | 0.46 | 0.93 | 1 | 0.84 | 0.94 | 1 | 0.05 | 0.81 | 1 | 0.15 | 0.71 | 1 | 0.0071 | 0.0071 | 0.0069 | 0.007 | 0.0071 | 0.0022 | 0.0082 | 0.0093 | 0.0029 | 0.23 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0066 | 0.0062 | 0.0019 | 0.0068 | 0.0067 | 0.0015 | 0.043 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.1.16 | Acetylornithine deacetylase. | 0.2 | 0.53 | 1 | 0.35 | 1 | 1 | 0.67 | 0.98 | 1 | 0.38 | 0.65 | 1 | 0.58 | 0.93 | 1 | 0.44 | 0.86 | 1 | 0.0015 | 0.0014 | 0.00099 | 0.0019 | 0.0018 | 0.0012 | 0.0016 | 0.00085 | 0.0015 | -0.25 | 34 / 36 (94%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0011 | 0.00097 | 0.00067 | 0.0014 | 0.00068 | 0.0017 | 0.35 | 11 / 11 (100%) | 9 / 11 (82%) | 1 | 0.818 |
| 5.1.1.- | 0.57 | 0.78 | 1 | 0.96 | 1 | 1 | 0.37 | 0.9 | 1 | 0.55 | 0.78 | 1 | 0.51 | 0.93 | 1 | 0.59 | 0.91 | 1 | 0.0051 | 0.0051 | 0.0048 | 0.005 | 0.0052 | 0.0016 | 0.005 | 0.005 | 0.0012 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0048 | 0.003 | 0.0038 | 0.0056 | 0.0061 | 0.0027 | 0.22 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 2.4.2.8 | Hypoxanthine phosphoribosyltransferase. | 0.061 | 0.49 | 1 | 0.55 | 1 | 1 | 0.59 | 0.96 | 1 | 0.091 | 0.45 | 1 | 0.67 | 0.95 | 1 | 0.11 | 0.66 | 1 | 0.012 | 0.012 | 0.012 | 0.011 | 0.01 | 0.0021 | 0.01 | 0.011 | 0.0023 | -0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.013 | 0.012 | 0.0035 | 0.012 | 0.013 | 0.0015 | -0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.21.102 | C-terminal processing peptidase. | 0.38 | 0.65 | 1 | 0.87 | 1 | 1 | 0.38 | 0.9 | 1 | 0.67 | 0.86 | 1 | 0.72 | 0.95 | 1 | 0.79 | 0.96 | 1 | 0.011 | 0.011 | 0.011 | 0.012 | 0.013 | 0.004 | 0.012 | 0.011 | 0.0043 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.01 | 0.011 | 0.0032 | 0.013 | 0.0094 | 0.0077 | 0.38 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.4.3.- | 0.15 | 0.5 | 1 | 0.14 | 1 | 1 | 0.64 | 0.97 | 1 | 0.14 | 0.47 | 1 | 0.22 | 0.88 | 1 | 0.73 | 0.94 | 1 | 0.00036 | 9.9e-05 | 0 | 0.00024 | 1.6e-05 | 0.00052 | 0.00017 | 0 | 4e-04 | -0.5 | 10 / 36 (28%) | 4 / 7 (57%) | 2 / 7 (29%) | 0.571 | 0.286 | 5.1e-05 | 0 | 0.00012 | 9.5e-06 | 0 | 2.3e-05 | -2.4 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 | |
| 4.2.1.20 | Tryptophan synthase. | 0.056 | 0.49 | 1 | 0.32 | 1 | 1 | 0.52 | 0.95 | 1 | 0.11 | 0.47 | 1 | 0.089 | 0.86 | 1 | 0.16 | 0.71 | 1 | 0.032 | 0.032 | 0.031 | 0.028 | 0.028 | 0.0034 | 0.03 | 0.03 | 0.0055 | 0.1 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.033 | 0.033 | 0.0086 | 0.033 | 0.035 | 0.0042 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.5.1.77 | Transferred entry: 2.5.1.147. | 0.93 | 0.97 | 1 | 0.73 | 1 | 1 | 0.15 | 0.84 | 1 | 0.96 | 0.99 | 1 | 0.9 | 0.99 | 1 | 0.24 | 0.76 | 1 | 0.00015 | 5.5e-05 | 0 | 3.9e-05 | 0 | 6.9e-05 | 2.1e-05 | 0 | 3.7e-05 | -0.89 | 13 / 36 (36%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 3.7e-05 | 0 | 7e-05 | 0.00011 | 7.5e-05 | 0.00014 | 1.6 | 3 / 11 (27%) | 6 / 11 (55%) | 0.273 | 0.545 |
| 1.3.1.54 | Precorrin-6A reductase. | 0.47 | 0.71 | 1 | 0.93 | 1 | 1 | 0.7 | 0.98 | 1 | 0.8 | 0.92 | 1 | 0.4 | 0.9 | 1 | 0.22 | 0.74 | 1 | 0.011 | 0.011 | 0.012 | 0.011 | 0.013 | 0.0032 | 0.012 | 0.013 | 0.004 | 0.13 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.01 | 0.01 | 0.0042 | 0.0097 | 0.0086 | 0.0041 | -0.044 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.11.1.1 | NADH peroxidase. | 0.33 | 0.6 | 1 | 0.19 | 1 | 1 | 0.31 | 0.89 | 1 | 0.92 | 0.97 | 1 | 0.56 | 0.93 | 1 | 0.87 | 0.97 | 1 | 0.015 | 0.015 | 0.015 | 0.017 | 0.016 | 0.0052 | 0.015 | 0.018 | 0.0059 | -0.18 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.015 | 0.015 | 0.003 | 0.015 | 0.015 | 0.0031 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.1.173 | 23S rRNA (guanine(2445)-N(2))-methyltransferase. | 0.17 | 0.51 | 1 | 0.94 | 1 | 1 | 0.57 | 0.96 | 1 | 0.22 | 0.52 | 1 | 0.69 | 0.95 | 1 | 0.34 | 0.8 | 1 | 0.018 | 0.018 | 0.018 | 0.016 | 0.016 | 0.0045 | 0.017 | 0.019 | 0.0084 | 0.087 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.02 | 0.023 | 0.0066 | 0.019 | 0.017 | 0.0042 | -0.074 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.5.3.2 | N-methyl-L-amino-acid oxidase. | 0.69 | 0.85 | 1 | 0.17 | 1 | 1 | 0.24 | 0.88 | 1 | 0.75 | 0.9 | 1 | 0.25 | 0.88 | 1 | 0.26 | 0.76 | 1 | 2e-04 | 2.3e-05 | 0 | 1.7e-05 | 0 | 4.5e-05 | 8.7e-05 | 0 | 0.00023 | 2.4 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 5.1e-06 | 0 | 1.7e-05 | 2.7e-06 | 0 | 8.9e-06 | -0.92 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.7.11.1 | Non-specific serine/threonine protein kinase. | 0.087 | 0.49 | 1 | 0.73 | 1 | 1 | 0.57 | 0.96 | 1 | 0.044 | 0.43 | 1 | 0.34 | 0.9 | 1 | 0.3 | 0.78 | 1 | 0.089 | 0.089 | 0.086 | 0.072 | 0.071 | 0.021 | 0.077 | 0.085 | 0.03 | 0.097 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.1 | 0.086 | 0.061 | 0.094 | 0.1 | 0.022 | -0.089 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.47 | Glucose 1-dehydrogenase (NAD(P)(+)). | 0.57 | 0.78 | 1 | 0.29 | 1 | 1 | 0.32 | 0.89 | 1 | 0.28 | 0.57 | 1 | 0.098 | 0.86 | 1 | 0.099 | 0.66 | 1 | 0.0027 | 0.0027 | 0.0023 | 0.0025 | 0.0029 | 0.0015 | 0.003 | 0.0028 | 0.002 | 0.26 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0027 | 0.0021 | 0.0013 | 0.0027 | 0.0022 | 0.0015 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.14.13.107 | Limonene 1.2-monooxygenase. | 0.57 | 0.79 | 1 | 0.88 | 1 | 1 | 0.61 | 0.96 | 1 | 0.97 | 0.99 | 1 | 0.92 | 0.99 | 1 | 0.56 | 0.9 | 1 | 0.00027 | 6.1e-05 | 0 | 0.00012 | 0 | 0.00031 | 0.00014 | 0 | 0.00036 | 0.22 | 8 / 36 (22%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.8e-05 | 0 | 4.9e-05 | 9.5e-06 | 0 | 2.3e-05 | -1.6 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 2.7.7.41 | Phosphatidate cytidylyltransferase. | 0.5 | 0.73 | 1 | 0.36 | 1 | 1 | 0.46 | 0.93 | 1 | 0.35 | 0.62 | 1 | 0.89 | 0.99 | 1 | 0.82 | 0.96 | 1 | 0.014 | 0.014 | 0.013 | 0.013 | 0.013 | 0.0021 | 0.012 | 0.012 | 0.0024 | -0.12 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.014 | 0.013 | 0.0043 | 0.014 | 0.014 | 0.002 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.6.5.- | 0.5 | 0.74 | 1 | 0.071 | 1 | 1 | 0.023 | 0.71 | 1 | 0.74 | 0.9 | 1 | 0.067 | 0.86 | 1 | 0.081 | 0.66 | 1 | 0.0039 | 0.0038 | 0.0026 | 0.0042 | 0.0029 | 0.0035 | 0.0027 | 0.0023 | 0.002 | -0.64 | 35 / 36 (97%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.0032 | 0.0024 | 0.0021 | 0.0048 | 0.004 | 0.0037 | 0.58 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 2.1.1.190 | 23S rRNA (uracil(1939)-C(5))-methyltransferase. | 0.52 | 0.75 | 1 | 0.46 | 1 | 1 | 0.55 | 0.95 | 1 | 0.37 | 0.64 | 1 | 0.11 | 0.87 | 1 | 0.15 | 0.71 | 1 | 0.032 | 0.032 | 0.032 | 0.03 | 0.032 | 0.0062 | 0.033 | 0.036 | 0.01 | 0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.033 | 0.034 | 0.0084 | 0.032 | 0.033 | 0.0043 | -0.044 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.6.1.66 | XTP/dITP diphosphatase. | 0.007 | 0.49 | 1 | 0.93 | 1 | 1 | 0.4 | 0.92 | 1 | 0.03 | 0.43 | 1 | 0.25 | 0.88 | 1 | 0.049 | 0.65 | 1 | 0.011 | 0.011 | 0.011 | 0.0091 | 0.0092 | 0.0022 | 0.009 | 0.0089 | 0.0018 | -0.016 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.012 | 0.012 | 0.0031 | 0.011 | 0.011 | 0.0019 | -0.13 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.89 | Thiamine kinase. | 0.13 | 0.5 | 1 | 0.32 | 1 | 1 | 0.44 | 0.93 | 1 | 0.12 | 0.47 | 1 | 0.25 | 0.88 | 1 | 0.33 | 0.8 | 1 | 0.00027 | 3e-05 | 0 | 7e-05 | 0 | 0.00018 | 8.6e-05 | 0 | 0.00018 | 0.3 | 4 / 36 (11%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 3.5.1.124 | Protein deglycase. | 0.27 | 0.56 | 1 | 0.26 | 1 | 1 | 0.59 | 0.96 | 1 | 0.53 | 0.77 | 1 | 0.71 | 0.95 | 1 | 0.86 | 0.96 | 1 | 0.00045 | 7.5e-05 | 0 | 0.00022 | 0 | 0.00058 | 0.00012 | 0 | 0.00031 | -0.87 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 1.9e-05 | 0 | 4.2e-05 | 1.1e-05 | 0 | 3.6e-05 | -0.79 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 3.6.4.13 | RNA helicase. | 0.18 | 0.52 | 1 | 0.46 | 1 | 1 | 0.59 | 0.96 | 1 | 0.055 | 0.43 | 1 | 0.4 | 0.9 | 1 | 0.38 | 0.82 | 1 | 0.032 | 0.032 | 0.024 | 0.023 | 0.019 | 0.0092 | 0.019 | 0.019 | 0.01 | -0.28 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.041 | 0.022 | 0.047 | 0.036 | 0.031 | 0.019 | -0.19 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.5.1.39 | FMN reductase (NAD(P)H). | 0.68 | 0.85 | 1 | 0.34 | 1 | 1 | 0.54 | 0.95 | 1 | 0.32 | 0.59 | 1 | 0.051 | 0.81 | 1 | 0.084 | 0.66 | 1 | 0.0076 | 0.0076 | 0.0071 | 0.0072 | 0.0071 | 0.0032 | 0.0079 | 0.007 | 0.0023 | 0.13 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0075 | 0.0071 | 0.0021 | 0.0078 | 0.0073 | 0.0027 | 0.057 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.85 | 6-phospho-beta-galactosidase. | 0.013 | 0.49 | 1 | 0.42 | 1 | 1 | 0.18 | 0.85 | 1 | 0.0088 | 0.38 | 1 | 0.21 | 0.88 | 1 | 0.17 | 0.71 | 1 | 0.0036 | 0.0036 | 0.0026 | 0.0014 | 0.00071 | 0.0014 | 0.0022 | 0.0023 | 0.0023 | 0.65 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0059 | 0.0035 | 0.0078 | 0.0037 | 0.0028 | 0.0025 | -0.67 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.8.1.3 | Haloacetate dehalogenase. | 0.017 | 0.49 | 1 | 0.32 | 1 | 1 | 0.41 | 0.92 | 1 | 0.0039 | 0.36 | 1 | 0.2 | 0.87 | 1 | 0.28 | 0.78 | 1 | 0.0018 | 0.0017 | 0.0012 | 0.00082 | 0.00035 | 0.001 | 0.0012 | 0.00051 | 0.0013 | 0.55 | 33 / 36 (92%) | 5 / 7 (71%) | 6 / 7 (86%) | 0.714 | 0.857 | 0.0021 | 0.0023 | 0.0013 | 0.0021 | 0.0013 | 0.0019 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.1.1.21 | Histidine--tRNA ligase. | 0.044 | 0.49 | 1 | 0.38 | 1 | 1 | 0.31 | 0.89 | 1 | 0.07 | 0.43 | 1 | 0.57 | 0.93 | 1 | 0.021 | 0.55 | 1 | 0.018 | 0.018 | 0.017 | 0.017 | 0.016 | 0.0028 | 0.016 | 0.015 | 0.0039 | -0.087 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.02 | 0.02 | 0.0041 | 0.018 | 0.018 | 0.0029 | -0.15 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.4.99.4 | 2-methyleneglutarate mutase. | 0.099 | 0.49 | 1 | 0.023 | 0.92 | 1 | 0.052 | 0.71 | 1 | 0.21 | 0.51 | 1 | 0.039 | 0.8 | 1 | 0.09 | 0.66 | 1 | 6.6e-05 | 7.3e-06 | 0 | 2.4e-05 | 0 | 5.2e-05 | 0 | 0 | 0 | -Inf | 4 / 36 (11%) | 2 / 7 (29%) | 0 / 7 (0%) | 0.286 | 0 | 3e-06 | 0 | 9.9e-06 | 5.4e-06 | 0 | 1.8e-05 | 0.85 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 4.2.1.136 | ADP-dependent NAD(P)H-hydrate dehydratase. | 0.094 | 0.49 | 1 | 0.46 | 1 | 1 | 0.19 | 0.86 | 1 | 0.11 | 0.47 | 1 | 0.063 | 0.86 | 1 | 0.02 | 0.55 | 1 | 0.017 | 0.017 | 0.017 | 0.016 | 0.016 | 0.0043 | 0.017 | 0.016 | 0.0053 | 0.087 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.019 | 0.017 | 0.0058 | 0.017 | 0.017 | 0.0018 | -0.16 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.6.1.77 | Taurine--pyruvate aminotransferase. | 1e-05 | 0.024 | 0.024 | 0.87 | 1 | 1 | 0.38 | 0.9 | 1 | 1.6e-06 | 0.0036 | 0.0036 | 0.63 | 0.95 | 1 | 0.29 | 0.78 | 1 | 0.00083 | 6e-04 | 0.00024 | 4e-05 | 0 | 7.1e-05 | 4.3e-05 | 0 | 7.8e-05 | 0.1 | 26 / 36 (72%) | 2 / 7 (29%) | 3 / 7 (43%) | 0.286 | 0.429 | 0.001 | 9e-04 | 0.00068 | 0.00088 | 0.00092 | 0.00064 | -0.18 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 5.4.99.19 | 16S rRNA pseudouridine(516) synthase. | 0.28 | 0.56 | 1 | 0.98 | 1 | 1 | 0.71 | 0.98 | 1 | 0.21 | 0.51 | 1 | 0.94 | 0.99 | 1 | 0.75 | 0.95 | 1 | 0.00015 | 9.3e-05 | 6.8e-05 | 4.8e-05 | 2.8e-05 | 5.8e-05 | 7.5e-05 | 0 | 0.00015 | 0.64 | 23 / 36 (64%) | 4 / 7 (57%) | 3 / 7 (43%) | 0.571 | 0.429 | 9.9e-05 | 9.5e-05 | 9.6e-05 | 0.00013 | 0.00012 | 1e-04 | 0.39 | 8 / 11 (73%) | 8 / 11 (73%) | 0.727 | 0.727 |
| 4.1.2.19 | Rhamnulose-1-phosphate aldolase. | 0.48 | 0.72 | 1 | 0.27 | 1 | 1 | 0.6 | 0.96 | 1 | 0.86 | 0.95 | 1 | 0.036 | 0.8 | 1 | 0.81 | 0.96 | 1 | 0.0056 | 0.0056 | 0.0056 | 0.0053 | 0.0049 | 0.0015 | 0.0062 | 0.0062 | 0.0017 | 0.23 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0047 | 0.0039 | 0.0014 | 0.0063 | 0.0055 | 0.0032 | 0.42 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.6.1.54 | UDP-2.3-diacylglucosamine diphosphatase. | 0.32 | 0.6 | 1 | 0.97 | 1 | 1 | 0.73 | 0.99 | 1 | 0.1 | 0.46 | 1 | 0.49 | 0.92 | 1 | 0.59 | 0.91 | 1 | 0.00072 | 0.00052 | 0.00013 | 0.00044 | 0.00047 | 0.00032 | 0.00067 | 0.00043 | 9e-04 | 0.61 | 26 / 36 (72%) | 7 / 7 (100%) | 5 / 7 (71%) | 1 | 0.714 | 0.00032 | 9.7e-05 | 0.00057 | 0.00066 | 7.1e-05 | 0.0015 | 1 | 7 / 11 (64%) | 7 / 11 (64%) | 0.636 | 0.636 |
| 3.1.1.81 | Quorum-quenching N-acyl-homoserine lactonase. | 0.012 | 0.49 | 1 | 0.16 | 1 | 1 | 0.38 | 0.9 | 1 | 0.039 | 0.43 | 1 | 0.32 | 0.9 | 1 | 0.61 | 0.91 | 1 | 0.00013 | 6.2e-05 | 0 | 0.00016 | 0.00014 | 0.00016 | 7.1e-05 | 6.8e-05 | 8e-05 | -1.2 | 17 / 36 (47%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 3.5e-05 | 0 | 5e-05 | 2.2e-05 | 0 | 3.7e-05 | -0.67 | 4 / 11 (36%) | 4 / 11 (36%) | 0.364 | 0.364 |
| 3.6.3.25 | Transferred entry: 7.3.2.3. | 0.13 | 0.5 | 1 | 0.83 | 1 | 1 | 0.69 | 0.98 | 1 | 0.05 | 0.43 | 1 | 0.46 | 0.92 | 1 | 0.57 | 0.9 | 1 | 0.025 | 0.025 | 0.017 | 0.014 | 0.011 | 0.007 | 0.015 | 0.013 | 0.0093 | 0.1 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.035 | 0.018 | 0.056 | 0.027 | 0.019 | 0.019 | -0.37 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.2.14 | Oleoyl-[acyl-carrier-protein] hydrolase. | 0.17 | 0.51 | 1 | 0.46 | 1 | 1 | 0.54 | 0.95 | 1 | 0.25 | 0.54 | 1 | 0.86 | 0.99 | 1 | 0.89 | 0.98 | 1 | 0.0017 | 0.0017 | 0.0015 | 0.0011 | 0.001 | 6e-04 | 0.0016 | 0.00094 | 0.0015 | 0.54 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.002 | 0.0019 | 0.0014 | 0.0018 | 0.0016 | 0.00098 | -0.15 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.1.11 | Glycogen(starch) synthase. | 0.52 | 0.75 | 1 | 0.7 | 1 | 1 | 0.59 | 0.96 | 1 | 0.33 | 0.6 | 1 | 0.24 | 0.88 | 1 | 0.13 | 0.68 | 1 | 0.0049 | 0.0049 | 0.0046 | 0.0046 | 0.0046 | 0.0017 | 0.0048 | 0.0045 | 0.0016 | 0.061 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0051 | 0.0046 | 0.0017 | 0.005 | 0.0048 | 0.0024 | -0.029 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.4.19 | Phosphoribosyl-AMP cyclohydrolase. | 0.38 | 0.65 | 1 | 0.57 | 1 | 1 | 0.71 | 0.98 | 1 | 0.26 | 0.56 | 1 | 0.18 | 0.87 | 1 | 0.72 | 0.94 | 1 | 0.011 | 0.011 | 0.011 | 0.0097 | 0.0095 | 0.0019 | 0.011 | 0.01 | 0.0034 | 0.18 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.012 | 0.0026 | 0.012 | 0.012 | 0.0025 | 0.13 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.5.2 | Quinoprotein glucose dehydrogenase (PQQ. quinone). | 0.17 | 0.5 | 1 | 0.25 | 1 | 1 | 0.14 | 0.83 | 1 | 0.29 | 0.58 | 1 | 0.98 | 0.99 | 1 | 0.5 | 0.89 | 1 | 0.0012 | 0.00016 | 0 | 5e-04 | 0 | 0.0013 | 0.00029 | 0 | 0.00077 | -0.79 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1.8e-05 | 0 | 4.1e-05 | 2.8 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 4.2.-.- | 0.059 | 0.49 | 1 | 0.76 | 1 | 1 | 0.34 | 0.9 | 1 | 0.068 | 0.43 | 1 | 0.23 | 0.88 | 1 | 0.069 | 0.66 | 1 | 0.0086 | 0.0086 | 0.0082 | 0.0078 | 0.0074 | 0.0014 | 0.008 | 0.0086 | 0.0018 | 0.037 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0094 | 0.0087 | 0.0022 | 0.0087 | 0.0084 | 0.0019 | -0.11 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 4.1.1.36 | Phosphopantothenoylcysteine decarboxylase. | 0.23 | 0.54 | 1 | 0.48 | 1 | 1 | 0.47 | 0.93 | 1 | 0.17 | 0.49 | 1 | 0.81 | 0.98 | 1 | 0.89 | 0.97 | 1 | 0.015 | 0.015 | 0.014 | 0.014 | 0.015 | 0.0041 | 0.013 | 0.011 | 0.0028 | -0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.016 | 0.014 | 0.0061 | 0.016 | 0.015 | 0.0037 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.13.3 | Transferred entry: 3.4.13.18 and 3.4.13.20. | 0.22 | 0.54 | 1 | 0.78 | 1 | 1 | 0.41 | 0.92 | 1 | 0.13 | 0.47 | 1 | 0.61 | 0.95 | 1 | 0.7 | 0.94 | 1 | 0.0053 | 0.0053 | 0.0048 | 0.0038 | 0.0029 | 0.0023 | 0.0042 | 0.0022 | 0.0031 | 0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0053 | 0.0057 | 0.0024 | 0.0071 | 0.0049 | 0.0045 | 0.42 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.4.99.9 | UDP-galactopyranose mutase. | 0.41 | 0.67 | 1 | 0.73 | 1 | 1 | 0.46 | 0.93 | 1 | 0.24 | 0.54 | 1 | 0.19 | 0.87 | 1 | 0.91 | 0.98 | 1 | 0.021 | 0.021 | 0.019 | 0.018 | 0.017 | 0.005 | 0.019 | 0.019 | 0.0071 | 0.078 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.022 | 0.019 | 0.01 | 0.023 | 0.024 | 0.0075 | 0.064 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.1.41 | Beta-ketoacyl-[acyl-carrier-protein] synthase I. | 0.75 | 0.88 | 1 | 0.9 | 1 | 1 | 0.28 | 0.89 | 1 | 0.78 | 0.92 | 1 | 0.31 | 0.9 | 1 | 0.75 | 0.95 | 1 | 0.0058 | 0.0058 | 0.0037 | 0.0062 | 0.0049 | 0.0078 | 0.006 | 0.0045 | 0.0071 | -0.047 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0047 | 0.0034 | 0.0055 | 0.0065 | 0.0036 | 0.0048 | 0.47 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.1.14 | 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase. | 0.083 | 0.49 | 1 | 0.62 | 1 | 1 | 0.69 | 0.98 | 1 | 0.069 | 0.43 | 1 | 0.36 | 0.9 | 1 | 0.53 | 0.9 | 1 | 0.015 | 0.015 | 0.014 | 0.011 | 0.014 | 0.0041 | 0.013 | 0.013 | 0.0061 | 0.24 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.018 | 0.012 | 0.012 | 0.017 | 0.016 | 0.004 | -0.082 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.7.70 | D-glycero-beta-D-manno-heptose 1-phosphate adenylyltransferase. | 0.39 | 0.66 | 1 | 0.42 | 1 | 1 | 0.84 | 1 | 1 | 0.42 | 0.68 | 1 | 0.18 | 0.87 | 1 | 0.6 | 0.91 | 1 | 0.00074 | 0.00068 | 0.00033 | 0.00096 | 0.00042 | 0.0014 | 9e-04 | 0.00019 | 0.0013 | -0.093 | 33 / 36 (92%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00057 | 0.00032 | 0.00061 | 0.00047 | 0.00028 | 0.00053 | -0.28 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 3.5.1.n3 | 4-deoxy-4-formamido-L-arabinose-phosphoundecaprenol deformylase. | 0.045 | 0.49 | 1 | 0.18 | 1 | 1 | 0.071 | 0.73 | 1 | 0.048 | 0.43 | 1 | 0.31 | 0.9 | 1 | 0.11 | 0.66 | 1 | 0.00037 | 5.1e-05 | 0 | 0.00014 | 0 | 0.00025 | 7.7e-05 | 0 | 2e-04 | -0.86 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.8e-05 | 0 | 6.6e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 4.1.1.50 | Adenosylmethionine decarboxylase. | 0.27 | 0.56 | 1 | 0.0084 | 0.7 | 1 | 0.005 | 0.6 | 1 | 0.47 | 0.72 | 1 | 0.015 | 0.76 | 1 | 0.016 | 0.55 | 1 | 0.001 | 0.001 | 0.00084 | 0.0013 | 0.0011 | 0.00092 | 0.00059 | 0.00043 | 0.00041 | -1.1 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 9e-04 | 9e-04 | 0.00046 | 0.0013 | 0.0011 | 0.00097 | 0.53 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.-.- | 0.47 | 0.72 | 1 | 0.73 | 1 | 1 | 0.96 | 1 | 1 | 0.32 | 0.59 | 1 | 0.97 | 0.99 | 1 | 0.83 | 0.96 | 1 | 0.02 | 0.02 | 0.02 | 0.019 | 0.017 | 0.0079 | 0.019 | 0.021 | 0.0085 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.022 | 0.019 | 0.011 | 0.02 | 0.021 | 0.0041 | -0.14 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 3.5.1.110 | Peroxyureidoacrylate/ureidoacrylate amidohydrolase. | 0.23 | 0.54 | 1 | 0.87 | 1 | 1 | 0.32 | 0.89 | 1 | 0.37 | 0.64 | 1 | 0.53 | 0.93 | 1 | 0.59 | 0.91 | 1 | 0.00047 | 9.1e-05 | 0 | 0.00022 | 0 | 0.00058 | 0.00019 | 0 | 0.00048 | -0.21 | 7 / 36 (19%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 2.6e-06 | 0 | 8.5e-06 | 3.6e-05 | 0 | 7.1e-05 | 3.8 | 1 / 11 (9.1%) | 3 / 11 (27%) | 0.0909 | 0.273 |
| 3.6.3.5 | Transferred entry: 7.2.2.12. | 0.046 | 0.49 | 1 | 0.89 | 1 | 1 | 0.82 | 1 | 1 | 0.063 | 0.43 | 1 | 0.96 | 0.99 | 1 | 0.98 | 0.99 | 1 | 0.012 | 0.012 | 0.013 | 0.0095 | 0.01 | 0.0045 | 0.0097 | 0.0085 | 0.0063 | 0.03 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.014 | 0.014 | 0.0039 | 0.014 | 0.014 | 0.0029 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.3.4.19 | tRNA(Ile)-lysidine synthetase. | 0.092 | 0.49 | 1 | 0.6 | 1 | 1 | 0.69 | 0.98 | 1 | 0.17 | 0.49 | 1 | 0.57 | 0.93 | 1 | 0.094 | 0.66 | 1 | 0.018 | 0.018 | 0.019 | 0.018 | 0.019 | 0.0036 | 0.017 | 0.017 | 0.0038 | -0.082 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.02 | 0.02 | 0.0034 | 0.018 | 0.018 | 0.0026 | -0.15 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.-.- | 0.26 | 0.56 | 1 | 0.88 | 1 | 1 | 0.46 | 0.93 | 1 | 0.12 | 0.47 | 1 | 0.31 | 0.9 | 1 | 0.75 | 0.95 | 1 | 0.17 | 0.17 | 0.17 | 0.16 | 0.16 | 0.031 | 0.16 | 0.16 | 0.033 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.18 | 0.17 | 0.061 | 0.19 | 0.19 | 0.04 | 0.078 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 1.2.1.51 | Pyruvate dehydrogenase (NADP(+)). | 0.98 | 0.99 | 1 | 0.44 | 1 | 1 | 0.35 | 0.9 | 1 | 0.79 | 0.92 | 1 | 0.35 | 0.9 | 1 | 0.22 | 0.74 | 1 | 0.001 | 0.00075 | 0.00015 | 0.00059 | 0.00042 | 0.00085 | 0.00033 | 0.00011 | 7e-04 | -0.84 | 26 / 36 (72%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 0.00086 | 8e-05 | 0.0015 | 0.001 | 0.00018 | 0.0017 | 0.22 | 8 / 11 (73%) | 9 / 11 (82%) | 0.727 | 0.818 |
| 1.12.-.- | 0.87 | 0.95 | 1 | 0.22 | 1 | 1 | 0.82 | 1 | 1 | 0.78 | 0.92 | 1 | 0.16 | 0.87 | 1 | 0.55 | 0.9 | 1 | 0.0088 | 0.0088 | 0.0087 | 0.0082 | 0.0087 | 0.0024 | 0.01 | 0.009 | 0.0048 | 0.29 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.008 | 0.0085 | 0.0024 | 0.0091 | 0.0097 | 0.0028 | 0.19 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 3.1.3.1 | Alkaline phosphatase. | 0.46 | 0.71 | 1 | 0.95 | 1 | 1 | 0.53 | 0.95 | 1 | 0.34 | 0.61 | 1 | 0.8 | 0.98 | 1 | 0.57 | 0.9 | 1 | 0.0068 | 0.0068 | 0.0045 | 0.005 | 0.0042 | 0.0032 | 0.0053 | 0.0046 | 0.0042 | 0.084 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0072 | 0.0045 | 0.0074 | 0.0084 | 0.0089 | 0.0056 | 0.22 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.7.40 | D-ribitol-5-phosphate cytidylyltransferase. | 0.15 | 0.5 | 1 | 0.42 | 1 | 1 | 0.6 | 0.96 | 1 | 0.17 | 0.49 | 1 | 0.13 | 0.87 | 1 | 0.22 | 0.74 | 1 | 0.00091 | 0.00091 | 0.00075 | 0.00064 | 0.00063 | 0.00037 | 0.00077 | 0.00071 | 0.00039 | 0.27 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.001 | 0.001 | 0.00057 | 0.0011 | 0.00071 | 0.00073 | 0.14 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.2.1.62 | 4-formylbenzenesulfonate dehydrogenase. | 0.41 | 0.67 | 1 | 0.75 | 1 | 1 | 0.99 | 1 | 1 | 0.33 | 0.6 | 1 | 0.55 | 0.93 | 1 | 0.86 | 0.96 | 1 | 0.00019 | 2.1e-05 | 0 | 0 | 0 | 0 | 3.9e-06 | 0 | 1e-05 | Inf | 4 / 36 (11%) | 0 / 7 (0%) | 1 / 7 (14%) | 0 | 0.143 | 3.3e-05 | 0 | 0.00011 | 3.4e-05 | 0 | 8.3e-05 | 0.043 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 1.1.1.- | 0.62 | 0.82 | 1 | 0.78 | 1 | 1 | 0.46 | 0.93 | 1 | 0.63 | 0.84 | 1 | 0.16 | 0.87 | 1 | 0.63 | 0.92 | 1 | 0.06 | 0.06 | 0.059 | 0.06 | 0.06 | 0.016 | 0.061 | 0.065 | 0.018 | 0.024 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.056 | 0.056 | 0.0059 | 0.062 | 0.064 | 0.011 | 0.15 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 2.7.6.3 | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine diphosphokinase. | 0.96 | 0.98 | 1 | 0.34 | 1 | 1 | 0.84 | 1 | 1 | 0.69 | 0.87 | 1 | 0.25 | 0.88 | 1 | 0.28 | 0.77 | 1 | 0.0019 | 0.0019 | 0.0017 | 0.0017 | 0.002 | 0.0011 | 0.002 | 0.0023 | 0.0011 | 0.23 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0017 | 0.0013 | 0.0013 | 0.002 | 0.0018 | 0.0013 | 0.23 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 5.3.1.13 | Arabinose-5-phosphate isomerase. | 0.11 | 0.5 | 1 | 0.18 | 1 | 1 | 0.067 | 0.73 | 1 | 0.091 | 0.45 | 1 | 0.14 | 0.87 | 1 | 0.14 | 0.68 | 1 | 0.001 | 0.00094 | 0.00045 | 0.0012 | 0.00091 | 0.00098 | 0.0011 | 0.00077 | 0.0011 | -0.13 | 33 / 36 (92%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00061 | 0.00026 | 0.00088 | 0.001 | 0.00038 | 0.0014 | 0.71 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 5.1.3.12 | UDP-glucuronate 5'-epimerase. | 0.27 | 0.56 | 1 | 0.023 | 0.92 | 1 | 0.069 | 0.73 | 1 | 0.35 | 0.62 | 1 | 0.016 | 0.76 | 1 | 0.04 | 0.63 | 1 | 0.00015 | 1.2e-05 | 0 | 2e-05 | 0 | 5.2e-05 | 4.4e-05 | 0 | 8.9e-05 | 1.1 | 3 / 36 (8.3%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 2.5.-.- | 0.85 | 0.93 | 1 | 0.091 | 1 | 1 | 0.16 | 0.85 | 1 | 0.42 | 0.68 | 1 | 0.13 | 0.87 | 1 | 0.18 | 0.72 | 1 | 0.00052 | 0.00048 | 0.00036 | 0.00063 | 0.00024 | 0.00093 | 2e-04 | 0.00019 | 0.00018 | -1.7 | 33 / 36 (92%) | 6 / 7 (86%) | 5 / 7 (71%) | 0.857 | 0.714 | 0.00053 | 0.00048 | 0.00048 | 5e-04 | 0.00044 | 0.00036 | -0.084 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 6.2.-.- | 0.12 | 0.5 | 1 | 0.32 | 1 | 1 | 0.83 | 1 | 1 | 0.11 | 0.47 | 1 | 0.49 | 0.92 | 1 | 0.56 | 0.9 | 1 | 0.0063 | 0.0063 | 0.0057 | 0.0058 | 0.0042 | 0.003 | 0.0049 | 0.004 | 0.003 | -0.24 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0076 | 0.0079 | 0.0023 | 0.0062 | 0.0055 | 0.0024 | -0.29 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 2.3.1.183 | Phosphinothricin acetyltransferase. | 0.29 | 0.57 | 1 | 0.67 | 1 | 1 | 0.96 | 1 | 1 | 0.17 | 0.49 | 1 | 0.75 | 0.97 | 1 | 0.47 | 0.87 | 1 | 0.0073 | 0.0073 | 0.0075 | 0.0066 | 0.0061 | 0.0026 | 0.0063 | 0.0071 | 0.002 | -0.067 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.008 | 0.0076 | 0.0031 | 0.0077 | 0.0081 | 0.0027 | -0.055 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.107 | Diacylglycerol kinase (ATP). | 0.21 | 0.53 | 1 | 0.67 | 1 | 1 | 0.62 | 0.96 | 1 | 0.14 | 0.47 | 1 | 0.3 | 0.9 | 1 | 0.42 | 0.84 | 1 | 0.017 | 0.017 | 0.015 | 0.013 | 0.013 | 0.0051 | 0.015 | 0.015 | 0.007 | 0.21 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.019 | 0.016 | 0.016 | 0.017 | 0.015 | 0.0053 | -0.16 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.31 | Beta-glucuronidase. | 0.079 | 0.49 | 1 | 0.7 | 1 | 1 | 0.18 | 0.85 | 1 | 0.14 | 0.47 | 1 | 1 | 1 | 1 | 0.31 | 0.79 | 1 | 0.013 | 0.013 | 0.012 | 0.014 | 0.015 | 0.0055 | 0.014 | 0.013 | 0.0069 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.01 | 0.0088 | 0.0047 | 0.013 | 0.013 | 0.006 | 0.38 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.5.1.64 | Transferred entry: 2.2.1.9 and 4.2.99.20. | 0.16 | 0.5 | 1 | 0.13 | 1 | 1 | 0.38 | 0.9 | 1 | 0.25 | 0.55 | 1 | 0.75 | 0.97 | 1 | 0.61 | 0.92 | 1 | 0.00044 | 3.6e-05 | 0 | 0.00013 | 0 | 0.00036 | 4.8e-05 | 0 | 0.00013 | -1.4 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 4.1.3.34 | Citryl-CoA lyase. | 0.34 | 0.62 | 1 | 0.81 | 1 | 1 | 0.99 | 1 | 1 | 0.78 | 0.92 | 1 | 0.49 | 0.92 | 1 | 0.36 | 0.81 | 1 | 0.00034 | 0.00011 | 0 | 0.00025 | 0 | 0.00044 | 0.00016 | 0 | 0.00026 | -0.64 | 12 / 36 (33%) | 2 / 7 (29%) | 3 / 7 (43%) | 0.286 | 0.429 | 5.3e-05 | 0 | 8.3e-05 | 5.9e-05 | 0 | 0.00012 | 0.15 | 4 / 11 (36%) | 3 / 11 (27%) | 0.364 | 0.273 |
| 3.5.1.3 | Omega-amidase. | 0.71 | 0.86 | 1 | 0.75 | 1 | 1 | 0.5 | 0.95 | 1 | 0.91 | 0.97 | 1 | 0.57 | 0.93 | 1 | 0.41 | 0.84 | 1 | 0.00015 | 3.3e-05 | 0 | 5.3e-05 | 0 | 0.00013 | 5.6e-05 | 0 | 1e-04 | 0.079 | 8 / 36 (22%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 3.1e-05 | 0 | 7.6e-05 | 8e-06 | 0 | 1.9e-05 | -2 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 3.8.1.- | 0.0023 | 0.49 | 1 | 0.26 | 1 | 1 | 0.13 | 0.82 | 1 | 0.0022 | 0.34 | 1 | 0.54 | 0.93 | 1 | 0.24 | 0.76 | 1 | 0.00012 | 3.9e-05 | 0 | 0.00011 | 0.00011 | 0.00014 | 6e-05 | 3.1e-05 | 7.6e-05 | -0.87 | 12 / 36 (33%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 2.6e-06 | 0 | 8.5e-06 | 1.5e-05 | 0 | 2.8e-05 | 2.5 | 1 / 11 (9.1%) | 3 / 11 (27%) | 0.0909 | 0.273 | |
| 4.1.1.23 | Orotidine-5'-phosphate decarboxylase. | 0.028 | 0.49 | 1 | 0.91 | 1 | 1 | 0.96 | 1 | 1 | 0.035 | 0.43 | 1 | 0.25 | 0.88 | 1 | 0.34 | 0.8 | 1 | 0.017 | 0.017 | 0.016 | 0.014 | 0.013 | 0.0038 | 0.014 | 0.014 | 0.0039 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.019 | 0.017 | 0.0076 | 0.019 | 0.018 | 0.0032 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.1.13 | Transferred entry: 4.3.1.17. | 0.02 | 0.49 | 1 | 0.0013 | 0.42 | 1 | 0.0023 | 0.51 | 1 | 0.0011 | 0.31 | 1 | 0.0052 | 0.7 | 1 | 0.0079 | 0.55 | 1 | 0.00071 | 0.00018 | 0 | 0.00051 | 8.1e-05 | 0.0011 | 0.00037 | 0 | 0.00097 | -0.46 | 9 / 36 (25%) | 5 / 7 (71%) | 1 / 7 (14%) | 0.714 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1.9e-05 | 0 | 4.7e-05 | 2.9 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 3.2.1.180 | Unsaturated chondroitin disaccharide hydrolase. | 0.16 | 0.5 | 1 | 0.66 | 1 | 1 | 0.56 | 0.96 | 1 | 0.079 | 0.44 | 1 | 0.96 | 0.99 | 1 | 0.68 | 0.93 | 1 | 0.0011 | 0.00091 | 0.00032 | 0.001 | 0.00047 | 0.0011 | 0.0015 | 0.00035 | 0.0021 | 0.58 | 30 / 36 (83%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00042 | 0.00019 | 0.00069 | 0.00097 | 0.00045 | 0.0014 | 1.2 | 8 / 11 (73%) | 8 / 11 (73%) | 0.727 | 0.727 |
| 7.1.1.- | 0.19 | 0.53 | 1 | 0.31 | 1 | 1 | 0.05 | 0.71 | 1 | 0.079 | 0.44 | 1 | 0.13 | 0.87 | 1 | 0.027 | 0.61 | 1 | 0.0075 | 0.0075 | 0.004 | 0.0082 | 0.0062 | 0.0049 | 0.0076 | 0.0039 | 0.0083 | -0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0053 | 0.0014 | 0.0073 | 0.0091 | 0.004 | 0.011 | 0.78 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 6.3.5.6 | Asparaginyl-tRNA synthase (glutamine-hydrolyzing). | 0.036 | 0.49 | 1 | 0.048 | 0.97 | 1 | 0.019 | 0.71 | 1 | 0.059 | 0.43 | 1 | 0.032 | 0.79 | 1 | 0.016 | 0.55 | 1 | 0.00047 | 4e-04 | 0.00026 | 0.00015 | 8.1e-05 | 0.00021 | 0.00044 | 0.00038 | 0.00034 | 1.6 | 31 / 36 (86%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00058 | 0.00029 | 0.00059 | 0.00037 | 0.00026 | 0.00044 | -0.65 | 10 / 11 (91%) | 9 / 11 (82%) | 0.909 | 0.818 |
| 2.6.1.52 | Phosphoserine transaminase. | 0.03 | 0.49 | 1 | 0.7 | 1 | 1 | 0.66 | 0.97 | 1 | 0.095 | 0.45 | 1 | 0.37 | 0.9 | 1 | 0.37 | 0.82 | 1 | 0.019 | 0.019 | 0.019 | 0.017 | 0.017 | 0.002 | 0.017 | 0.018 | 0.0028 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.02 | 0.02 | 0.0034 | 0.02 | 0.02 | 0.0021 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.3.12 | Trehalose-phosphatase. | 0.22 | 0.54 | 1 | 0.8 | 1 | 1 | 0.17 | 0.85 | 1 | 0.065 | 0.43 | 1 | 0.7 | 0.95 | 1 | 0.33 | 0.8 | 1 | 0.001 | 0.00082 | 0.00026 | 0.00077 | 0.00059 | 0.00074 | 0.00072 | 0.00068 | 0.00061 | -0.097 | 29 / 36 (81%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00043 | 0.00013 | 0.00091 | 0.0013 | 0.00036 | 0.0026 | 1.6 | 8 / 11 (73%) | 8 / 11 (73%) | 0.727 | 0.727 |
| 2.3.1.190 | Acetoin dehydrogenase. | 0.86 | 0.94 | 1 | 0.76 | 1 | 1 | 0.76 | 0.99 | 1 | 0.94 | 0.98 | 1 | 0.96 | 0.99 | 1 | 0.93 | 0.98 | 1 | 0.00031 | 0.00015 | 2.4e-05 | 0.00017 | 4.8e-05 | 0.00031 | 1e-04 | 6.8e-05 | 0.00013 | -0.77 | 18 / 36 (50%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.00014 | 0 | 0.00021 | 0.00018 | 0 | 0.00034 | 0.36 | 5 / 11 (45%) | 5 / 11 (45%) | 0.455 | 0.455 |
| 2.7.8.14 | CDP-ribitol ribitolphosphotransferase. | 0.51 | 0.74 | 1 | 0.75 | 1 | 1 | 0.79 | 1 | 1 | 0.44 | 0.69 | 1 | 0.55 | 0.93 | 1 | 0.85 | 0.96 | 1 | 0.00036 | 6e-05 | 0 | 4.1e-06 | 0 | 1.1e-05 | 1.8e-05 | 0 | 4.7e-05 | 2.1 | 6 / 36 (17%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 6e-05 | 0 | 0.00017 | 0.00012 | 0 | 0.00028 | 1 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 3.5.1.1 | Asparaginase. | 0.76 | 0.89 | 1 | 0.37 | 1 | 1 | 0.16 | 0.85 | 1 | 0.97 | 0.99 | 1 | 0.8 | 0.97 | 1 | 0.44 | 0.85 | 1 | 0.0098 | 0.0098 | 0.0091 | 0.0097 | 0.0097 | 0.0022 | 0.0087 | 0.0089 | 0.0023 | -0.16 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0096 | 0.0079 | 0.0057 | 0.011 | 0.01 | 0.0043 | 0.2 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.2.- | 0.15 | 0.5 | 1 | 0.078 | 1 | 1 | 0.088 | 0.79 | 1 | 0.11 | 0.47 | 1 | 0.023 | 0.76 | 1 | 0.034 | 0.63 | 1 | 0.0042 | 0.0042 | 0.0039 | 0.0027 | 0.0023 | 0.0021 | 0.0046 | 0.0046 | 0.0027 | 0.77 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.005 | 0.0042 | 0.0048 | 0.0041 | 0.0043 | 0.0025 | -0.29 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 1.10.9.1 | Transferred entry: 7.1.1.6. | 0.67 | 0.85 | 1 | 0.55 | 1 | 1 | 0.58 | 0.96 | 1 | 0.93 | 0.98 | 1 | 0.94 | 0.99 | 1 | 0.95 | 0.99 | 1 | 0.00014 | 9.6e-05 | 8.6e-05 | 0.00014 | 8e-05 | 0.00016 | 6.7e-05 | 9.3e-05 | 5.6e-05 | -1.1 | 25 / 36 (69%) | 4 / 7 (57%) | 5 / 7 (71%) | 0.571 | 0.714 | 8.3e-05 | 9.7e-05 | 7.8e-05 | 9.6e-05 | 5.9e-05 | 1e-04 | 0.21 | 8 / 11 (73%) | 8 / 11 (73%) | 0.727 | 0.727 |
| 1.1.1.100 | 3-oxoacyl-[acyl-carrier-protein] reductase. | 0.22 | 0.54 | 1 | 0.75 | 1 | 1 | 0.89 | 1 | 1 | 0.095 | 0.45 | 1 | 0.29 | 0.89 | 1 | 0.44 | 0.85 | 1 | 0.035 | 0.035 | 0.034 | 0.031 | 0.035 | 0.0071 | 0.032 | 0.033 | 0.0096 | 0.046 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.037 | 0.033 | 0.015 | 0.037 | 0.038 | 0.0048 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.3.1.1 | Dihydrouracil dehydrogenase (NAD(+)). | 0.58 | 0.79 | 1 | 0.37 | 1 | 1 | 0.54 | 0.95 | 1 | 0.8 | 0.93 | 1 | 0.32 | 0.9 | 1 | 0.22 | 0.74 | 1 | 0.0032 | 0.0032 | 0.0028 | 0.0032 | 0.0028 | 0.0017 | 0.0038 | 0.0029 | 0.0022 | 0.25 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0028 | 0.0033 | 0.0013 | 0.0031 | 0.0024 | 0.0022 | 0.15 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.7.63 | Transferred entry: 6.3.1.20. | 0.11 | 0.5 | 1 | 0.42 | 1 | 1 | 0.22 | 0.88 | 1 | 0.04 | 0.43 | 1 | 0.18 | 0.87 | 1 | 0.11 | 0.66 | 1 | 0.0025 | 0.0023 | 0.0011 | 0.0011 | 0.00048 | 0.0015 | 0.0014 | 0.00085 | 0.0014 | 0.35 | 33 / 36 (92%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.0037 | 0.0016 | 0.0074 | 0.0022 | 0.0015 | 0.0021 | -0.75 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 2.1.1.166 | 23S rRNA (uridine(2552)-2'-O)-methyltransferase. | 0.26 | 0.56 | 1 | 0.73 | 1 | 1 | 0.59 | 0.96 | 1 | 0.3 | 0.58 | 1 | 0.78 | 0.97 | 1 | 0.65 | 0.92 | 1 | 0.00019 | 5.2e-05 | 0 | 9.7e-05 | 0 | 0.00022 | 0.00011 | 0 | 0.00025 | 0.18 | 10 / 36 (28%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 2e-05 | 0 | 4.5e-05 | 1.8e-05 | 0 | 3.7e-05 | -0.15 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 3.5.1.14 | N-acyl-aliphatic-L-amino acid amidohydrolase. | 0.094 | 0.49 | 1 | 0.072 | 1 | 1 | 0.23 | 0.88 | 1 | 0.032 | 0.43 | 1 | 0.0039 | 0.69 | 1 | 0.088 | 0.66 | 1 | 0.0034 | 0.0034 | 0.0025 | 0.0016 | 0.0015 | 0.00077 | 0.003 | 0.0032 | 0.0011 | 0.91 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0045 | 0.0023 | 0.0072 | 0.0038 | 0.0031 | 0.0023 | -0.24 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.3.1.- | 0.22 | 0.54 | 1 | 0.75 | 1 | 1 | 0.23 | 0.88 | 1 | 0.14 | 0.47 | 1 | 0.35 | 0.9 | 1 | 0.15 | 0.71 | 1 | 0.001 | 0.00075 | 0.00022 | 0.00076 | 0.00034 | 0.00072 | 0.0011 | 2e-04 | 0.0021 | 0.53 | 27 / 36 (75%) | 6 / 7 (86%) | 5 / 7 (71%) | 0.857 | 0.714 | 0.00032 | 9.5e-05 | 0.00064 | 0.00097 | 0.00014 | 0.0018 | 1.6 | 7 / 11 (64%) | 9 / 11 (82%) | 0.636 | 0.818 | |
| 2.3.3.10 | Hydroxymethylglutaryl-CoA synthase. | 0.94 | 0.97 | 1 | 0.57 | 1 | 1 | 0.41 | 0.92 | 1 | 0.82 | 0.93 | 1 | 0.36 | 0.9 | 1 | 0.35 | 0.81 | 1 | 4e-04 | 0.00027 | 0.00012 | 0.00026 | 8.1e-05 | 0.00033 | 0.00034 | 0.00016 | 0.00037 | 0.39 | 24 / 36 (67%) | 5 / 7 (71%) | 6 / 7 (86%) | 0.714 | 0.857 | 0.00031 | 0.00014 | 0.00047 | 0.00018 | 0.00011 | 3e-04 | -0.78 | 6 / 11 (55%) | 7 / 11 (64%) | 0.545 | 0.636 |
| 2.7.1.193 | Protein-N(pi)-phosphohistidine--N-acetyl-D-glucosamine | 0.14 | 0.5 | 1 | 0.51 | 1 | 1 | 0.76 | 0.99 | 1 | 0.22 | 0.52 | 1 | 0.16 | 0.87 | 1 | 0.34 | 0.8 | 1 | 0.00035 | 6.8e-05 | 0 | 0.00019 | 0 | 0.00049 | 0.00014 | 0 | 0.00027 | -0.44 | 7 / 36 (19%) | 2 / 7 (29%) | 3 / 7 (43%) | 0.286 | 0.429 | 2.6e-06 | 0 | 8.5e-06 | 1.1e-05 | 0 | 3.6e-05 | 2.1 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 1.13.11.24 | Quercetin 2.3-dioxygenase. | 0.15 | 0.5 | 1 | 0.25 | 1 | 1 | 0.12 | 0.82 | 1 | 0.11 | 0.47 | 1 | 0.34 | 0.9 | 1 | 0.3 | 0.78 | 1 | 0.00064 | 0.00053 | 0.00033 | 0.00065 | 0.00072 | 0.00044 | 0.00039 | 0.00043 | 0.00025 | -0.74 | 30 / 36 (83%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00035 | 0.00023 | 0.00046 | 0.00071 | 0.00014 | 0.0012 | 1 | 8 / 11 (73%) | 9 / 11 (82%) | 0.727 | 0.818 |
| 3.1.3.89 | 5'-deoxynucleotidase. | 0.14 | 0.5 | 1 | 0.21 | 1 | 1 | 0.2 | 0.87 | 1 | 0.24 | 0.53 | 1 | 0.33 | 0.9 | 1 | 0.25 | 0.76 | 1 | 0.0014 | 0.0014 | 0.0013 | 0.0017 | 0.0018 | 0.00049 | 0.0014 | 0.0013 | 0.00077 | -0.28 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0013 | 0.0012 | 0.00085 | 0.0013 | 0.0012 | 0.00065 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.1.61 | tRNA (5-methylaminomethyl-2-thiouridylate)-methyltransferase. | 0.047 | 0.49 | 1 | 0.25 | 1 | 1 | 0.01 | 0.61 | 1 | 0.0042 | 0.36 | 1 | 0.46 | 0.92 | 1 | 0.03 | 0.61 | 1 | 0.00076 | 7e-04 | 0.00036 | 0.00087 | 0.00069 | 7e-04 | 0.00064 | 0.00038 | 0.00074 | -0.44 | 33 / 36 (92%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00037 | 0.00016 | 6e-04 | 0.00095 | 0.00023 | 0.0013 | 1.4 | 8 / 11 (73%) | 11 / 11 (100%) | 0.727 | 1 |
| 2.7.6.2 | Thiamine diphosphokinase. | 0.16 | 0.5 | 1 | 0.83 | 1 | 1 | 0.87 | 1 | 1 | 0.12 | 0.47 | 1 | 0.26 | 0.88 | 1 | 0.54 | 0.9 | 1 | 0.0088 | 0.0088 | 0.0086 | 0.0076 | 0.0081 | 0.002 | 0.0078 | 0.0075 | 0.0022 | 0.037 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0094 | 0.0089 | 0.0038 | 0.0095 | 0.009 | 0.0018 | 0.015 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.3.2.28 | Transferred entry: 6.3.2.49. | 0.66 | 0.84 | 1 | 0.37 | 1 | 1 | 0.18 | 0.86 | 1 | 0.97 | 0.99 | 1 | 0.61 | 0.95 | 1 | 0.32 | 0.8 | 1 | 2e-04 | 1e-04 | 1.5e-05 | 0.00015 | 0 | 0.00031 | 3.3e-05 | 0 | 4.8e-05 | -2.2 | 18 / 36 (50%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 7.4e-05 | 0 | 0.00012 | 0.00014 | 5.9e-05 | 0.00018 | 0.92 | 5 / 11 (45%) | 7 / 11 (64%) | 0.455 | 0.636 |
| 1.1.99.28 | Glucose-fructose oxidoreductase. | 0.79 | 0.9 | 1 | 0.7 | 1 | 1 | 0.76 | 0.99 | 1 | 0.37 | 0.64 | 1 | 0.76 | 0.97 | 1 | 0.53 | 0.9 | 1 | 0.0062 | 0.0062 | 0.0061 | 0.0063 | 0.006 | 0.0035 | 0.0063 | 0.0076 | 0.0041 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0063 | 0.0063 | 0.0017 | 0.0058 | 0.0055 | 0.0021 | -0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.1.303 | UDP-Gal:alpha-D-GlcNAc-diphosphoundecaprenol beta-1.3- | 0.54 | 0.77 | 1 | 0.33 | 1 | 1 | 0.85 | 1 | 1 | 0.74 | 0.9 | 1 | 0.48 | 0.92 | 1 | 0.58 | 0.91 | 1 | 0.00013 | 3.3e-05 | 0 | 7.4e-05 | 0 | 0.00015 | 2.6e-05 | 0 | 5.8e-05 | -1.5 | 9 / 36 (25%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 4.3e-05 | 0 | 8.4e-05 | 2.7e-06 | 0 | 8.9e-06 | -4 | 3 / 11 (27%) | 1 / 11 (9.1%) | 0.273 | 0.0909 |
| 2.4.1.291 | N-acetylgalactosamine-N.N'-diacetylbacillosaminyl-diphospho-undecaprenol | 0.85 | 0.94 | 1 | 0.83 | 1 | 1 | 0.38 | 0.9 | 1 | 0.85 | 0.95 | 1 | 0.97 | 0.99 | 1 | 0.71 | 0.94 | 1 | 0.00064 | 0.00047 | 0.00017 | 0.00038 | 0.00034 | 0.00041 | 0.00035 | 0.00022 | 0.00041 | -0.12 | 26 / 36 (72%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.00041 | 0.00017 | 0.00068 | 0.00065 | 0.00015 | 0.001 | 0.66 | 8 / 11 (73%) | 8 / 11 (73%) | 0.727 | 0.727 |
| 2.2.1.2 | Transaldolase. | 0.046 | 0.49 | 1 | 0.3 | 1 | 1 | 0.16 | 0.85 | 1 | 0.04 | 0.43 | 1 | 0.044 | 0.8 | 1 | 0.023 | 0.56 | 1 | 0.012 | 0.012 | 0.012 | 0.011 | 0.01 | 0.0021 | 0.012 | 0.011 | 0.0032 | 0.13 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.014 | 0.013 | 0.0051 | 0.013 | 0.012 | 0.0022 | -0.11 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.2.19 | Chondroitin B lyase. | 0.24 | 0.55 | 1 | 0.84 | 1 | 1 | 0.16 | 0.85 | 1 | 0.25 | 0.54 | 1 | 0.63 | 0.95 | 1 | 0.12 | 0.66 | 1 | 0.00025 | 7.7e-05 | 0 | 8.9e-05 | 0 | 0.00014 | 0.00012 | 0 | 0.00028 | 0.43 | 11 / 36 (31%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 1.6e-05 | 0 | 3.6e-05 | 1e-04 | 0 | 0.00019 | 2.6 | 2 / 11 (18%) | 4 / 11 (36%) | 0.182 | 0.364 |
| 2.1.1.200 | tRNA (cytidine(32)/uridine(32)-2'-O)-methyltransferase. | 0.15 | 0.5 | 1 | 0.67 | 1 | 1 | 0.48 | 0.93 | 1 | 0.11 | 0.47 | 1 | 0.5 | 0.93 | 1 | 0.37 | 0.82 | 1 | 0.00034 | 5.6e-05 | 0 | 0.00012 | 0 | 0.00026 | 0.00015 | 0 | 0.00041 | 0.32 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 8e-06 | 0 | 1.9e-05 | 1.6 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 4.2.2.8 | Heparin-sulfate lyase. | 0.16 | 0.5 | 1 | 1 | 1 | 1 | 0.44 | 0.93 | 1 | 0.49 | 0.74 | 1 | 0.45 | 0.92 | 1 | 0.95 | 0.99 | 1 | 0.00086 | 0.00055 | 9.6e-05 | 0.0011 | 8e-05 | 0.0023 | 0.00064 | 0.00059 | 0.00069 | -0.78 | 23 / 36 (64%) | 4 / 7 (57%) | 5 / 7 (71%) | 0.571 | 0.714 | 0.00013 | 3.1e-05 | 0.00019 | 0.00057 | 1e-04 | 0.0011 | 2.1 | 7 / 11 (64%) | 7 / 11 (64%) | 0.636 | 0.636 |
| 2.5.1.30 | Heptaprenyl diphosphate synthase. | 0.86 | 0.94 | 1 | 0.83 | 1 | 1 | 0.6 | 0.96 | 1 | 0.95 | 0.98 | 1 | 0.96 | 0.99 | 1 | 0.49 | 0.88 | 1 | 0.00099 | 0.00096 | 0.00082 | 0.00087 | 6e-04 | 0.00068 | 0.00079 | 0.00083 | 0.00059 | -0.14 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0011 | 0.00052 | 0.0011 | 0.001 | 0.00097 | 0.00066 | -0.14 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 1.18.1.6 | Adrenodoxin-NADP(+) reductase. | 0.1 | 0.49 | 1 | 0.96 | 1 | 1 | 0.95 | 1 | 1 | 0.043 | 0.43 | 1 | 0.48 | 0.92 | 1 | 0.76 | 0.95 | 1 | 0.0029 | 0.0021 | 0.00061 | 0.00079 | 0 | 0.0014 | 0.00081 | 6.8e-05 | 0.0017 | 0.036 | 26 / 36 (72%) | 3 / 7 (43%) | 4 / 7 (57%) | 0.429 | 0.571 | 0.0032 | 0.0012 | 0.0063 | 0.0025 | 0.0017 | 0.0029 | -0.36 | 9 / 11 (82%) | 10 / 11 (91%) | 0.818 | 0.909 |
| 5.4.99.29 | 23S rRNA pseudouridine(746) synthase. | 0.15 | 0.5 | 1 | 0.92 | 1 | 1 | 0.69 | 0.98 | 1 | 0.18 | 0.49 | 1 | 0.39 | 0.9 | 1 | 0.73 | 0.94 | 1 | 0.0018 | 0.0015 | 8e-04 | 0.00063 | 0.00041 | 0.00059 | 0.00091 | 0.00072 | 0.0011 | 0.53 | 30 / 36 (83%) | 6 / 7 (86%) | 4 / 7 (57%) | 0.857 | 0.571 | 0.0024 | 0.00097 | 0.0045 | 0.0017 | 0.0011 | 0.0018 | -0.5 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 2.7.7.71 | D-glycero-alpha-D-manno-heptose 1-phosphate guanylyltransferase. | 0.25 | 0.55 | 1 | 0.69 | 1 | 1 | 0.1 | 0.82 | 1 | 0.14 | 0.47 | 1 | 0.86 | 0.99 | 1 | 0.17 | 0.71 | 1 | 0.00057 | 0.00025 | 0 | 0.00025 | 0.00027 | 0.00025 | 0.00019 | 0.00011 | 0.00025 | -0.4 | 16 / 36 (44%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.00014 | 0 | 4e-04 | 4e-04 | 7.5e-05 | 0.00073 | 1.5 | 2 / 11 (18%) | 6 / 11 (55%) | 0.182 | 0.545 |
| 5.1.3.13 | dTDP-4-dehydrorhamnose 3.5-epimerase. | 0.14 | 0.5 | 1 | 0.26 | 1 | 1 | 0.053 | 0.71 | 1 | 0.55 | 0.78 | 1 | 0.65 | 0.95 | 1 | 0.16 | 0.71 | 1 | 0.009 | 0.009 | 0.0092 | 0.0097 | 0.0094 | 0.0022 | 0.0084 | 0.0089 | 0.0019 | -0.21 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0083 | 0.0079 | 0.0026 | 0.0097 | 0.0095 | 0.0015 | 0.22 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.13.21 | Dipeptidase E. | 0.22 | 0.54 | 1 | 0.29 | 1 | 1 | 0.5 | 0.95 | 1 | 0.51 | 0.75 | 1 | 0.27 | 0.88 | 1 | 0.58 | 0.91 | 1 | 0.0031 | 0.0031 | 0.0026 | 0.0039 | 0.0025 | 0.0024 | 0.0036 | 0.0027 | 0.0037 | -0.12 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0027 | 0.0026 | 0.0011 | 0.0026 | 0.0031 | 0.00076 | -0.054 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.1.38 | Glutamin-(asparagin-)ase. | 0.34 | 0.62 | 1 | 0.23 | 1 | 1 | 0.42 | 0.92 | 1 | 0.59 | 0.81 | 1 | 0.16 | 0.87 | 1 | 0.42 | 0.85 | 1 | 0.00062 | 0.00043 | 0.00026 | 0.00018 | 2.8e-05 | 0.00022 | 0.00041 | 0.00027 | 0.00039 | 1.2 | 25 / 36 (69%) | 5 / 7 (71%) | 6 / 7 (86%) | 0.714 | 0.857 | 0.00052 | 0.00017 | 0.00063 | 0.00051 | 0.00028 | 0.00064 | -0.028 | 6 / 11 (55%) | 8 / 11 (73%) | 0.545 | 0.727 |
| 3.1.4.12 | Sphingomyelin phosphodiesterase. | 0.077 | 0.49 | 1 | 0.2 | 1 | 1 | 0.69 | 0.98 | 1 | 0.011 | 0.42 | 1 | 0.41 | 0.9 | 1 | 0.99 | 1 | 1 | 6e-04 | 0.00027 | 0 | 0.00025 | 0.00022 | 0.00029 | 0.00087 | 0.00011 | 0.0019 | 1.8 | 16 / 36 (44%) | 5 / 7 (71%) | 6 / 7 (86%) | 0.714 | 0.857 | 1.7e-05 | 0 | 3.8e-05 | 0.00014 | 0 | 0.00029 | 3 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 4.99.1.11 | Sirohydrochlorin nickelchelatase. | 1 | 1 | 1 | 0.98 | 1 | 1 | 1 | 1 | 1 | 0.93 | 0.98 | 1 | 0.81 | 0.98 | 1 | 0.82 | 0.96 | 1 | 0.00015 | 1.7e-05 | 0 | 1.2e-05 | 0 | 3.2e-05 | 1.3e-05 | 0 | 3.5e-05 | 0.12 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 1.9e-05 | 0 | 6.3e-05 | 2.1e-05 | 0 | 6.8e-05 | 0.14 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.1.4.3 | Phospholipase C. | 0.29 | 0.57 | 1 | 0.19 | 1 | 1 | 0.16 | 0.85 | 1 | 0.22 | 0.52 | 1 | 0.26 | 0.88 | 1 | 0.27 | 0.77 | 1 | 0.00052 | 0.00036 | 1e-04 | 0.00046 | 0.00024 | 0.00047 | 0.00014 | 8.1e-05 | 0.00017 | -1.7 | 25 / 36 (69%) | 7 / 7 (100%) | 5 / 7 (71%) | 1 | 0.714 | 3e-04 | 8.7e-05 | 0.00046 | 5e-04 | 5.9e-05 | 0.00086 | 0.74 | 6 / 11 (55%) | 7 / 11 (64%) | 0.545 | 0.636 |
| 3.1.4.55 | Phosphoribosyl 1.2-cyclic phosphate phosphodiesterase. | 0.4 | 0.66 | 1 | 0.45 | 1 | 1 | 0.21 | 0.87 | 1 | 0.53 | 0.77 | 1 | 0.8 | 0.98 | 1 | 0.73 | 0.94 | 1 | 0.00045 | 0.00024 | 4.3e-05 | 0.00029 | 0.00011 | 0.00037 | 0.00016 | 0.00011 | 0.00017 | -0.86 | 19 / 36 (53%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.00013 | 2.8e-05 | 0.00025 | 0.00035 | 0 | 0.00072 | 1.4 | 6 / 11 (55%) | 5 / 11 (45%) | 0.545 | 0.455 |
| 6.3.2.45 | UDP-N-acetylmuramate L-alanyl-gamma-D-glutamyl-meso-2.6- | 0.51 | 0.74 | 1 | 0.52 | 1 | 1 | 0.88 | 1 | 1 | 0.65 | 0.85 | 1 | 0.63 | 0.95 | 1 | 0.39 | 0.83 | 1 | 0.00029 | 7.2e-05 | 0 | 0.00013 | 0 | 0.00031 | 9.5e-05 | 0 | 0.00023 | -0.45 | 9 / 36 (25%) | 2 / 7 (29%) | 3 / 7 (43%) | 0.286 | 0.429 | 5.7e-05 | 0 | 0.00013 | 3.4e-05 | 0 | 7.8e-05 | -0.75 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 1.2.1.68 | Coniferyl-aldehyde dehydrogenase. | 0.13 | 0.5 | 1 | 0.25 | 1 | 1 | 0.25 | 0.89 | 1 | 0.15 | 0.47 | 1 | 0.12 | 0.87 | 1 | 0.17 | 0.71 | 1 | 0.00035 | 0.00017 | 1.3e-05 | 5.3e-05 | 0 | 9.9e-05 | 0.00013 | 2.7e-05 | 0.00019 | 1.3 | 18 / 36 (50%) | 2 / 7 (29%) | 4 / 7 (57%) | 0.286 | 0.571 | 0.00026 | 9.8e-05 | 0.00037 | 2e-04 | 0.00013 | 0.00026 | -0.38 | 6 / 11 (55%) | 6 / 11 (55%) | 0.545 | 0.545 |
| 2.7.10.2 | Non-specific protein-tyrosine kinase. | 0.48 | 0.72 | 1 | 0.006 | 0.7 | 1 | 0.25 | 0.89 | 1 | 0.31 | 0.59 | 1 | 0.0024 | 0.62 | 1 | 0.057 | 0.66 | 1 | 0.023 | 0.023 | 0.021 | 0.019 | 0.018 | 0.0055 | 0.025 | 0.022 | 0.0094 | 0.4 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.022 | 0.019 | 0.0076 | 0.025 | 0.023 | 0.0096 | 0.18 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.8.3.9 | Butyrate--acetoacetate CoA-transferase. | 0.22 | 0.54 | 1 | 0.005 | 0.7 | 1 | 0.037 | 0.71 | 1 | 0.039 | 0.43 | 1 | 0.003 | 0.62 | 1 | 0.021 | 0.55 | 1 | 0.00051 | 0.00048 | 0.00039 | 0.00037 | 0.00024 | 0.00054 | 0.00075 | 0.00083 | 0.00044 | 1 | 34 / 36 (94%) | 5 / 7 (71%) | 7 / 7 (100%) | 0.714 | 1 | 0.00042 | 0.00039 | 0.00025 | 0.00045 | 0.00045 | 0.00022 | 0.1 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.2.6 | Nucleoside deoxyribosyltransferase. | 0.41 | 0.67 | 1 | 0.2 | 1 | 1 | 0.89 | 1 | 1 | 0.24 | 0.53 | 1 | 0.13 | 0.87 | 1 | 0.83 | 0.96 | 1 | 0.00054 | 4e-04 | 0.00031 | 0.00027 | 0.00011 | 0.00039 | 0.00038 | 0.00037 | 0.00038 | 0.49 | 27 / 36 (75%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.00042 | 0.00029 | 0.00065 | 0.00049 | 0.00045 | 0.00036 | 0.22 | 9 / 11 (82%) | 10 / 11 (91%) | 0.818 | 0.909 |
| 2.7.3.3 | Arginine kinase. | 0.83 | 0.92 | 1 | 0.63 | 1 | 1 | 0.81 | 1 | 1 | 0.96 | 0.99 | 1 | 0.27 | 0.88 | 1 | 0.99 | 1 | 1 | 0.0011 | 0.00077 | 0.00032 | 0.00058 | 4e-04 | 0.00084 | 0.00072 | 0.00024 | 0.0013 | 0.31 | 26 / 36 (72%) | 5 / 7 (71%) | 6 / 7 (86%) | 0.714 | 0.857 | 0.00081 | 0.00029 | 0.0013 | 0.00086 | 0.00059 | 0.00083 | 0.086 | 7 / 11 (64%) | 8 / 11 (73%) | 0.636 | 0.727 |
| 1.3.1.84 | Acrylyl-CoA reductase (NADPH). | 0.048 | 0.49 | 1 | 0.4 | 1 | 1 | 0.72 | 0.98 | 1 | 0.012 | 0.42 | 1 | 0.24 | 0.88 | 1 | 0.73 | 0.94 | 1 | 0.0014 | 0.00095 | 0.00029 | 0.00014 | 0 | 0.00031 | 0.00039 | 0 | 0.00058 | 1.5 | 25 / 36 (69%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 0.0016 | 0.00034 | 0.0035 | 0.0012 | 0.00075 | 0.0012 | -0.42 | 9 / 11 (82%) | 10 / 11 (91%) | 0.818 | 0.909 |
| 1.1.1.276 | Serine 3-dehydrogenase (NADP(+)). | 0.057 | 0.49 | 1 | 0.62 | 1 | 1 | 0.98 | 1 | 1 | 0.011 | 0.42 | 1 | 0.21 | 0.88 | 1 | 0.82 | 0.96 | 1 | 0.003 | 0.003 | 0.0026 | 0.0017 | 0.0024 | 0.0011 | 0.002 | 0.0016 | 0.0015 | 0.23 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0039 | 0.0026 | 0.0043 | 0.0037 | 0.0035 | 0.0015 | -0.076 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.9.1.1 | L-seryl-tRNA(Sec) selenium transferase. | 0.34 | 0.62 | 1 | 0.84 | 1 | 1 | 0.39 | 0.91 | 1 | 0.3 | 0.58 | 1 | 0.48 | 0.92 | 1 | 0.72 | 0.94 | 1 | 0.002 | 0.002 | 0.0018 | 0.0018 | 0.0019 | 0.00099 | 0.002 | 0.0026 | 0.0015 | 0.15 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0025 | 0.0017 | 0.0018 | 0.0017 | 0.0018 | 0.00071 | -0.56 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.1.198 | Phosphatidylinositol N-acetylglucosaminyltransferase. | 0.26 | 0.56 | 1 | 0.39 | 1 | 1 | 0.34 | 0.89 | 1 | 0.25 | 0.54 | 1 | 0.23 | 0.88 | 1 | 0.31 | 0.79 | 1 | 0.00039 | 0.00021 | 6.5e-05 | 0.00012 | 0 | 0.00023 | 2e-04 | 0.00016 | 0.00023 | 0.74 | 19 / 36 (53%) | 2 / 7 (29%) | 4 / 7 (57%) | 0.286 | 0.571 | 0.00031 | 0.00011 | 0.00058 | 0.00017 | 7.1e-05 | 0.00024 | -0.87 | 6 / 11 (55%) | 7 / 11 (64%) | 0.545 | 0.636 |
| 2.3.2.16 | Lipid II:glycine glycyltransferase. | 0.28 | 0.56 | 1 | 0.55 | 1 | 1 | 0.9 | 1 | 1 | 0.52 | 0.76 | 1 | 0.17 | 0.87 | 1 | 0.29 | 0.78 | 1 | 0.0013 | 0.0013 | 0.00073 | 0.00078 | 0.00082 | 0.00043 | 0.00081 | 0.00071 | 8e-04 | 0.054 | 34 / 36 (94%) | 7 / 7 (100%) | 5 / 7 (71%) | 1 | 0.714 | 0.0017 | 0.00064 | 0.0017 | 0.0014 | 0.00076 | 0.0014 | -0.28 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.3.2.1 | Argininosuccinate lyase. | 0.19 | 0.53 | 1 | 0.25 | 1 | 1 | 0.091 | 0.79 | 1 | 0.22 | 0.52 | 1 | 0.96 | 0.99 | 1 | 0.67 | 0.93 | 1 | 0.023 | 0.023 | 0.023 | 0.021 | 0.021 | 0.0026 | 0.019 | 0.022 | 0.0039 | -0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.023 | 0.023 | 0.0044 | 0.025 | 0.025 | 0.0036 | 0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.1.3 | Triacylglycerol lipase. | 0.46 | 0.71 | 1 | 0.17 | 1 | 1 | 0.28 | 0.89 | 1 | 0.31 | 0.59 | 1 | 0.03 | 0.78 | 1 | 0.085 | 0.66 | 1 | 0.0095 | 0.0095 | 0.0081 | 0.008 | 0.006 | 0.0038 | 0.01 | 0.008 | 0.0048 | 0.32 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.01 | 0.0091 | 0.0075 | 0.0095 | 0.0078 | 0.0042 | -0.074 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.2.1.13 | Acetate--CoA ligase (ADP-forming). | 0.36 | 0.64 | 1 | 0.39 | 1 | 1 | 0.98 | 1 | 1 | 0.3 | 0.58 | 1 | 0.62 | 0.95 | 1 | 0.82 | 0.96 | 1 | 0.00094 | 0.00078 | 0.00037 | 0.00063 | 0.00045 | 0.00068 | 0.00038 | 0.00032 | 0.00045 | -0.73 | 30 / 36 (83%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.001 | 0.00072 | 0.0011 | 0.00086 | 0.00022 | 0.0012 | -0.22 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 1.2.7.3 | 2-oxoglutarate synthase. | 0.41 | 0.67 | 1 | 0.63 | 1 | 1 | 0.37 | 0.9 | 1 | 0.32 | 0.59 | 1 | 0.37 | 0.9 | 1 | 0.55 | 0.9 | 1 | 0.003 | 0.0028 | 0.0012 | 0.0028 | 0.002 | 0.0025 | 0.0027 | 0.0012 | 0.0031 | -0.052 | 34 / 36 (94%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.002 | 0.00064 | 0.003 | 0.0037 | 0.0011 | 0.0066 | 0.89 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 3.4.15.5 | Peptidyl-dipeptidase Dcp. | 0.13 | 0.5 | 1 | 0.78 | 1 | 1 | 0.2 | 0.87 | 1 | 0.023 | 0.42 | 1 | 0.65 | 0.95 | 1 | 0.23 | 0.75 | 1 | 0.0032 | 0.003 | 0.001 | 0.0031 | 0.0024 | 0.0021 | 0.0034 | 0.0011 | 0.004 | 0.13 | 33 / 36 (92%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0016 | 0.00042 | 0.0033 | 0.004 | 9e-04 | 0.0072 | 1.3 | 9 / 11 (82%) | 10 / 11 (91%) | 0.818 | 0.909 |
| 3.5.99.10 | 2-iminobutanoate/2-iminopropanoate deaminase. | 0.62 | 0.81 | 1 | 0.57 | 1 | 1 | 0.29 | 0.89 | 1 | 0.93 | 0.98 | 1 | 0.44 | 0.91 | 1 | 0.22 | 0.74 | 1 | 2e-04 | 0.00012 | 7.8e-05 | 0.00019 | 6.4e-05 | 0.00034 | 0.00019 | 0.00013 | 0.00026 | 0 | 21 / 36 (58%) | 4 / 7 (57%) | 5 / 7 (71%) | 0.571 | 0.714 | 8.4e-05 | 9.5e-05 | 7.3e-05 | 5.1e-05 | 0 | 6.8e-05 | -0.72 | 7 / 11 (64%) | 5 / 11 (45%) | 0.636 | 0.455 |
| 1.5.1.42 | FMN reductase (NADH). | 0.29 | 0.57 | 1 | 0.45 | 1 | 1 | 0.88 | 1 | 1 | 0.27 | 0.56 | 1 | 0.43 | 0.91 | 1 | 0.91 | 0.98 | 1 | 0.00024 | 2e-05 | 0 | 3.4e-05 | 0 | 8.9e-05 | 5.8e-05 | 0 | 0.00015 | 0.77 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 8.1e-06 | 0 | 2.7e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 4.1.2.42 | D-threonine aldolase. | 0.19 | 0.53 | 1 | 0.3 | 1 | 1 | 0.47 | 0.93 | 1 | 0.12 | 0.47 | 1 | 0.19 | 0.87 | 1 | 0.3 | 0.78 | 1 | 0.00013 | 2.8e-05 | 0 | 0 | 0 | 0 | 2.3e-05 | 0 | 4.2e-05 | Inf | 8 / 36 (22%) | 0 / 7 (0%) | 2 / 7 (29%) | 0 | 0.286 | 3.4e-05 | 0 | 6.6e-05 | 4.4e-05 | 0 | 9.9e-05 | 0.37 | 3 / 11 (27%) | 3 / 11 (27%) | 0.273 | 0.273 |
| 2.7.8.6 | Undecaprenyl-phosphate galactose phosphotransferase. | 0.075 | 0.49 | 1 | 0.4 | 1 | 1 | 0.96 | 1 | 1 | 0.035 | 0.43 | 1 | 0.16 | 0.87 | 1 | 0.77 | 0.95 | 1 | 0.0092 | 0.0092 | 0.0082 | 0.0062 | 0.0061 | 0.003 | 0.0077 | 0.0062 | 0.0049 | 0.31 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.01 | 0.0082 | 0.0078 | 0.011 | 0.011 | 0.0029 | 0.14 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.5.1.5 | Methylenetetrahydrofolate dehydrogenase (NADP(+)). | 0.2 | 0.53 | 1 | 0.2 | 1 | 1 | 0.54 | 0.95 | 1 | 0.12 | 0.47 | 1 | 0.81 | 0.98 | 1 | 0.34 | 0.8 | 1 | 0.014 | 0.014 | 0.014 | 0.013 | 0.014 | 0.0028 | 0.012 | 0.012 | 0.0028 | -0.12 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.015 | 0.014 | 0.0027 | 0.014 | 0.014 | 0.0019 | -0.1 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.1.40 | Acyl-[acyl-carrier-protein]--phospholipid O-acyltransferase. | 0.28 | 0.56 | 1 | 0.82 | 1 | 1 | 0.68 | 0.98 | 1 | 0.44 | 0.69 | 1 | 0.84 | 0.98 | 1 | 0.92 | 0.98 | 1 | 0.00034 | 8.6e-05 | 0 | 0.00021 | 0 | 0.00053 | 0.00018 | 0 | 0.00046 | -0.22 | 9 / 36 (25%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 1.8e-05 | 0 | 4.9e-05 | 1.8e-05 | 0 | 3.7e-05 | 0 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 3.5.3.26 | (S)-ureidoglycine aminohydrolase. | 0.13 | 0.5 | 1 | 0.95 | 1 | 1 | 0.51 | 0.95 | 1 | 0.19 | 0.5 | 1 | 0.78 | 0.97 | 1 | 0.32 | 0.79 | 1 | 0.00023 | 0.00015 | 7.8e-05 | 0.00024 | 0.00012 | 0.00025 | 0.00023 | 0.00013 | 0.00023 | -0.061 | 23 / 36 (64%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00012 | 5.7e-05 | 0.00015 | 6.6e-05 | 0 | 0.00013 | -0.86 | 6 / 11 (55%) | 5 / 11 (45%) | 0.545 | 0.455 |
| 6.3.2.8 | UDP-N-acetylmuramate--L-alanine ligase. | 0.062 | 0.49 | 1 | 0.24 | 1 | 1 | 0.52 | 0.95 | 1 | 0.1 | 0.46 | 1 | 0.8 | 0.98 | 1 | 0.73 | 0.94 | 1 | 0.024 | 0.024 | 0.024 | 0.022 | 0.021 | 0.0037 | 0.02 | 0.02 | 0.0061 | -0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.026 | 0.026 | 0.0046 | 0.025 | 0.027 | 0.0032 | -0.057 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.46 | L-arabinose 1-dehydrogenase. | 0.16 | 0.5 | 1 | 0.41 | 1 | 1 | 0.18 | 0.85 | 1 | 0.2 | 0.5 | 1 | 0.91 | 0.99 | 1 | 0.53 | 0.9 | 1 | 0.00056 | 4.6e-05 | 0 | 0.00013 | 0 | 0.00036 | 8.7e-05 | 0 | 0.00023 | -0.58 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 1e-05 | 0 | 3.4e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 3.2.1.136 | Glucuronoarabinoxylan endo-1.4-beta-xylanase. | 0.35 | 0.62 | 1 | 0.99 | 1 | 1 | 0.38 | 0.9 | 1 | 0.13 | 0.47 | 1 | 0.83 | 0.98 | 1 | 0.33 | 0.8 | 1 | 0.0012 | 0.0011 | 0.00046 | 0.00099 | 0.00071 | 0.001 | 0.0012 | 0.00036 | 0.0016 | 0.28 | 32 / 36 (89%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00067 | 0.00029 | 0.00095 | 0.0014 | 3e-04 | 0.002 | 1.1 | 8 / 11 (73%) | 10 / 11 (91%) | 0.727 | 0.909 |
| 2.3.1.243 | Lauroyl-Kdo(2)-lipid IV(A) myristoyltransferase. | 0.2 | 0.53 | 1 | 0.77 | 1 | 1 | 0.084 | 0.77 | 1 | 0.24 | 0.54 | 1 | 0.85 | 0.98 | 1 | 0.16 | 0.71 | 1 | 0.00026 | 3.6e-05 | 0 | 8.4e-05 | 0 | 0.00022 | 6.8e-05 | 0 | 0.00018 | -0.3 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 2.2e-05 | 0 | 4.1e-05 | Inf | 0 / 11 (0%) | 3 / 11 (27%) | 0 | 0.273 |
| 3.4.21.72 | IgA-specific serine endopeptidase. | 0.44 | 0.69 | 1 | 0.4 | 1 | 1 | 0.94 | 1 | 1 | 0.66 | 0.85 | 1 | 0.2 | 0.87 | 1 | 0.71 | 0.94 | 1 | 4.4e-05 | 4.9e-06 | 0 | 4.1e-06 | 0 | 1.1e-05 | 8.3e-06 | 0 | 1.4e-05 | 1 | 4 / 36 (11%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 0 | 0 | 0 | 8.1e-06 | 0 | 2.7e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 6.1.1.4 | Leucine--tRNA ligase. | 0.24 | 0.55 | 1 | 0.63 | 1 | 1 | 0.56 | 0.95 | 1 | 0.24 | 0.54 | 1 | 0.49 | 0.92 | 1 | 0.49 | 0.88 | 1 | 0.039 | 0.039 | 0.037 | 0.037 | 0.036 | 0.004 | 0.036 | 0.037 | 0.0062 | -0.04 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.04 | 0.039 | 0.0065 | 0.041 | 0.04 | 0.0067 | 0.036 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.6.1.44 | Alanine--glyoxylate transaminase. | 0.95 | 0.98 | 1 | 0.29 | 1 | 1 | 0.83 | 1 | 1 | 0.96 | 0.99 | 1 | 0.19 | 0.87 | 1 | 0.7 | 0.94 | 1 | 0.00059 | 4e-04 | 0.00012 | 0.00043 | 0.00035 | 0.00042 | 0.00025 | 5.4e-05 | 0.00037 | -0.78 | 24 / 36 (67%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 0.00054 | 9.5e-05 | 0.00073 | 0.00032 | 0.00011 | 0.00051 | -0.75 | 8 / 11 (73%) | 7 / 11 (64%) | 0.727 | 0.636 |
| 3.6.1.8 | ATP diphosphatase. | 0.47 | 0.72 | 1 | 0.041 | 0.97 | 1 | 0.82 | 1 | 1 | 0.16 | 0.49 | 1 | 0.016 | 0.76 | 1 | 0.33 | 0.8 | 1 | 0.0026 | 0.0026 | 0.0026 | 0.0022 | 0.0025 | 0.00093 | 0.0027 | 0.0027 | 0.00086 | 0.3 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0025 | 0.0025 | 0.0013 | 0.003 | 0.0026 | 0.0011 | 0.26 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.5.1.41 | Riboflavin reductase (NAD(P)H). | 0.22 | 0.54 | 1 | 0.99 | 1 | 1 | 0.41 | 0.92 | 1 | 0.24 | 0.53 | 1 | 0.72 | 0.95 | 1 | 0.63 | 0.92 | 1 | 0.00036 | 3e-05 | 0 | 6.7e-05 | 0 | 0.00018 | 6.8e-05 | 0 | 0.00018 | 0.021 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 1.3e-05 | 0 | 4.5e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.1.2.3 | L-lactate dehydrogenase (cytochrome). | 0.64 | 0.83 | 1 | 0.98 | 1 | 1 | 0.65 | 0.97 | 1 | 0.49 | 0.74 | 1 | 0.51 | 0.93 | 1 | 0.89 | 0.97 | 1 | 0.004 | 0.004 | 0.004 | 0.0037 | 0.0041 | 0.0013 | 0.0036 | 0.0033 | 0.0014 | -0.04 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0041 | 0.0041 | 0.0019 | 0.0044 | 0.0038 | 0.0023 | 0.1 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.1.46 | Monogalactosyldiacylglycerol synthase. | 0.28 | 0.57 | 1 | 0.22 | 1 | 1 | 0.19 | 0.86 | 1 | 0.25 | 0.55 | 1 | 0.17 | 0.87 | 1 | 0.12 | 0.66 | 1 | 0.00015 | 1.6e-05 | 0 | 0 | 0 | 0 | 1.8e-05 | 0 | 4.8e-05 | Inf | 4 / 36 (11%) | 0 / 7 (0%) | 1 / 7 (14%) | 0 | 0.143 | 2.2e-05 | 0 | 5e-05 | 2e-05 | 0 | 6.7e-05 | -0.14 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 2.7.7.58 | (2.3-dihydroxybenzoyl)adenylate synthase. | 0.26 | 0.56 | 1 | 0.73 | 1 | 1 | 0.19 | 0.87 | 1 | 0.39 | 0.66 | 1 | 0.92 | 0.99 | 1 | 0.4 | 0.83 | 1 | 0.0014 | 0.0011 | 0.00025 | 0.0017 | 0.00056 | 0.0024 | 0.002 | 0.00086 | 0.0029 | 0.23 | 29 / 36 (81%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 8e-04 | 0.00019 | 0.0016 | 0.00048 | 0.00023 | 0.00092 | -0.74 | 9 / 11 (82%) | 8 / 11 (73%) | 0.818 | 0.727 |
| 3.5.4.23 | Blasticidin-S deaminase. | 0.75 | 0.88 | 1 | 0.073 | 1 | 1 | 0.22 | 0.88 | 1 | 0.89 | 0.96 | 1 | 0.11 | 0.87 | 1 | 0.27 | 0.77 | 1 | 9.7e-05 | 3e-05 | 0 | 4.8e-05 | 0 | 7.2e-05 | 0 | 0 | 0 | -Inf | 11 / 36 (31%) | 3 / 7 (43%) | 0 / 7 (0%) | 0.429 | 0 | 4.1e-05 | 0 | 7.3e-05 | 2.6e-05 | 0 | 4e-05 | -0.66 | 4 / 11 (36%) | 4 / 11 (36%) | 0.364 | 0.364 |
| 2.4.2.18 | Anthranilate phosphoribosyltransferase. | 0.64 | 0.83 | 1 | 0.6 | 1 | 1 | 0.54 | 0.95 | 1 | 0.43 | 0.69 | 1 | 0.83 | 0.98 | 1 | 0.86 | 0.96 | 1 | 0.014 | 0.014 | 0.014 | 0.013 | 0.013 | 0.0025 | 0.013 | 0.012 | 0.0041 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.014 | 0.014 | 0.0032 | 0.014 | 0.014 | 0.0025 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.2.99.3 | Transferred entry: 1.2.5.2. | 0.23 | 0.55 | 1 | 0.46 | 1 | 1 | 0.64 | 0.97 | 1 | 0.31 | 0.58 | 1 | 0.31 | 0.9 | 1 | 0.53 | 0.9 | 1 | 0.00018 | 0.00013 | 9.3e-05 | 0.00017 | 0.00016 | 0.00012 | 0.00018 | 0.00011 | 0.00026 | 0.082 | 25 / 36 (69%) | 6 / 7 (86%) | 5 / 7 (71%) | 0.857 | 0.714 | 1e-04 | 6.5e-05 | 0.00012 | 9.4e-05 | 7.1e-05 | 0.00011 | -0.089 | 7 / 11 (64%) | 7 / 11 (64%) | 0.636 | 0.636 |
| 3.4.13.19 | Membrane dipeptidase. | 0.15 | 0.5 | 1 | 0.23 | 1 | 1 | 0.21 | 0.87 | 1 | 0.23 | 0.53 | 1 | 0.46 | 0.92 | 1 | 0.47 | 0.87 | 1 | 0.00094 | 0.00094 | 0.00066 | 0.0012 | 0.00081 | 0.00085 | 0.00097 | 0.00064 | 0.00079 | -0.31 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00078 | 0.00053 | 0.00069 | 0.00088 | 0.00075 | 7e-04 | 0.17 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.1.156 | Glycine/sarcosine N-methyltransferase. | 0.77 | 0.89 | 1 | 0.85 | 1 | 1 | 0.74 | 0.99 | 1 | 0.97 | 0.99 | 1 | 0.32 | 0.9 | 1 | 0.26 | 0.76 | 1 | 0.0076 | 0.0076 | 0.0076 | 0.0077 | 0.0079 | 0.0021 | 0.008 | 0.0087 | 0.0024 | 0.055 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0076 | 0.008 | 0.0033 | 0.0073 | 0.0074 | 0.0028 | -0.058 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.1.226 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase. | 0.12 | 0.5 | 1 | 0.28 | 1 | 1 | 0.3 | 0.89 | 1 | 0.055 | 0.43 | 1 | 0.071 | 0.86 | 1 | 0.084 | 0.66 | 1 | 0.0047 | 0.0042 | 0.0027 | 0.0019 | 6e-04 | 0.0023 | 0.0032 | 0.0031 | 0.0032 | 0.75 | 32 / 36 (89%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.0061 | 0.0034 | 0.011 | 0.0044 | 0.0035 | 0.0039 | -0.47 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.2.1.3 | Formaldehyde transketolase. | 0.28 | 0.57 | 1 | 0.76 | 1 | 1 | 0.59 | 0.96 | 1 | 0.24 | 0.53 | 1 | 0.65 | 0.95 | 1 | 0.47 | 0.87 | 1 | 0.00012 | 2.4e-05 | 0 | 4.6e-06 | 0 | 1.2e-05 | 1.5e-05 | 0 | 4e-05 | 1.7 | 7 / 36 (19%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3.7e-05 | 0 | 7.6e-05 | 2.8e-05 | 0 | 6.7e-05 | -0.4 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 1.4.1.16 | Diaminopimelate dehydrogenase. | 0.024 | 0.49 | 1 | 0.15 | 1 | 1 | 0.32 | 0.89 | 1 | 0.16 | 0.48 | 1 | 0.39 | 0.9 | 1 | 0.78 | 0.95 | 1 | 0.0062 | 0.0062 | 0.0055 | 0.0083 | 0.0085 | 0.003 | 0.0068 | 0.0054 | 0.0033 | -0.29 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0054 | 0.0056 | 0.0015 | 0.0054 | 0.0049 | 0.0022 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.1.91 | Arogenate dehydratase. | 0.1 | 0.49 | 1 | 0.79 | 1 | 1 | 0.74 | 0.99 | 1 | 0.072 | 0.43 | 1 | 0.51 | 0.93 | 1 | 0.27 | 0.77 | 1 | 0.002 | 0.0015 | 0.00081 | 0.00045 | 8.1e-05 | 0.00058 | 0.00041 | 0 | 0.00063 | -0.13 | 26 / 36 (72%) | 4 / 7 (57%) | 3 / 7 (43%) | 0.571 | 0.429 | 0.0025 | 9e-04 | 0.0057 | 0.0017 | 0.0012 | 0.0017 | -0.56 | 9 / 11 (82%) | 10 / 11 (91%) | 0.818 | 0.909 |
| 3.5.3.8 | Formimidoylglutamase. | 0.91 | 0.96 | 1 | 0.97 | 1 | 1 | 0.53 | 0.95 | 1 | 0.91 | 0.97 | 1 | 1 | 1 | 1 | 0.4 | 0.83 | 1 | 0.00025 | 0.00016 | 0.00011 | 0.00015 | 0.00014 | 0.00012 | 0.00017 | 0.00012 | 2e-04 | 0.18 | 23 / 36 (64%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.00017 | 9.5e-05 | 0.00019 | 0.00016 | 0 | 3e-04 | -0.087 | 8 / 11 (73%) | 5 / 11 (45%) | 0.727 | 0.455 |
| 4.1.1.12 | Aspartate 4-decarboxylase. | 0.04 | 0.49 | 1 | 0.78 | 1 | 1 | 0.3 | 0.89 | 1 | 0.0077 | 0.38 | 1 | 0.57 | 0.93 | 1 | 0.14 | 0.68 | 1 | 0.00082 | 5e-04 | 1e-04 | 0.00061 | 0.00024 | 0.00078 | 0.0012 | 0.00043 | 0.0023 | 0.98 | 22 / 36 (61%) | 7 / 7 (100%) | 5 / 7 (71%) | 1 | 0.714 | 6.3e-05 | 0 | 0.00012 | 0.00045 | 0.00011 | 0.00073 | 2.8 | 4 / 11 (36%) | 6 / 11 (55%) | 0.364 | 0.545 |
| 2.7.1.16 | Ribulokinase. | 0.64 | 0.83 | 1 | 0.33 | 1 | 1 | 0.37 | 0.9 | 1 | 0.66 | 0.86 | 1 | 0.13 | 0.87 | 1 | 0.19 | 0.72 | 1 | 0.0048 | 0.0048 | 0.0039 | 0.0039 | 0.0038 | 0.0013 | 0.005 | 0.0045 | 0.0018 | 0.36 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0054 | 0.0034 | 0.0066 | 0.0047 | 0.0036 | 0.0028 | -0.2 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.1.52 | Peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase. | 0.018 | 0.49 | 1 | 0.59 | 1 | 1 | 0.35 | 0.9 | 1 | 0.0062 | 0.37 | 1 | 0.34 | 0.9 | 1 | 0.16 | 0.71 | 1 | 0.0018 | 0.001 | 0.00017 | 6.2e-05 | 0 | 0.00012 | 2e-04 | 0 | 0.00034 | 1.7 | 21 / 36 (58%) | 2 / 7 (29%) | 3 / 7 (43%) | 0.286 | 0.429 | 0.0019 | 0.00041 | 0.004 | 0.0013 | 0.00044 | 0.0016 | -0.55 | 9 / 11 (82%) | 7 / 11 (64%) | 0.818 | 0.636 |
| 2.3.1.266 | [Ribosomal protein S18]-alanine N-acetyltransferase. | 0.25 | 0.56 | 1 | 0.76 | 1 | 1 | 0.45 | 0.93 | 1 | 0.31 | 0.58 | 1 | 0.41 | 0.9 | 1 | 0.2 | 0.73 | 1 | 0.0047 | 0.0047 | 0.0049 | 0.0042 | 0.0043 | 0.00093 | 0.0044 | 0.0055 | 0.0016 | 0.067 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0051 | 0.0056 | 0.0015 | 0.0047 | 0.0039 | 0.0017 | -0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.30 | Glycerol kinase. | 0.32 | 0.6 | 1 | 0.77 | 1 | 1 | 0.35 | 0.9 | 1 | 0.22 | 0.52 | 1 | 0.43 | 0.91 | 1 | 0.15 | 0.71 | 1 | 0.025 | 0.025 | 0.024 | 0.024 | 0.023 | 0.0057 | 0.026 | 0.022 | 0.01 | 0.12 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.027 | 0.025 | 0.0087 | 0.024 | 0.022 | 0.0039 | -0.17 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.99.3 | Deoxyribodipyrimidine photo-lyase. | 0.38 | 0.65 | 1 | 0.23 | 1 | 1 | 0.67 | 0.98 | 1 | 0.55 | 0.78 | 1 | 0.25 | 0.88 | 1 | 0.63 | 0.92 | 1 | 0.00024 | 4.1e-05 | 0 | 6.7e-05 | 0 | 0.00018 | 0.00011 | 0 | 0.00023 | 0.72 | 6 / 36 (17%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 5.1e-06 | 0 | 1.7e-05 | 1.8e-05 | 0 | 4e-05 | 1.8 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 3.4.24.76 | Flavastacin. | 0.071 | 0.49 | 1 | 0.55 | 1 | 1 | 0.93 | 1 | 1 | 0.061 | 0.43 | 1 | 0.38 | 0.9 | 1 | 0.95 | 0.99 | 1 | 0.00039 | 0.00017 | 0 | 2e-05 | 0 | 5.2e-05 | 6.1e-05 | 0 | 0.00012 | 1.6 | 16 / 36 (44%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 0.00025 | 0 | 0.00034 | 0.00027 | 0.00011 | 0.00042 | 0.11 | 5 / 11 (45%) | 8 / 11 (73%) | 0.455 | 0.727 |
| 2.1.1.144 | Trans-aconitate 2-methyltransferase. | 0.36 | 0.64 | 1 | 0.2 | 1 | 1 | 0.17 | 0.85 | 1 | 0.16 | 0.48 | 1 | 0.092 | 0.86 | 1 | 0.11 | 0.66 | 1 | 0.00062 | 0.00048 | 0.00034 | 0.00042 | 8.1e-05 | 0.00064 | 0.00052 | 0.00024 | 0.00061 | 0.31 | 28 / 36 (78%) | 4 / 7 (57%) | 6 / 7 (86%) | 0.571 | 0.857 | 0.00052 | 0.00039 | 0.00051 | 0.00045 | 0.00032 | 0.00048 | -0.21 | 9 / 11 (82%) | 9 / 11 (82%) | 0.818 | 0.818 |
| 3.6.1.17 | Bis(5'-nucleosyl)-tetraphosphatase (asymmetrical). | 0.1 | 0.49 | 1 | 0.68 | 1 | 1 | 0.54 | 0.95 | 1 | 0.099 | 0.46 | 1 | 0.43 | 0.91 | 1 | 0.23 | 0.75 | 1 | 0.0011 | 0.001 | 0.00053 | 0.00044 | 0.00028 | 0.00043 | 0.00052 | 0.00036 | 4e-04 | 0.24 | 34 / 36 (94%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0015 | 0.00057 | 0.0023 | 0.0012 | 0.0011 | 0.0011 | -0.32 | 11 / 11 (100%) | 9 / 11 (82%) | 1 | 0.818 |
| 6.3.1.12 | D-aspartate ligase. | 0.023 | 0.49 | 1 | 0.68 | 1 | 1 | 0.55 | 0.95 | 1 | 0.0018 | 0.31 | 1 | 0.69 | 0.95 | 1 | 0.47 | 0.87 | 1 | 0.0022 | 0.0016 | 0.00024 | 8.5e-05 | 0 | 0.00011 | 0.00032 | 0 | 0.00062 | 1.9 | 26 / 36 (72%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 0.0031 | 0.00097 | 0.0073 | 0.0019 | 0.0012 | 0.0021 | -0.71 | 11 / 11 (100%) | 9 / 11 (82%) | 1 | 0.818 |
| 1.1.1.85 | 3-isopropylmalate dehydrogenase. | 0.46 | 0.71 | 1 | 0.52 | 1 | 1 | 0.36 | 0.9 | 1 | 0.24 | 0.53 | 1 | 0.59 | 0.93 | 1 | 0.78 | 0.95 | 1 | 0.017 | 0.017 | 0.017 | 0.016 | 0.017 | 0.0027 | 0.016 | 0.015 | 0.0033 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.018 | 0.017 | 0.0044 | 0.018 | 0.017 | 0.0032 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.1.64 | Alpha.alpha-trehalose phosphorylase. | 0.92 | 0.96 | 1 | 0.8 | 1 | 1 | 0.35 | 0.9 | 1 | 0.8 | 0.93 | 1 | 0.59 | 0.93 | 1 | 0.66 | 0.92 | 1 | 0.00034 | 0.00014 | 0 | 0.00012 | 0 | 0.00031 | 0.00012 | 0 | 0.00028 | 0 | 15 / 36 (42%) | 2 / 7 (29%) | 3 / 7 (43%) | 0.286 | 0.429 | 7.1e-05 | 0 | 0.00017 | 0.00024 | 3.3e-05 | 0.00046 | 1.8 | 4 / 11 (36%) | 6 / 11 (55%) | 0.364 | 0.545 |
| 3.2.2.28 | Double-stranded uracil-DNA glycosylase. | 0.95 | 0.98 | 1 | 0.23 | 1 | 1 | 0.23 | 0.88 | 1 | 0.78 | 0.92 | 1 | 0.14 | 0.87 | 1 | 0.11 | 0.66 | 1 | 0.00011 | 8.8e-06 | 0 | 2.3e-06 | 0 | 6.1e-06 | 3.9e-05 | 0 | 1e-04 | 4.1 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 1.1.1.154 | Ureidoglycolate dehydrogenase. | 0.37 | 0.65 | 1 | 0.83 | 1 | 1 | 0.95 | 1 | 1 | 0.54 | 0.77 | 1 | 0.96 | 0.99 | 1 | 0.98 | 1 | 1 | 0.00033 | 0.00022 | 0.00012 | 0.00028 | 0.00014 | 0.00037 | 0.00041 | 0.00012 | 0.00078 | 0.55 | 24 / 36 (67%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.00013 | 0.00011 | 0.00013 | 0.00015 | 1e-04 | 0.00018 | 0.21 | 7 / 11 (64%) | 7 / 11 (64%) | 0.636 | 0.636 |
| 3.1.3.11 | Fructose-bisphosphatase. | 0.16 | 0.5 | 1 | 0.45 | 1 | 1 | 0.43 | 0.93 | 1 | 0.18 | 0.5 | 1 | 0.85 | 0.98 | 1 | 0.9 | 0.98 | 1 | 0.027 | 0.027 | 0.026 | 0.024 | 0.024 | 0.0025 | 0.023 | 0.022 | 0.0074 | -0.061 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.029 | 0.029 | 0.0072 | 0.029 | 0.027 | 0.0072 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.11.33 | [Pyruvate. water dikinase] kinase. | 0.11 | 0.5 | 1 | 0.15 | 1 | 1 | 0.058 | 0.71 | 1 | 0.12 | 0.47 | 1 | 0.26 | 0.88 | 1 | 0.12 | 0.66 | 1 | 2e-04 | 4.9e-05 | 0 | 0.00012 | 0 | 0.00026 | 7.7e-05 | 0 | 2e-04 | -0.64 | 9 / 36 (25%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 5.5e-06 | 0 | 1.2e-05 | 3.2e-05 | 0 | 6.7e-05 | 2.5 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 3.6.1.31 | Phosphoribosyl-ATP diphosphatase. | 0.42 | 0.68 | 1 | 0.87 | 1 | 1 | 0.49 | 0.95 | 1 | 0.3 | 0.58 | 1 | 0.45 | 0.92 | 1 | 0.91 | 0.98 | 1 | 0.0092 | 0.0092 | 0.0093 | 0.0082 | 0.0086 | 0.0017 | 0.0085 | 0.0072 | 0.0032 | 0.052 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0091 | 0.0091 | 0.0024 | 0.01 | 0.011 | 0.0027 | 0.14 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.3.3 | Ornithine carbamoyltransferase. | 0.44 | 0.69 | 1 | 0.8 | 1 | 1 | 0.98 | 1 | 1 | 0.27 | 0.56 | 1 | 0.13 | 0.87 | 1 | 0.19 | 0.72 | 1 | 0.022 | 0.022 | 0.022 | 0.021 | 0.022 | 0.0036 | 0.022 | 0.021 | 0.0058 | 0.067 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.023 | 0.023 | 0.0033 | 0.023 | 0.023 | 0.0039 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.3.99.3 | 7-carboxy-7-deazaguanine synthase. | 0.055 | 0.49 | 1 | 0.4 | 1 | 1 | 0.28 | 0.89 | 1 | 0.23 | 0.53 | 1 | 0.92 | 0.99 | 1 | 0.77 | 0.95 | 1 | 0.0028 | 0.0028 | 0.0027 | 0.0034 | 0.0034 | 0.0013 | 0.0031 | 0.0033 | 0.0011 | -0.13 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0024 | 0.0024 | 0.00091 | 0.0026 | 0.0026 | 0.0012 | 0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.2.4.4 | 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring). | 0.78 | 0.9 | 1 | 0.52 | 1 | 1 | 0.82 | 1 | 1 | 0.62 | 0.83 | 1 | 0.21 | 0.88 | 1 | 0.33 | 0.8 | 1 | 0.00074 | 7e-04 | 0.00048 | 0.00054 | 0.00048 | 0.00043 | 0.00064 | 5e-04 | 0.00055 | 0.25 | 34 / 36 (94%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.00062 | 0.00065 | 0.00046 | 9e-04 | 0.00036 | 0.0014 | 0.54 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 3.6.3.32 | Transferred entry: 7.6.2.9. | 0.45 | 0.7 | 1 | 0.41 | 1 | 1 | 0.14 | 0.83 | 1 | 0.82 | 0.93 | 1 | 0.51 | 0.93 | 1 | 0.04 | 0.63 | 1 | 0.0024 | 0.0024 | 0.0016 | 0.0015 | 0.0012 | 0.001 | 0.0011 | 0.00077 | 0.00093 | -0.45 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0031 | 0.0017 | 0.005 | 0.0031 | 0.003 | 0.0022 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.2.14 | Amidophosphoribosyltransferase. | 0.089 | 0.49 | 1 | 0.79 | 1 | 1 | 0.66 | 0.97 | 1 | 0.11 | 0.47 | 1 | 0.15 | 0.87 | 1 | 0.14 | 0.7 | 1 | 0.035 | 0.035 | 0.034 | 0.031 | 0.03 | 0.0068 | 0.031 | 0.031 | 0.0072 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.038 | 0.036 | 0.014 | 0.036 | 0.036 | 0.0046 | -0.078 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.14 | L-iditol 2-dehydrogenase. | 0.97 | 0.99 | 1 | 0.24 | 1 | 1 | 0.71 | 0.98 | 1 | 0.62 | 0.83 | 1 | 0.055 | 0.81 | 1 | 0.26 | 0.76 | 1 | 0.022 | 0.022 | 0.022 | 0.021 | 0.02 | 0.0053 | 0.024 | 0.024 | 0.011 | 0.19 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.021 | 0.022 | 0.0049 | 0.022 | 0.023 | 0.0037 | 0.067 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.39 | Homoserine kinase. | 0.21 | 0.54 | 1 | 0.83 | 1 | 1 | 0.87 | 1 | 1 | 0.15 | 0.47 | 1 | 0.35 | 0.9 | 1 | 0.45 | 0.86 | 1 | 0.0089 | 0.0089 | 0.0085 | 0.0077 | 0.0074 | 0.0025 | 0.008 | 0.0077 | 0.0026 | 0.055 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0098 | 0.0081 | 0.0047 | 0.0095 | 0.0096 | 0.0023 | -0.045 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.8.3.21 | L-carnitine CoA-transferase. | 0.12 | 0.5 | 1 | 0.35 | 1 | 1 | 0.07 | 0.73 | 1 | 0.11 | 0.47 | 1 | 0.63 | 0.95 | 1 | 0.15 | 0.71 | 1 | 3e-04 | 4.2e-05 | 0 | 0.00011 | 0 | 0.00027 | 7.7e-05 | 0 | 2e-04 | -0.51 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.1e-05 | 0 | 4.7e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 3.2.1.70 | Glucan 1.6-alpha-glucosidase. | 0.24 | 0.55 | 1 | 0.81 | 1 | 1 | 0.54 | 0.95 | 1 | 0.38 | 0.65 | 1 | 0.36 | 0.9 | 1 | 0.56 | 0.9 | 1 | 0.003 | 0.0029 | 0.0021 | 0.0021 | 0.0018 | 0.0014 | 0.0026 | 0.0019 | 0.0023 | 0.31 | 35 / 36 (97%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.0035 | 0.0033 | 0.0025 | 0.003 | 0.0028 | 0.0019 | -0.22 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.89 | Arabinogalactan endo-beta-1.4-galactanase. | 0.22 | 0.54 | 1 | 0.81 | 1 | 1 | 0.64 | 0.97 | 1 | 0.17 | 0.49 | 1 | 0.97 | 0.99 | 1 | 0.76 | 0.95 | 1 | 0.015 | 0.015 | 0.013 | 0.012 | 0.012 | 0.0057 | 0.012 | 0.008 | 0.0084 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.016 | 0.015 | 0.0086 | 0.017 | 0.014 | 0.011 | 0.087 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.1.3 | Aconitate hydratase. | 0.011 | 0.49 | 1 | 0.22 | 1 | 1 | 0.19 | 0.87 | 1 | 0.0046 | 0.36 | 1 | 0.033 | 0.79 | 1 | 0.04 | 0.63 | 1 | 0.013 | 0.013 | 0.012 | 0.0092 | 0.0087 | 0.0018 | 0.011 | 0.012 | 0.0023 | 0.26 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.016 | 0.014 | 0.011 | 0.014 | 0.015 | 0.0043 | -0.19 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.1.3.24 | N-acetylneuraminate epimerase. | 0.39 | 0.65 | 1 | 0.066 | 1 | 1 | 0.044 | 0.71 | 1 | 0.27 | 0.56 | 1 | 0.032 | 0.79 | 1 | 0.071 | 0.66 | 1 | 0.00061 | 0.00041 | 0.00011 | 0.00041 | 0.00041 | 0.00029 | 0.00025 | 0 | 0.00041 | -0.71 | 24 / 36 (67%) | 6 / 7 (86%) | 3 / 7 (43%) | 0.857 | 0.429 | 0.00031 | 9.5e-05 | 0.00061 | 0.00061 | 0.00011 | 0.0012 | 0.98 | 8 / 11 (73%) | 7 / 11 (64%) | 0.727 | 0.636 |
| 2.1.1.177 | 23S rRNA (pseudouridine(1915)-N(3))-methyltransferase. | 0.13 | 0.5 | 1 | 0.16 | 1 | 1 | 0.31 | 0.89 | 1 | 0.1 | 0.46 | 1 | 0.57 | 0.93 | 1 | 0.81 | 0.96 | 1 | 0.0085 | 0.0085 | 0.0082 | 0.0078 | 0.0071 | 0.0019 | 0.0068 | 0.0067 | 0.0017 | -0.2 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0094 | 0.0088 | 0.0031 | 0.0091 | 0.0093 | 0.0011 | -0.047 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.7.n1 | Protein adenylyltransferase. | 0.052 | 0.49 | 1 | 0.17 | 1 | 1 | 0.25 | 0.89 | 1 | 0.019 | 0.42 | 1 | 0.33 | 0.9 | 1 | 0.42 | 0.85 | 1 | 5e-04 | 0.00015 | 0 | 4e-04 | 2.8e-05 | 0.00087 | 0.00036 | 0 | 0.00089 | -0.15 | 11 / 36 (31%) | 4 / 7 (57%) | 3 / 7 (43%) | 0.571 | 0.429 | 9.9e-06 | 0 | 2.5e-05 | 1.2e-05 | 0 | 2.6e-05 | 0.28 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 3.1.3.31 | Nucleotidase. | 0.14 | 0.5 | 1 | 0.64 | 1 | 1 | 0.31 | 0.89 | 1 | 0.14 | 0.47 | 1 | 0.49 | 0.92 | 1 | 0.25 | 0.76 | 1 | 9.4e-05 | 1e-05 | 0 | 0 | 0 | 0 | 3.9e-06 | 0 | 1e-05 | Inf | 4 / 36 (11%) | 0 / 7 (0%) | 1 / 7 (14%) | 0 | 0.143 | 2.5e-05 | 0 | 5.5e-05 | 6.8e-06 | 0 | 2.3e-05 | -1.9 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 3.5.1.32 | Hippurate hydrolase. | 0.099 | 0.49 | 1 | 0.18 | 1 | 1 | 0.36 | 0.9 | 1 | 0.03 | 0.43 | 1 | 0.019 | 0.76 | 1 | 0.19 | 0.72 | 1 | 0.0033 | 0.0033 | 0.0023 | 0.0016 | 0.0015 | 0.0012 | 0.0025 | 0.0028 | 0.0014 | 0.64 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0044 | 0.0023 | 0.0072 | 0.0036 | 0.0028 | 0.0023 | -0.29 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.21.108 | HtrA2 peptidase. | 0.18 | 0.52 | 1 | 0.23 | 1 | 1 | 0.94 | 1 | 1 | 0.2 | 0.51 | 1 | 0.82 | 0.98 | 1 | 0.35 | 0.81 | 1 | 0.00058 | 0.00034 | 4.4e-05 | 0.00041 | 5.7e-05 | 0.00062 | 9e-04 | 2.7e-05 | 0.0015 | 1.1 | 21 / 36 (58%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 5.5e-05 | 0 | 7.5e-05 | 0.00022 | 0.00011 | 0.00039 | 2 | 5 / 11 (45%) | 7 / 11 (64%) | 0.455 | 0.636 |
| 2.7.7.21 | Transferred entry: 2.7.7.72. | 0.25 | 0.56 | 1 | 0.49 | 1 | 1 | 0.73 | 0.99 | 1 | 0.23 | 0.53 | 1 | 0.78 | 0.97 | 1 | 0.99 | 1 | 1 | 0.0048 | 0.0048 | 0.0041 | 0.0038 | 0.0035 | 0.0027 | 0.0047 | 0.0029 | 0.0038 | 0.31 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.005 | 0.0043 | 0.0024 | 0.0051 | 0.0041 | 0.0017 | 0.029 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.1.132 | Precorrin-6B C(5.15)-methyltransferase (decarboxylating). | 0.56 | 0.78 | 1 | 0.041 | 0.97 | 1 | 0.51 | 0.95 | 1 | 0.35 | 0.62 | 1 | 0.083 | 0.86 | 1 | 0.86 | 0.96 | 1 | 0.0034 | 0.0034 | 0.0033 | 0.0035 | 0.0038 | 0.00082 | 0.0027 | 0.0025 | 0.0014 | -0.37 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0038 | 0.0036 | 0.00097 | 0.0033 | 0.0032 | 0.0014 | -0.2 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.1.19 | Phosphate butyryltransferase. | 0.044 | 0.49 | 1 | 0.83 | 1 | 1 | 0.54 | 0.95 | 1 | 0.057 | 0.43 | 1 | 0.97 | 0.99 | 1 | 0.87 | 0.97 | 1 | 0.0018 | 0.0018 | 0.0015 | 0.0024 | 0.0018 | 0.0015 | 0.0023 | 0.0022 | 0.0014 | -0.061 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0012 | 0.001 | 0.00068 | 0.0016 | 0.00091 | 0.0015 | 0.42 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.6.99.2 | Pyridoxine 5'-phosphate synthase. | 0.31 | 0.59 | 1 | 0.92 | 1 | 1 | 0.83 | 1 | 1 | 0.29 | 0.57 | 1 | 0.97 | 0.99 | 1 | 0.86 | 0.96 | 1 | 0.00081 | 0.00063 | 0.00028 | 0.00071 | 8e-04 | 0.00046 | 0.00074 | 0.00043 | 7e-04 | 0.06 | 28 / 36 (78%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00049 | 0.00016 | 0.00081 | 0.00066 | 0.00015 | 0.0011 | 0.43 | 9 / 11 (82%) | 7 / 11 (64%) | 0.818 | 0.636 |
| 2.4.2.52 | Triphosphoribosyl-dephospho-CoA synthase. | 0.71 | 0.87 | 1 | 0.82 | 1 | 1 | 0.56 | 0.96 | 1 | 0.75 | 0.9 | 1 | 0.89 | 0.99 | 1 | 0.39 | 0.83 | 1 | 0.0012 | 0.0012 | 0.0011 | 0.0011 | 0.0012 | 0.00054 | 0.0011 | 0.001 | 0.00056 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0013 | 0.0015 | 0.00084 | 0.0011 | 0.0011 | 0.00087 | -0.24 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.1.182 | (R)-citramalate synthase. | 0.55 | 0.77 | 1 | 0.4 | 1 | 1 | 0.97 | 1 | 1 | 0.38 | 0.65 | 1 | 0.68 | 0.95 | 1 | 0.96 | 0.99 | 1 | 0.001 | 0.00084 | 3e-04 | 0.00062 | 0.00045 | 0.00059 | 0.0012 | 0.00043 | 0.0016 | 0.95 | 29 / 36 (81%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 5e-04 | 0.00013 | 0.00082 | 0.0011 | 0.00011 | 0.0019 | 1.1 | 8 / 11 (73%) | 9 / 11 (82%) | 0.727 | 0.818 |
| 5.4.99.13 | Isobutyryl-CoA mutase. | 0.8 | 0.9 | 1 | 0.22 | 1 | 1 | 0.25 | 0.89 | 1 | 0.8 | 0.93 | 1 | 0.22 | 0.88 | 1 | 0.38 | 0.83 | 1 | 0.00058 | 0.00031 | 6e-05 | 3e-04 | 0.00024 | 0.00035 | 0.00018 | 0 | 0.00026 | -0.74 | 19 / 36 (53%) | 4 / 7 (57%) | 3 / 7 (43%) | 0.571 | 0.429 | 3e-04 | 8.7e-05 | 0.00053 | 4e-04 | 3.3e-05 | 0.00077 | 0.42 | 6 / 11 (55%) | 6 / 11 (55%) | 0.545 | 0.545 |
| 5.4.99.22 | 23S rRNA pseudouridine(2605) synthase. | 0.28 | 0.57 | 1 | 0.63 | 1 | 1 | 0.27 | 0.89 | 1 | 0.16 | 0.49 | 1 | 0.91 | 0.99 | 1 | 0.17 | 0.71 | 1 | 0.00085 | 0.00061 | 0.00033 | 0.00058 | 0.00034 | 0.00075 | 0.00045 | 0.00032 | 0.00067 | -0.37 | 26 / 36 (72%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.00084 | 7e-04 | 0.00076 | 0.00052 | 0.00021 | 0.00081 | -0.69 | 10 / 11 (91%) | 8 / 11 (73%) | 0.909 | 0.727 |
| 3.4.11.19 | D-stereospecific aminopeptidase. | 0.57 | 0.79 | 1 | 0.59 | 1 | 1 | 0.52 | 0.95 | 1 | 0.84 | 0.94 | 1 | 0.96 | 0.99 | 1 | 0.82 | 0.96 | 1 | 0.00063 | 0.00056 | 0.00043 | 0.00065 | 0.00051 | 6e-04 | 0.00045 | 0.00025 | 0.00041 | -0.53 | 32 / 36 (89%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.00048 | 0.00039 | 0.00043 | 0.00066 | 7e-04 | 0.00073 | 0.46 | 10 / 11 (91%) | 9 / 11 (82%) | 0.909 | 0.818 |
| 2.7.4.22 | UMP kinase. | 0.11 | 0.5 | 1 | 0.48 | 1 | 1 | 0.32 | 0.89 | 1 | 0.16 | 0.48 | 1 | 0.69 | 0.95 | 1 | 0.86 | 0.96 | 1 | 0.012 | 0.012 | 0.012 | 0.011 | 0.011 | 0.0018 | 0.01 | 0.01 | 0.002 | -0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.013 | 0.012 | 0.0024 | 0.013 | 0.013 | 0.0018 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.2.3 | Uridine nucleosidase. | 0.11 | 0.5 | 1 | 0.36 | 1 | 1 | 0.44 | 0.93 | 1 | 0.053 | 0.43 | 1 | 0.22 | 0.88 | 1 | 0.61 | 0.92 | 1 | 0.0038 | 0.003 | 0.002 | 0.0012 | 0.00091 | 0.0016 | 0.0022 | 0.0027 | 0.0025 | 0.87 | 29 / 36 (81%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.0044 | 0.002 | 0.0079 | 0.0033 | 0.0031 | 0.0029 | -0.42 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 3.2.1.187 | (Ara-f)(3)-Hyp beta-L-arabinobiosidase. | 0.61 | 0.81 | 1 | 0.96 | 1 | 1 | 0.3 | 0.89 | 1 | 0.7 | 0.88 | 1 | 0.79 | 0.97 | 1 | 0.2 | 0.73 | 1 | 0.0053 | 0.0052 | 0.004 | 0.004 | 0.0059 | 0.0034 | 0.0044 | 0.0038 | 0.0044 | 0.14 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0051 | 0.0038 | 0.0054 | 0.0064 | 0.0051 | 0.005 | 0.33 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 2.7.4.26 | Isopentenyl phosphate kinase. | 0.97 | 0.99 | 1 | 0.8 | 1 | 1 | 0.83 | 1 | 1 | 0.98 | 0.99 | 1 | 0.97 | 0.99 | 1 | 1 | 1 | 1 | 0.00021 | 2.4e-05 | 0 | 2e-05 | 0 | 5.4e-05 | 8.8e-06 | 0 | 2.3e-05 | -1.2 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.9e-05 | 0 | 9.5e-05 | 3.1e-05 | 0 | 1e-04 | 0.096 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.6.3.42 | Transferred entry: 7.5.2.3. | 0.14 | 0.5 | 1 | 0.56 | 1 | 1 | 0.5 | 0.95 | 1 | 0.14 | 0.47 | 1 | 0.78 | 0.97 | 1 | 0.94 | 0.98 | 1 | 0.0058 | 0.0053 | 0.004 | 0.0029 | 0.002 | 0.003 | 0.004 | 0.0038 | 0.0044 | 0.46 | 33 / 36 (92%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.0072 | 0.004 | 0.01 | 0.0058 | 0.0057 | 0.0045 | -0.31 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 1.2.1.21 | Glycolaldehyde dehydrogenase. | 0.3 | 0.58 | 1 | 0.15 | 1 | 1 | 0.021 | 0.71 | 1 | 0.049 | 0.43 | 1 | 0.091 | 0.86 | 1 | 0.017 | 0.55 | 1 | 0.00067 | 0.00048 | 0.00025 | 0.00041 | 0.00035 | 3e-04 | 0.00023 | 0.00027 | 0.00026 | -0.83 | 26 / 36 (72%) | 7 / 7 (100%) | 4 / 7 (57%) | 1 | 0.571 | 0.00038 | 3.1e-05 | 0.00074 | 8e-04 | 0.00034 | 0.0014 | 1.1 | 6 / 11 (55%) | 9 / 11 (82%) | 0.545 | 0.818 |
| 2.4.2.29 | tRNA-guanine(34) transglycosylase. | 0.012 | 0.49 | 1 | 0.66 | 1 | 1 | 0.26 | 0.89 | 1 | 0.036 | 0.43 | 1 | 0.16 | 0.87 | 1 | 0.051 | 0.65 | 1 | 0.02 | 0.02 | 0.02 | 0.017 | 0.017 | 0.0024 | 0.018 | 0.019 | 0.0043 | 0.082 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.022 | 0.021 | 0.0056 | 0.021 | 0.021 | 0.0026 | -0.067 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.68 | 1-phosphatidylinositol-4-phosphate 5-kinase. | 0.24 | 0.55 | 1 | 0.44 | 1 | 1 | 0.31 | 0.89 | 1 | 0.22 | 0.52 | 1 | 0.35 | 0.9 | 1 | 0.28 | 0.77 | 1 | 9e-05 | 2e-05 | 0 | 2.3e-06 | 0 | 6.1e-06 | 1.9e-05 | 0 | 4e-05 | 3 | 8 / 36 (22%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 3.5e-05 | 0 | 7.8e-05 | 1.7e-05 | 0 | 3.9e-05 | -1 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 3.5.99.5 | 2-aminomuconate deaminase. | 0.8 | 0.9 | 1 | 0.9 | 1 | 1 | 0.52 | 0.95 | 1 | 0.92 | 0.98 | 1 | 0.8 | 0.97 | 1 | 0.59 | 0.91 | 1 | 0.00014 | 2.4e-05 | 0 | 2e-05 | 0 | 5.4e-05 | 2.6e-05 | 0 | 7e-05 | 0.38 | 6 / 36 (17%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 1.5e-05 | 0 | 4.9e-05 | 3.4e-05 | 0 | 5.9e-05 | 1.2 | 1 / 11 (9.1%) | 3 / 11 (27%) | 0.0909 | 0.273 |
| 5.4.99.62 | D-ribose pyranase. | 0.72 | 0.87 | 1 | 0.28 | 1 | 1 | 0.65 | 0.97 | 1 | 0.58 | 0.8 | 1 | 0.15 | 0.87 | 1 | 0.5 | 0.88 | 1 | 0.00045 | 0.00043 | 0.00037 | 0.00037 | 0.00047 | 0.00019 | 0.00045 | 0.00037 | 2e-04 | 0.28 | 34 / 36 (94%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.00042 | 0.00032 | 0.00029 | 0.00046 | 0.00034 | 0.00028 | 0.13 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 2.5.1.114 | tRNA(Phe) (4-demethylwyosine(37)-C(7)) aminocarboxypropyltransferase. | 0.69 | 0.85 | 1 | 0.43 | 1 | 1 | 0.87 | 1 | 1 | 0.62 | 0.83 | 1 | 0.92 | 0.99 | 1 | 0.55 | 0.9 | 1 | 0.00013 | 2.2e-05 | 0 | 1.6e-05 | 0 | 4.3e-05 | 8.8e-06 | 0 | 2.3e-05 | -0.86 | 6 / 36 (17%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3.1e-05 | 0 | 7.6e-05 | 2.3e-05 | 0 | 5.2e-05 | -0.43 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 3.5.2.7 | Imidazolonepropionase. | 0.8 | 0.91 | 1 | 0.71 | 1 | 1 | 0.66 | 0.97 | 1 | 0.72 | 0.89 | 1 | 0.77 | 0.97 | 1 | 0.41 | 0.83 | 1 | 0.0016 | 0.0016 | 0.0012 | 0.0014 | 0.0012 | 0.00088 | 0.0017 | 0.0012 | 0.0019 | 0.28 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0017 | 0.0011 | 0.0017 | 0.0017 | 0.0011 | 0.0016 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.8.3.8 | Acetate CoA-transferase. | 0.25 | 0.55 | 1 | 0.071 | 1 | 1 | 0.19 | 0.87 | 1 | 0.66 | 0.86 | 1 | 0.026 | 0.76 | 1 | 0.084 | 0.66 | 1 | 0.0027 | 0.0027 | 0.0025 | 0.003 | 0.0026 | 0.0019 | 0.0038 | 0.0036 | 0.0014 | 0.34 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0022 | 0.0025 | 0.00076 | 0.0023 | 0.002 | 9e-04 | 0.064 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.3.19 | Transferred entry: 3.5.1.116. | 0.61 | 0.81 | 1 | 0.47 | 1 | 1 | 0.87 | 1 | 1 | 0.56 | 0.79 | 1 | 0.55 | 0.93 | 1 | 0.61 | 0.91 | 1 | 0.00023 | 0.00015 | 9.8e-05 | 0.00014 | 0.00011 | 0.00014 | 9.5e-05 | 0.00011 | 9.8e-05 | -0.56 | 24 / 36 (67%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 0.00019 | 9.7e-05 | 0.00024 | 0.00016 | 7.5e-05 | 0.00024 | -0.25 | 9 / 11 (82%) | 6 / 11 (55%) | 0.818 | 0.545 |
| 1.3.5.1 | Succinate dehydrogenase (quinone). | 0.89 | 0.96 | 1 | 0.9 | 1 | 1 | 0.58 | 0.96 | 1 | 0.74 | 0.9 | 1 | 0.82 | 0.98 | 1 | 0.78 | 0.95 | 1 | 0.0053 | 0.0053 | 0.0047 | 0.0053 | 0.0064 | 0.0033 | 0.0055 | 0.0047 | 0.0044 | 0.053 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0048 | 0.0046 | 0.0023 | 0.0058 | 0.0036 | 0.004 | 0.27 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.5.1.108 | 2-(3-amino-3-carboxypropyl)histidine synthase. | 0.49 | 0.72 | 1 | 0.24 | 1 | 1 | 0.35 | 0.9 | 1 | 0.73 | 0.89 | 1 | 0.38 | 0.9 | 1 | 0.51 | 0.89 | 1 | 0.00028 | 2.3e-05 | 0 | 5.7e-05 | 0 | 0.00015 | 0 | 0 | 0 | -Inf | 3 / 36 (8.3%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 1.9e-05 | 0 | 6.3e-05 | 2.1e-05 | 0 | 6.8e-05 | 0.14 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.1.3.2 | Aspartate carbamoyltransferase. | 0.15 | 0.5 | 1 | 0.75 | 1 | 1 | 0.71 | 0.98 | 1 | 0.14 | 0.47 | 1 | 0.38 | 0.9 | 1 | 0.41 | 0.84 | 1 | 0.019 | 0.019 | 0.019 | 0.018 | 0.018 | 0.0031 | 0.018 | 0.015 | 0.0045 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.021 | 0.019 | 0.0048 | 0.021 | 0.021 | 0.0026 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.88 | Non-reducing end beta-L-arabinopyranosidase. | 0.015 | 0.49 | 1 | 0.52 | 1 | 1 | 0.45 | 0.93 | 1 | 0.0013 | 0.31 | 1 | 0.11 | 0.87 | 1 | 0.19 | 0.72 | 1 | 0.0044 | 0.0026 | 0.00055 | 8.1e-05 | 0 | 0.00022 | 0.00049 | 0 | 0.00092 | 2.6 | 21 / 36 (58%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 0.0051 | 0.0012 | 0.012 | 0.003 | 0.0014 | 0.0034 | -0.77 | 9 / 11 (82%) | 9 / 11 (82%) | 0.818 | 0.818 |
| 2.1.1.230 | 23S rRNA (adenosine(1067)-2'-O)-methyltransferase. | 0.86 | 0.94 | 1 | 0.96 | 1 | 1 | 0.27 | 0.89 | 1 | 0.79 | 0.92 | 1 | 0.8 | 0.98 | 1 | 0.14 | 0.69 | 1 | 0.00015 | 7.4e-05 | 8e-06 | 0.00011 | 1.6e-05 | 0.00015 | 9.8e-05 | 5.4e-05 | 0.00014 | -0.17 | 18 / 36 (50%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 7e-05 | 7.2e-05 | 6.8e-05 | 3.9e-05 | 0 | 7.4e-05 | -0.84 | 7 / 11 (64%) | 3 / 11 (27%) | 0.636 | 0.273 |
| 1.21.4.4 | Betaine reductase. | 0.43 | 0.69 | 1 | 0.16 | 1 | 1 | 0.0019 | 0.51 | 1 | 0.2 | 0.5 | 1 | 0.15 | 0.87 | 1 | 0.0025 | 0.48 | 1 | 0.00066 | 0.00061 | 0.00029 | 0.00063 | 0.00022 | 0.00075 | 0.00084 | 0.00059 | 0.00083 | 0.42 | 33 / 36 (92%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00074 | 0.00087 | 0.00056 | 0.00032 | 0.00022 | 0.00031 | -1.2 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 1.13.11.- | 0.15 | 0.5 | 1 | 0.41 | 1 | 1 | 0.71 | 0.98 | 1 | 0.21 | 0.52 | 1 | 0.98 | 0.99 | 1 | 0.77 | 0.95 | 1 | 0.00035 | 5.8e-05 | 0 | 0.00018 | 0 | 0.00044 | 9.5e-05 | 0 | 0.00023 | -0.92 | 6 / 36 (17%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 1.3e-05 | 0 | 4.3e-05 | 2.7e-06 | 0 | 8.9e-06 | -2.3 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 | |
| 5.4.99.20 | 23S rRNA pseudouridine(2457) synthase. | 0.13 | 0.5 | 1 | 0.069 | 1 | 1 | 0.37 | 0.9 | 1 | 0.15 | 0.47 | 1 | 0.42 | 0.91 | 1 | 0.92 | 0.98 | 1 | 0.00015 | 1.7e-05 | 0 | 5.3e-05 | 0 | 0.00013 | 2.9e-05 | 0 | 7.7e-05 | -0.87 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 3.1.11.2 | Exodeoxyribonuclease III. | 0.51 | 0.74 | 1 | 0.56 | 1 | 1 | 0.6 | 0.96 | 1 | 0.34 | 0.61 | 1 | 0.97 | 0.99 | 1 | 0.96 | 0.99 | 1 | 0.011 | 0.011 | 0.011 | 0.01 | 0.011 | 0.0018 | 0.0096 | 0.009 | 0.0038 | -0.059 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.01 | 0.0053 | 0.011 | 0.012 | 0.0022 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.14.13.- | 0.15 | 0.5 | 1 | 0.64 | 1 | 1 | 0.42 | 0.93 | 1 | 0.19 | 0.5 | 1 | 0.38 | 0.9 | 1 | 0.62 | 0.92 | 1 | 0.00029 | 0.00019 | 8e-05 | 0.00031 | 0.00011 | 0.00055 | 0.00029 | 0.00013 | 0.00048 | -0.096 | 23 / 36 (64%) | 5 / 7 (71%) | 6 / 7 (86%) | 0.714 | 0.857 | 9e-05 | 0 | 0.00016 | 0.00014 | 8.9e-05 | 0.00018 | 0.64 | 4 / 11 (36%) | 8 / 11 (73%) | 0.364 | 0.727 | |
| 2.7.1.95 | Kanamycin kinase. | 0.87 | 0.95 | 1 | 0.88 | 1 | 1 | 0.63 | 0.96 | 1 | 0.82 | 0.93 | 1 | 0.68 | 0.95 | 1 | 0.41 | 0.84 | 1 | 0.00052 | 0.00032 | 1e-04 | 0.00031 | 0.00011 | 0.00055 | 0.00024 | 0.00012 | 0.00041 | -0.37 | 22 / 36 (61%) | 4 / 7 (57%) | 5 / 7 (71%) | 0.571 | 0.714 | 0.00038 | 9.5e-05 | 0.00072 | 0.00031 | 7.1e-05 | 0.00077 | -0.29 | 7 / 11 (64%) | 6 / 11 (55%) | 0.636 | 0.545 |
| 2.4.1.57 | Transferred entry: 2.4.1.345. | 0.54 | 0.77 | 1 | 0.27 | 1 | 1 | 0.22 | 0.88 | 1 | 0.27 | 0.56 | 1 | 0.3 | 0.9 | 1 | 0.17 | 0.71 | 1 | 0.0072 | 0.0072 | 0.0061 | 0.0064 | 0.0048 | 0.0035 | 0.0054 | 0.0038 | 0.0034 | -0.25 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0081 | 0.0065 | 0.007 | 0.008 | 0.0083 | 0.0031 | -0.018 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.7.22 | Mannose-1-phosphate guanylyltransferase (GDP). | 0.26 | 0.56 | 1 | 0.29 | 1 | 1 | 0.17 | 0.85 | 1 | 0.075 | 0.43 | 1 | 0.34 | 0.9 | 1 | 0.17 | 0.71 | 1 | 0.0012 | 0.00094 | 0.00041 | 0.001 | 0.0011 | 0.00065 | 0.00072 | 0.00043 | 0.00082 | -0.47 | 29 / 36 (81%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00087 | 0.00024 | 0.0015 | 0.0011 | 0.00043 | 0.0017 | 0.34 | 7 / 11 (64%) | 9 / 11 (82%) | 0.636 | 0.818 |
| 5.2.1.8 | Peptidylprolyl isomerase. | 0.27 | 0.56 | 1 | 0.55 | 1 | 1 | 0.2 | 0.87 | 1 | 0.17 | 0.49 | 1 | 0.17 | 0.87 | 1 | 0.48 | 0.87 | 1 | 0.086 | 0.086 | 0.086 | 0.082 | 0.081 | 0.0093 | 0.08 | 0.082 | 0.016 | -0.036 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.088 | 0.086 | 0.013 | 0.092 | 0.089 | 0.0098 | 0.064 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.1.245 | 3-hydroxy-5-phosphonooxypentane-2.4-dione thiolase. | 0.098 | 0.49 | 1 | 0.085 | 1 | 1 | 0.049 | 0.71 | 1 | 0.11 | 0.47 | 1 | 0.12 | 0.87 | 1 | 0.071 | 0.66 | 1 | 0.00015 | 5.1e-05 | 0 | 9e-05 | 6.4e-05 | 0.00011 | 3.4e-05 | 0 | 6.1e-05 | -1.4 | 12 / 36 (33%) | 4 / 7 (57%) | 2 / 7 (29%) | 0.571 | 0.286 | 3e-05 | 0 | 7.2e-05 | 5.7e-05 | 0 | 0.00012 | 0.93 | 2 / 11 (18%) | 4 / 11 (36%) | 0.182 | 0.364 |
| 3.6.3.16 | Transferred entry: 7.3.2.7. | 0.81 | 0.91 | 1 | 0.25 | 1 | 1 | 0.54 | 0.95 | 1 | 0.77 | 0.92 | 1 | 0.24 | 0.88 | 1 | 0.72 | 0.94 | 1 | 0.00032 | 0.00024 | 0.00013 | 0.00031 | 0.00016 | 0.00046 | 0.00022 | 0.00011 | 0.00032 | -0.49 | 27 / 36 (75%) | 6 / 7 (86%) | 4 / 7 (57%) | 0.857 | 0.571 | 0.00033 | 0.00026 | 0.00033 | 0.00012 | 0.00012 | 0.00013 | -1.5 | 10 / 11 (91%) | 7 / 11 (64%) | 0.909 | 0.636 |
| 1.2.1.60 | 5-carboxymethyl-2-hydroxymuconic-semialdehyde dehydrogenase. | 0.62 | 0.82 | 1 | 0.096 | 1 | 1 | 0.18 | 0.85 | 1 | 0.31 | 0.58 | 1 | 0.045 | 0.8 | 1 | 0.087 | 0.66 | 1 | 6.6e-05 | 5.5e-06 | 0 | 0 | 0 | 0 | 1.9e-05 | 0 | 5.1e-05 | Inf | 3 / 36 (8.3%) | 0 / 7 (0%) | 1 / 7 (14%) | 0 | 0.143 | 3e-06 | 0 | 9.9e-06 | 2.7e-06 | 0 | 8.9e-06 | -0.15 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 1.2.99.7 | Aldehyde dehydrogenase (FAD-independent). | 0.86 | 0.94 | 1 | 0.11 | 1 | 1 | 0.94 | 1 | 1 | 0.75 | 0.9 | 1 | 0.22 | 0.88 | 1 | 0.76 | 0.95 | 1 | 0.0099 | 0.0099 | 0.0092 | 0.011 | 0.012 | 0.0041 | 0.0091 | 0.0083 | 0.0048 | -0.27 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.0097 | 0.0035 | 0.0086 | 0.009 | 0.0034 | -0.36 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 7.5.2.8 | ABC-type D-allose transporter. | 0.067 | 0.49 | 1 | 0.034 | 0.97 | 1 | 0.089 | 0.79 | 1 | 0.031 | 0.43 | 1 | 0.14 | 0.87 | 1 | 0.28 | 0.78 | 1 | 0.00075 | 6.3e-05 | 0 | 0.00019 | 0 | 0.00044 | 0.00014 | 0 | 0.00036 | -0.44 | 3 / 36 (8.3%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 3.7.1.14 | 2-hydroxy-6-oxonona-2.4-dienedioate hydrolase. | 0.15 | 0.5 | 1 | 0.59 | 1 | 1 | 0.097 | 0.81 | 1 | 0.21 | 0.52 | 1 | 0.81 | 0.98 | 1 | 0.2 | 0.73 | 1 | 0.00016 | 5.6e-05 | 0 | 7.7e-05 | 0 | 1e-04 | 4.3e-05 | 0 | 5.5e-05 | -0.84 | 13 / 36 (36%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 1.3e-05 | 0 | 3e-05 | 9.4e-05 | 0 | 0.00015 | 2.9 | 2 / 11 (18%) | 5 / 11 (45%) | 0.182 | 0.455 |
| 2.3.1.51 | 1-acylglycerol-3-phosphate O-acyltransferase. | 0.3 | 0.58 | 1 | 0.92 | 1 | 1 | 0.76 | 0.99 | 1 | 0.18 | 0.49 | 1 | 0.66 | 0.95 | 1 | 0.88 | 0.97 | 1 | 0.017 | 0.017 | 0.017 | 0.015 | 0.013 | 0.0056 | 0.016 | 0.016 | 0.0074 | 0.093 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.018 | 0.016 | 0.0078 | 0.018 | 0.018 | 0.0034 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.6.1.7 | Acylphosphatase. | 0.73 | 0.87 | 1 | 0.91 | 1 | 1 | 0.6 | 0.96 | 1 | 0.39 | 0.65 | 1 | 0.62 | 0.95 | 1 | 0.22 | 0.74 | 1 | 0.0052 | 0.0052 | 0.005 | 0.0052 | 0.0054 | 0.0016 | 0.0051 | 0.005 | 0.0018 | -0.028 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0054 | 0.0055 | 0.0015 | 0.0049 | 0.0049 | 0.0016 | -0.14 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.4.2.11 | Phosphoglycerate mutase (2.3-diphosphoglycerate-dependent). | 0.18 | 0.52 | 1 | 0.54 | 1 | 1 | 0.63 | 0.96 | 1 | 0.04 | 0.43 | 1 | 0.18 | 0.87 | 1 | 0.33 | 0.8 | 1 | 0.0051 | 0.0051 | 0.0041 | 0.0033 | 0.0036 | 0.0018 | 0.0039 | 0.0026 | 0.0022 | 0.24 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0064 | 0.004 | 0.0081 | 0.0057 | 0.0061 | 0.0037 | -0.17 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.14.-.- | 0.24 | 0.55 | 1 | 0.38 | 1 | 1 | 0.99 | 1 | 1 | 0.28 | 0.57 | 1 | 0.58 | 0.93 | 1 | 0.86 | 0.96 | 1 | 0.00041 | 0.00018 | 0 | 0.00027 | 1.6e-05 | 0.00061 | 0.00043 | 2.7e-05 | 0.00087 | 0.67 | 16 / 36 (44%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 4.1e-05 | 0 | 7.2e-05 | 0.00011 | 0 | 0.00023 | 1.4 | 3 / 11 (27%) | 5 / 11 (45%) | 0.273 | 0.455 | |
| 1.3.8.8 | Long-chain-acyl-CoA dehydrogenase. | 0.26 | 0.56 | 1 | 0.25 | 1 | 1 | 0.4 | 0.92 | 1 | 0.43 | 0.69 | 1 | 0.095 | 0.86 | 1 | 0.21 | 0.74 | 1 | 0.00057 | 9.6e-05 | 0 | 0.00024 | 0 | 0.00062 | 0.00023 | 0 | 0.00053 | -0.061 | 6 / 36 (17%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 8.9e-06 | 0 | 3e-05 | 5.7e-06 | 0 | 1.3e-05 | -0.64 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 3.1.3.26 | 4-phytase. | 0.19 | 0.53 | 1 | 0.32 | 1 | 1 | 0.69 | 0.98 | 1 | 0.11 | 0.47 | 1 | 0.27 | 0.88 | 1 | 0.53 | 0.9 | 1 | 0.00052 | 0.00025 | 0 | 5e-04 | 0.00012 | 0.0011 | 0.00034 | 0 | 0.00064 | -0.56 | 17 / 36 (47%) | 6 / 7 (86%) | 3 / 7 (43%) | 0.857 | 0.429 | 0.00019 | 0 | 0.00047 | 7.9e-05 | 0 | 0.00015 | -1.3 | 4 / 11 (36%) | 4 / 11 (36%) | 0.364 | 0.364 |
| 6.3.4.21 | Nicotinate phosphoribosyltransferase. | 0.14 | 0.5 | 1 | 0.32 | 1 | 1 | 0.19 | 0.87 | 1 | 0.14 | 0.47 | 1 | 0.066 | 0.86 | 1 | 0.033 | 0.62 | 1 | 0.026 | 0.026 | 0.025 | 0.023 | 0.022 | 0.0039 | 0.025 | 0.023 | 0.0062 | 0.12 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.027 | 0.026 | 0.0063 | 0.026 | 0.026 | 0.0057 | -0.054 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 7.6.2.2 | ABC-type xenobiotic transporter. | 0.23 | 0.54 | 1 | 0.17 | 1 | 1 | 0.15 | 0.84 | 1 | 0.4 | 0.67 | 1 | 0.67 | 0.95 | 1 | 0.57 | 0.9 | 1 | 0.00048 | 9.4e-05 | 0 | 0.00025 | 0 | 0.00067 | 0.00018 | 0 | 0.00049 | -0.47 | 7 / 36 (19%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 9.9e-06 | 0 | 2.5e-05 | 1.9e-05 | 0 | 4.7e-05 | 0.94 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 1.7.2.2 | Nitrite reductase (cytochrome; ammonia-forming). | 0.2 | 0.53 | 1 | 0.22 | 1 | 1 | 0.72 | 0.98 | 1 | 0.12 | 0.47 | 1 | 0.55 | 0.93 | 1 | 0.85 | 0.96 | 1 | 0.00083 | 0.00076 | 0.00034 | 0.00079 | 5e-04 | 0.00096 | 0.0015 | 0.00048 | 0.0021 | 0.93 | 33 / 36 (92%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00035 | 0.00017 | 0.00051 | 0.00066 | 0.00034 | 0.001 | 0.92 | 10 / 11 (91%) | 9 / 11 (82%) | 0.909 | 0.818 |
| 2.7.12.2 | Mitogen-activated protein kinase kinase. | 0.1 | 0.49 | 1 | 0.29 | 1 | 1 | 0.23 | 0.88 | 1 | 0.048 | 0.43 | 1 | 0.15 | 0.87 | 1 | 0.078 | 0.66 | 1 | 0.0012 | 0.00092 | 0.00086 | 0.00052 | 4e-04 | 6e-04 | 0.00079 | 0.00048 | 0.00088 | 0.6 | 28 / 36 (78%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.0011 | 0.00087 | 0.0013 | 0.001 | 0.001 | 0.00082 | -0.14 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 4.2.2.n1 | Peptidoglycan lytic exotransglycosylase. | 0.29 | 0.58 | 1 | 0.11 | 1 | 1 | 0.48 | 0.94 | 1 | 0.59 | 0.81 | 1 | 0.15 | 0.87 | 1 | 0.49 | 0.87 | 1 | 0.001 | 0.00034 | 0 | 0.00082 | 0 | 0.0021 | 0.00077 | 0 | 0.002 | -0.091 | 12 / 36 (33%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 7e-05 | 0 | 0.00012 | 3.8e-05 | 0 | 6.3e-05 | -0.88 | 4 / 11 (36%) | 4 / 11 (36%) | 0.364 | 0.364 |
| 3.5.2.17 | Hydroxyisourate hydrolase. | 0.25 | 0.56 | 1 | 0.89 | 1 | 1 | 0.2 | 0.87 | 1 | 0.3 | 0.58 | 1 | 0.81 | 0.98 | 1 | 0.33 | 0.8 | 1 | 0.00016 | 1.8e-05 | 0 | 3.4e-05 | 0 | 8.9e-05 | 2.9e-05 | 0 | 7.7e-05 | -0.23 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 1.9e-05 | 0 | 4.6e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 2.7.1.191 | Protein-N(pi)-phosphohistidine--D-mannose phosphotransferase. | 0.13 | 0.5 | 1 | 0.39 | 1 | 1 | 0.17 | 0.85 | 1 | 0.22 | 0.52 | 1 | 0.68 | 0.95 | 1 | 0.38 | 0.82 | 1 | 0.007 | 0.007 | 0.0044 | 0.011 | 0.0047 | 0.014 | 0.0088 | 0.0053 | 0.0097 | -0.32 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0047 | 0.004 | 0.0023 | 0.0057 | 0.0053 | 0.0026 | 0.28 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.18.6.1 | Nitrogenase. | 0.38 | 0.65 | 1 | 0.61 | 1 | 1 | 0.89 | 1 | 1 | 0.28 | 0.57 | 1 | 0.93 | 0.99 | 1 | 0.65 | 0.92 | 1 | 0.01 | 0.01 | 0.01 | 0.0097 | 0.0086 | 0.0033 | 0.0091 | 0.0071 | 0.0044 | -0.092 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.01 | 0.0031 | 0.011 | 0.011 | 0.0034 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.167 | D-glycero-beta-D-manno-heptose-7-phosphate kinase. | 0.39 | 0.66 | 1 | 0.42 | 1 | 1 | 0.84 | 1 | 1 | 0.42 | 0.68 | 1 | 0.18 | 0.87 | 1 | 0.6 | 0.91 | 1 | 0.00074 | 0.00068 | 0.00033 | 0.00096 | 0.00042 | 0.0014 | 9e-04 | 0.00019 | 0.0013 | -0.093 | 33 / 36 (92%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00057 | 0.00032 | 0.00061 | 0.00047 | 0.00028 | 0.00053 | -0.28 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 3.1.1.6 | Acetylesterase. | 0.099 | 0.49 | 1 | 0.12 | 1 | 1 | 0.44 | 0.93 | 1 | 0.1 | 0.46 | 1 | 0.25 | 0.88 | 1 | 0.76 | 0.95 | 1 | 0.00026 | 2.9e-05 | 0 | 8.4e-05 | 0 | 0.00018 | 5.8e-05 | 0 | 0.00015 | -0.53 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 5.1e-06 | 0 | 1.7e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.1.1.174 | 23S rRNA (guanine(1835)-N(2))-methyltransferase. | 0.99 | 1 | 1 | 0.33 | 1 | 1 | 0.54 | 0.95 | 1 | 0.59 | 0.81 | 1 | 0.094 | 0.86 | 1 | 0.26 | 0.76 | 1 | 2e-04 | 1e-04 | 1.6e-05 | 0.00015 | 0 | 0.00035 | 0.00012 | 0.00012 | 1e-04 | -0.32 | 18 / 36 (50%) | 2 / 7 (29%) | 5 / 7 (71%) | 0.286 | 0.714 | 8e-05 | 0 | 0.00013 | 7.7e-05 | 5.9e-05 | 9.1e-05 | -0.055 | 5 / 11 (45%) | 6 / 11 (55%) | 0.455 | 0.545 |
| 2.4.1.301 | 2'-deamino-2'-hydroxyneamine 1-alpha-D-kanosaminyltransferase. | 0.4 | 0.66 | 1 | 0.91 | 1 | 1 | 0.88 | 1 | 1 | 0.32 | 0.59 | 1 | 0.99 | 1 | 1 | 0.99 | 1 | 1 | 0.00018 | 6.5e-05 | 0 | 7.4e-05 | 3.2e-05 | 9.5e-05 | 1e-04 | 0 | 0.00015 | 0.43 | 13 / 36 (36%) | 4 / 7 (57%) | 3 / 7 (43%) | 0.571 | 0.429 | 5.8e-05 | 0 | 0.00015 | 4.1e-05 | 0 | 7.6e-05 | -0.5 | 3 / 11 (27%) | 3 / 11 (27%) | 0.273 | 0.273 |
| 6.3.4.13 | Phosphoribosylamine--glycine ligase. | 0.46 | 0.71 | 1 | 0.49 | 1 | 1 | 0.62 | 0.96 | 1 | 0.25 | 0.55 | 1 | 0.79 | 0.97 | 1 | 0.64 | 0.92 | 1 | 0.019 | 0.019 | 0.019 | 0.018 | 0.02 | 0.0039 | 0.017 | 0.015 | 0.0039 | -0.082 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.02 | 0.019 | 0.0049 | 0.019 | 0.019 | 0.0029 | -0.074 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.4.1.13 | Cysteine-S-conjugate beta-lyase. | 0.28 | 0.57 | 1 | 0.93 | 1 | 1 | 0.94 | 1 | 1 | 0.44 | 0.69 | 1 | 0.74 | 0.96 | 1 | 0.86 | 0.96 | 1 | 0.00032 | 7e-05 | 0 | 0.00016 | 0 | 4e-04 | 0.00015 | 0 | 0.00038 | -0.093 | 8 / 36 (22%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 1.4e-05 | 0 | 3.9e-05 | 1.8e-05 | 0 | 4.1e-05 | 0.36 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 2.3.1.94 | 6-deoxyerythronolide-B synthase. | 0.59 | 0.79 | 1 | 0.7 | 1 | 1 | 0.53 | 0.95 | 1 | 0.23 | 0.53 | 1 | 0.39 | 0.9 | 1 | 0.2 | 0.73 | 1 | 0.00085 | 0.00062 | 0.00036 | 0.00066 | 0.00012 | 0.0012 | 0.00062 | 6e-04 | 0.00064 | -0.09 | 26 / 36 (72%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.00067 | 0.00036 | 0.00097 | 0.00053 | 0.00057 | 5e-04 | -0.34 | 10 / 11 (91%) | 8 / 11 (73%) | 0.909 | 0.727 |
| 2.4.1.12 | Cellulose synthase (UDP-forming). | 0.24 | 0.55 | 1 | 0.048 | 0.97 | 1 | 0.16 | 0.85 | 1 | 0.3 | 0.58 | 1 | 0.076 | 0.86 | 1 | 0.11 | 0.66 | 1 | 0.0015 | 0.0013 | 0.00027 | 0.0022 | 6e-04 | 0.0027 | 0.0013 | 0.00012 | 0.0019 | -0.76 | 30 / 36 (83%) | 7 / 7 (100%) | 5 / 7 (71%) | 1 | 0.714 | 0.0011 | 9.7e-05 | 0.0017 | 0.00081 | 0.00041 | 0.001 | -0.44 | 9 / 11 (82%) | 9 / 11 (82%) | 0.818 | 0.818 |
| 4.2.1.22 | Cystathionine beta-synthase. | 0.064 | 0.49 | 1 | 0.45 | 1 | 1 | 0.32 | 0.89 | 1 | 0.012 | 0.42 | 1 | 0.052 | 0.81 | 1 | 0.054 | 0.65 | 1 | 0.0015 | 0.0012 | 0.00033 | 0.00037 | 0.00012 | 0.00058 | 0.00062 | 2e-04 | 0.0011 | 0.74 | 31 / 36 (86%) | 4 / 7 (57%) | 6 / 7 (86%) | 0.571 | 0.857 | 0.0022 | 0.00032 | 0.0044 | 0.0013 | 0.00075 | 0.0015 | -0.76 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 6.3.1.- | 0.32 | 0.6 | 1 | 0.14 | 1 | 1 | 0.039 | 0.71 | 1 | 0.46 | 0.71 | 1 | 0.26 | 0.88 | 1 | 0.18 | 0.72 | 1 | 0.0025 | 0.0025 | 0.0019 | 0.0028 | 0.0019 | 0.0019 | 0.0018 | 0.0017 | 0.00099 | -0.64 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.002 | 0.0019 | 0.0011 | 0.0032 | 0.0031 | 0.0024 | 0.68 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 1.2.1.9 | Glyceraldehyde-3-phosphate dehydrogenase (NADP(+)). | 0.34 | 0.61 | 1 | 0.043 | 0.97 | 1 | 0.47 | 0.93 | 1 | 0.81 | 0.93 | 1 | 0.11 | 0.87 | 1 | 0.65 | 0.92 | 1 | 0.00064 | 0.00046 | 0.00013 | 0.0011 | 0.00026 | 0.002 | 0.00045 | 3.1e-05 | 0.001 | -1.3 | 26 / 36 (72%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 0.00041 | 0.00016 | 0.00074 | 0.00014 | 9.9e-05 | 0.00016 | -1.6 | 9 / 11 (82%) | 8 / 11 (73%) | 0.818 | 0.727 |
| 1.8.1.8 | Protein-disulfide reductase. | 0.88 | 0.95 | 1 | 0.26 | 1 | 1 | 0.2 | 0.87 | 1 | 0.93 | 0.98 | 1 | 0.28 | 0.88 | 1 | 0.4 | 0.83 | 1 | 0.0021 | 0.0021 | 0.0013 | 0.0018 | 0.0021 | 0.00086 | 0.0013 | 0.0011 | 0.00097 | -0.47 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0022 | 0.0015 | 0.0024 | 0.0028 | 0.0011 | 0.0037 | 0.35 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.7.1.17 | 4.5-9.10-diseco-3-hydroxy-5.9.17-trioxoandrosta-1(10).2-diene-4-oate | 0.61 | 0.81 | 1 | 0.23 | 1 | 1 | 0.76 | 0.99 | 1 | 0.45 | 0.7 | 1 | 0.16 | 0.87 | 1 | 0.57 | 0.9 | 1 | 9.6e-05 | 1.1e-05 | 0 | 0 | 0 | 0 | 1.7e-05 | 0 | 4.5e-05 | Inf | 4 / 36 (11%) | 0 / 7 (0%) | 1 / 7 (14%) | 0 | 0.143 | 6e-06 | 0 | 2e-05 | 1.8e-05 | 0 | 4e-05 | 1.6 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 1.3.1.9 | Enoyl-[acyl-carrier-protein] reductase (NADH). | 0.45 | 0.7 | 1 | 0.34 | 1 | 1 | 0.79 | 1 | 1 | 0.31 | 0.59 | 1 | 0.18 | 0.87 | 1 | 0.97 | 0.99 | 1 | 0.0043 | 0.0043 | 0.004 | 0.0028 | 0.0031 | 0.0017 | 0.004 | 0.004 | 0.003 | 0.51 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0041 | 0.0032 | 0.0041 | 0.0056 | 0.0051 | 0.0035 | 0.45 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.3.99.3 | Transferred entry: 1.3.8.7. 1.3.8.8 and 1.3.8.9. | 0.97 | 0.99 | 1 | 0.94 | 1 | 1 | 0.52 | 0.95 | 1 | 0.77 | 0.91 | 1 | 0.68 | 0.95 | 1 | 0.96 | 0.99 | 1 | 0.00073 | 0.00067 | 0.00054 | 0.00065 | 0.00055 | 0.00046 | 0.00079 | 0.00065 | 0.00063 | 0.28 | 33 / 36 (92%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00072 | 0.00058 | 0.00053 | 0.00054 | 0.00043 | 0.00044 | -0.42 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 4.2.1.33 | 3-isopropylmalate dehydratase. | 0.85 | 0.93 | 1 | 0.28 | 1 | 1 | 0.41 | 0.92 | 1 | 0.46 | 0.71 | 1 | 0.054 | 0.81 | 1 | 0.079 | 0.66 | 1 | 0.04 | 0.04 | 0.04 | 0.039 | 0.038 | 0.0075 | 0.043 | 0.041 | 0.015 | 0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.04 | 0.038 | 0.0075 | 0.04 | 0.041 | 0.0046 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.37 | Xylan 1.4-beta-xylosidase. | 0.066 | 0.49 | 1 | 0.68 | 1 | 1 | 0.61 | 0.96 | 1 | 0.017 | 0.42 | 1 | 0.88 | 0.99 | 1 | 0.86 | 0.96 | 1 | 0.041 | 0.041 | 0.035 | 0.028 | 0.033 | 0.017 | 0.025 | 0.019 | 0.014 | -0.16 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.051 | 0.038 | 0.047 | 0.05 | 0.049 | 0.024 | -0.029 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.4.27 | ([Pyruvate. phosphate dikinase] phosphate) phosphotransferase. | 0.67 | 0.84 | 1 | 0.87 | 1 | 1 | 0.99 | 1 | 1 | 0.99 | 1 | 1 | 0.76 | 0.97 | 1 | 1 | 1 | 1 | 0.00046 | 0.00044 | 0.00036 | 0.00033 | 0.00036 | 2e-04 | 0.00035 | 0.00035 | 0.00019 | 0.085 | 34 / 36 (94%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00052 | 0.00022 | 0.00063 | 0.00047 | 0.00045 | 0.00033 | -0.15 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 2.1.1.242 | 16S rRNA (guanine(1516)-N(2))-methyltransferase. | 0.42 | 0.68 | 1 | 0.95 | 1 | 1 | 0.24 | 0.89 | 1 | 0.27 | 0.56 | 1 | 0.82 | 0.98 | 1 | 0.12 | 0.67 | 1 | 0.0016 | 0.0016 | 0.0014 | 0.0016 | 0.0012 | 0.00092 | 0.0016 | 0.0011 | 0.0013 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0019 | 0.0017 | 0.00088 | 0.0013 | 0.0011 | 5e-04 | -0.55 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.17 | Mannitol-1-phosphate 5-dehydrogenase. | 0.92 | 0.96 | 1 | 0.8 | 1 | 1 | 0.32 | 0.89 | 1 | 0.82 | 0.93 | 1 | 0.89 | 0.99 | 1 | 0.31 | 0.79 | 1 | 0.003 | 0.003 | 0.0028 | 0.0026 | 0.0023 | 0.0013 | 0.0032 | 0.0025 | 0.0028 | 0.3 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0025 | 0.0028 | 9e-04 | 0.0035 | 0.0029 | 0.0014 | 0.49 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.20.4.4 | Arsenate reductase (thioredoxin). | 0.56 | 0.78 | 1 | 0.34 | 1 | 1 | 0.67 | 0.98 | 1 | 0.96 | 0.99 | 1 | 0.59 | 0.93 | 1 | 0.99 | 1 | 1 | 0.00012 | 1.7e-05 | 0 | 5e-05 | 0 | 0.00013 | 3.9e-06 | 0 | 1e-05 | -3.7 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 1.5e-05 | 0 | 3.3e-05 | 6.8e-06 | 0 | 2.3e-05 | -1.1 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 3.4.24.15 | Thimet oligopeptidase. | 0.077 | 0.49 | 1 | 1 | 1 | 1 | 0.29 | 0.89 | 1 | 0.047 | 0.43 | 1 | 0.71 | 0.95 | 1 | 0.17 | 0.71 | 1 | 8.5e-05 | 1.2e-05 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 5 / 36 (14%) | 0 / 7 (0%) | 0 / 7 (0%) | 0 | 0 | 2.9e-05 | 0 | 5.8e-05 | 9.8e-06 | 0 | 2.4e-05 | -1.6 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 3.2.1.3 | Glucan 1.4-alpha-glucosidase. | 0.48 | 0.72 | 1 | 0.17 | 1 | 1 | 0.38 | 0.9 | 1 | 0.25 | 0.55 | 1 | 0.97 | 0.99 | 1 | 0.85 | 0.96 | 1 | 0.0017 | 0.0015 | 7e-04 | 0.0015 | 0.00041 | 0.0024 | 0.00057 | 0.00062 | 0.00057 | -1.4 | 31 / 36 (86%) | 5 / 7 (71%) | 6 / 7 (86%) | 0.714 | 0.857 | 0.0018 | 0.00097 | 0.0019 | 0.0017 | 0.00083 | 0.0019 | -0.082 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 6.3.2.49 | L-alanine--L-anticapsin ligase. | 0.48 | 0.72 | 1 | 0.35 | 1 | 1 | 0.11 | 0.82 | 1 | 0.28 | 0.57 | 1 | 0.23 | 0.88 | 1 | 0.064 | 0.66 | 1 | 0.002 | 0.002 | 0.0016 | 0.0018 | 0.0016 | 0.0015 | 0.0022 | 0.0017 | 0.0017 | 0.29 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0021 | 0.0016 | 0.0011 | 0.0017 | 0.0012 | 0.0011 | -0.3 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.4.42 | N-isopropylammelide isopropylaminohydrolase. | 0.16 | 0.5 | 1 | 0.56 | 1 | 1 | 0.49 | 0.95 | 1 | 0.14 | 0.47 | 1 | 0.49 | 0.92 | 1 | 0.57 | 0.9 | 1 | 0.00021 | 3.5e-05 | 0 | 7e-05 | 0 | 0.00018 | 9.1e-05 | 0 | 0.00023 | 0.38 | 6 / 36 (17%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0 | 0 | 0 | 1.3e-05 | 0 | 3.5e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 3.2.2.19 | [Protein ADP-ribosylarginine] hydrolase. | 0.85 | 0.93 | 1 | 0.53 | 1 | 1 | 0.49 | 0.94 | 1 | 0.94 | 0.98 | 1 | 0.59 | 0.93 | 1 | 0.38 | 0.83 | 1 | 0.00024 | 6.7e-05 | 0 | 0.00012 | 0 | 0.00021 | 8.1e-05 | 0 | 2e-04 | -0.57 | 10 / 36 (28%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 9e-05 | 0 | 0.00015 | 3e-06 | 0 | 1e-05 | -4.9 | 4 / 11 (36%) | 1 / 11 (9.1%) | 0.364 | 0.0909 |
| 1.97.1.12 | Photosystem I. | 0.9 | 0.96 | 1 | 0.8 | 1 | 1 | 0.97 | 1 | 1 | 0.64 | 0.84 | 1 | 0.59 | 0.93 | 1 | 0.75 | 0.95 | 1 | 0.00018 | 4.6e-05 | 0 | 5.3e-05 | 0 | 0.00014 | 4.8e-05 | 0 | 0.00012 | -0.14 | 9 / 36 (25%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 3.6e-05 | 0 | 6.7e-05 | 5.1e-05 | 0 | 9.2e-05 | 0.5 | 3 / 11 (27%) | 3 / 11 (27%) | 0.273 | 0.273 |
| 1.10.3.14 | Transferred entry: 7.1.1.7. | 0.29 | 0.57 | 1 | 0.86 | 1 | 1 | 0.2 | 0.87 | 1 | 0.42 | 0.68 | 1 | 0.64 | 0.95 | 1 | 0.56 | 0.9 | 1 | 5e-04 | 9.6e-05 | 0 | 2e-04 | 0 | 0.00053 | 0.00018 | 0 | 0.00046 | -0.15 | 7 / 36 (19%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 1.8e-05 | 0 | 5.9e-05 | 5.5e-05 | 0 | 0.00014 | 1.6 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 2.1.2.2 | Phosphoribosylglycinamide formyltransferase. | 0.97 | 0.99 | 1 | 0.13 | 1 | 1 | 0.28 | 0.89 | 1 | 0.58 | 0.8 | 1 | 0.41 | 0.9 | 1 | 0.79 | 0.95 | 1 | 0.0092 | 0.0092 | 0.0093 | 0.0095 | 0.0095 | 0.0019 | 0.0081 | 0.0084 | 0.0016 | -0.23 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0095 | 0.0097 | 0.0019 | 0.0094 | 0.0095 | 0.0018 | -0.015 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.6.1.98 | UDP-2-acetamido-2-deoxy-ribo-hexuluronate aminotransferase. | 0.075 | 0.49 | 1 | 0.97 | 1 | 1 | 0.44 | 0.93 | 1 | 0.15 | 0.47 | 1 | 0.54 | 0.93 | 1 | 0.69 | 0.94 | 1 | 0.00019 | 3.6e-05 | 0 | 9.4e-05 | 0 | 0.00022 | 5.3e-05 | 0 | 8.4e-05 | -0.83 | 7 / 36 (19%) | 2 / 7 (29%) | 3 / 7 (43%) | 0.286 | 0.429 | 0 | 0 | 0 | 2.5e-05 | 0 | 5.6e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 2.1.1.208 | 23S rRNA (uridine(2479)-2'-O)-methyltransferase. | 0.054 | 0.49 | 1 | 0.67 | 1 | 1 | 0.67 | 0.98 | 1 | 0.079 | 0.44 | 1 | 0.27 | 0.88 | 1 | 0.74 | 0.94 | 1 | 0.0017 | 0.0017 | 0.0012 | 9e-04 | 0.00069 | 0.00059 | 0.00095 | 0.00047 | 0.001 | 0.078 | 35 / 36 (97%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.0025 | 0.0019 | 0.0031 | 0.0018 | 0.0014 | 0.0015 | -0.47 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.17.99.6 | Epoxyqueuosine reductase. | 0.49 | 0.73 | 1 | 0.74 | 1 | 1 | 0.58 | 0.96 | 1 | 0.4 | 0.67 | 1 | 0.67 | 0.95 | 1 | 0.87 | 0.97 | 1 | 0.0086 | 0.0086 | 0.0085 | 0.0081 | 0.008 | 0.00091 | 0.0079 | 0.0077 | 0.0015 | -0.036 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0088 | 0.0097 | 0.0023 | 0.009 | 0.0086 | 0.0015 | 0.032 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.4.99.6 | D-arginine dehydrogenase. | 0.28 | 0.57 | 1 | 0.16 | 1 | 1 | 0.91 | 1 | 1 | 0.51 | 0.75 | 1 | 0.58 | 0.93 | 1 | 0.48 | 0.87 | 1 | 0.00024 | 3.4e-05 | 0 | 1e-04 | 0 | 0.00027 | 4.8e-05 | 0 | 0.00013 | -1.1 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 1.4e-05 | 0 | 3.2e-05 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 3.5.4.16 | GTP cyclohydrolase I. | 0.47 | 0.71 | 1 | 0.65 | 1 | 1 | 0.31 | 0.89 | 1 | 0.23 | 0.53 | 1 | 0.16 | 0.87 | 1 | 0.052 | 0.65 | 1 | 0.0068 | 0.0068 | 0.0064 | 0.0064 | 0.0064 | 0.0018 | 0.0067 | 0.0068 | 0.002 | 0.066 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0073 | 0.0064 | 0.0036 | 0.0065 | 0.0064 | 0.0018 | -0.17 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.4.99.28 | tRNA pseudouridine(32) synthase. | 0.65 | 0.84 | 1 | 0.6 | 1 | 1 | 0.26 | 0.89 | 1 | 0.73 | 0.89 | 1 | 0.22 | 0.88 | 1 | 0.12 | 0.66 | 1 | 0.003 | 0.003 | 0.0021 | 0.0023 | 0.002 | 0.0017 | 0.0022 | 0.0021 | 0.0019 | -0.064 | 35 / 36 (97%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.0033 | 0.002 | 0.0045 | 0.0035 | 0.0026 | 0.0024 | 0.085 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.5.1.10 | (2E.6E)-farnesyl diphosphate synthase. | 0.78 | 0.9 | 1 | 0.87 | 1 | 1 | 0.046 | 0.71 | 1 | 0.43 | 0.69 | 1 | 0.69 | 0.95 | 1 | 0.14 | 0.69 | 1 | 0.0099 | 0.0099 | 0.0096 | 0.009 | 0.0084 | 0.0019 | 0.0091 | 0.0096 | 0.0037 | 0.016 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0093 | 0.0092 | 0.0019 | 0.012 | 0.011 | 0.0025 | 0.37 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.3.98.3 | Coproporphyrinogen dehydrogenase. | 0.96 | 0.98 | 1 | 0.55 | 1 | 1 | 0.52 | 0.95 | 1 | 0.78 | 0.92 | 1 | 0.99 | 1 | 1 | 0.99 | 1 | 1 | 0.014 | 0.014 | 0.014 | 0.014 | 0.014 | 0.0025 | 0.013 | 0.016 | 0.004 | -0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.014 | 0.014 | 0.0049 | 0.015 | 0.015 | 0.0045 | 0.1 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.1.3.21 | Maltose epimerase. | 0.59 | 0.8 | 1 | 1 | 1 | 1 | 0.4 | 0.92 | 1 | 0.34 | 0.61 | 1 | 0.86 | 0.99 | 1 | 0.59 | 0.91 | 1 | 0.00021 | 2.9e-05 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 5 / 36 (14%) | 0 / 7 (0%) | 0 / 7 (0%) | 0 | 0 | 9.1e-06 | 0 | 2.3e-05 | 8.5e-05 | 0 | 0.00024 | 3.2 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 4.1.1.41 | Transferred entry: 7.2.4.3. | 0.062 | 0.49 | 1 | 0.16 | 1 | 1 | 0.021 | 0.71 | 1 | 0.041 | 0.43 | 1 | 0.27 | 0.88 | 1 | 0.044 | 0.65 | 1 | 0.0027 | 0.0026 | 0.0018 | 0.0033 | 0.0022 | 0.0017 | 0.0022 | 0.0017 | 0.0015 | -0.58 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0016 | 0.0014 | 0.0015 | 0.0034 | 0.0021 | 0.0039 | 1.1 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 1.1.1.289 | Sorbose reductase. | 0.12 | 0.5 | 1 | 0.69 | 1 | 1 | 0.55 | 0.95 | 1 | 0.089 | 0.45 | 1 | 0.66 | 0.95 | 1 | 0.66 | 0.92 | 1 | 0.0015 | 0.00077 | 5.7e-05 | 0.00015 | 0 | 0.00031 | 0.00032 | 0 | 0.00075 | 1.1 | 18 / 36 (50%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0.0014 | 0.00032 | 0.0033 | 0.00084 | 0.00023 | 0.0012 | -0.74 | 7 / 11 (64%) | 7 / 11 (64%) | 0.636 | 0.636 |
| 2.7.7.62 | Adenosylcobinamide-phosphate guanylyltransferase. | 0.15 | 0.5 | 1 | 0.86 | 1 | 1 | 0.98 | 1 | 1 | 0.16 | 0.48 | 1 | 0.73 | 0.96 | 1 | 0.77 | 0.95 | 1 | 0.0028 | 0.0028 | 0.0024 | 0.0022 | 0.002 | 9e-04 | 0.0024 | 0.0018 | 0.0016 | 0.13 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0031 | 0.0033 | 0.0013 | 0.0032 | 0.0032 | 0.0015 | 0.046 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.17.11 | Glutamate carboxypeptidase. | 0.92 | 0.96 | 1 | 0.34 | 1 | 1 | 0.31 | 0.89 | 1 | 1 | 1 | 1 | 0.4 | 0.9 | 1 | 0.7 | 0.94 | 1 | 0.0011 | 0.0011 | 0.00062 | 0.0011 | 0.00079 | 0.0012 | 0.0012 | 0.0016 | 0.001 | 0.13 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0012 | 0.00043 | 0.0012 | 0.001 | 6e-04 | 0.00098 | -0.26 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 3.5.1.5 | Urease. | 0.78 | 0.9 | 1 | 0.99 | 1 | 1 | 0.65 | 0.97 | 1 | 0.95 | 0.98 | 1 | 0.96 | 0.99 | 1 | 0.56 | 0.9 | 1 | 0.0044 | 0.0044 | 0.0044 | 0.0046 | 0.0045 | 0.0015 | 0.0048 | 0.0047 | 0.0024 | 0.061 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0044 | 0.0045 | 0.002 | 0.0039 | 0.0032 | 0.002 | -0.17 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.94 | Glycerol-3-phosphate dehydrogenase (NAD(P)(+)). | 0.24 | 0.55 | 1 | 0.83 | 1 | 1 | 0.61 | 0.96 | 1 | 0.17 | 0.49 | 1 | 0.48 | 0.92 | 1 | 0.62 | 0.92 | 1 | 0.017 | 0.017 | 0.016 | 0.015 | 0.015 | 0.0029 | 0.015 | 0.014 | 0.0038 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.017 | 0.016 | 0.003 | 0.018 | 0.018 | 0.0027 | 0.082 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.5.1.9 | Riboflavin synthase. | 0.62 | 0.82 | 1 | 0.39 | 1 | 1 | 0.69 | 0.98 | 1 | 0.96 | 0.99 | 1 | 0.2 | 0.87 | 1 | 0.4 | 0.83 | 1 | 0.0056 | 0.0056 | 0.0055 | 0.0055 | 0.0055 | 0.00083 | 0.0063 | 0.0062 | 0.0013 | 0.2 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0053 | 0.0054 | 0.0015 | 0.0056 | 0.0053 | 0.002 | 0.079 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.4.2.10 | Phosphoglucosamine mutase. | 0.09 | 0.49 | 1 | 0.41 | 1 | 1 | 0.86 | 1 | 1 | 0.12 | 0.47 | 1 | 0.75 | 0.97 | 1 | 0.3 | 0.78 | 1 | 0.029 | 0.029 | 0.028 | 0.027 | 0.025 | 0.0051 | 0.025 | 0.025 | 0.0052 | -0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.031 | 0.033 | 0.0054 | 0.03 | 0.029 | 0.0075 | -0.047 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.192 | Protein-N(pi)-phosphohistidine--N-acetylmuramate phosphotransferase. | 0.15 | 0.5 | 1 | 0.82 | 1 | 1 | 0.92 | 1 | 1 | 0.063 | 0.43 | 1 | 0.61 | 0.95 | 1 | 0.47 | 0.87 | 1 | 0.00042 | 4.6e-05 | 0 | 7.9e-05 | 0 | 0.00018 | 0.00015 | 0 | 0.00041 | 0.93 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.4.7.1 | Glutamate synthase (ferredoxin). | 0.33 | 0.61 | 1 | 0.53 | 1 | 1 | 0.41 | 0.92 | 1 | 0.26 | 0.56 | 1 | 0.74 | 0.96 | 1 | 0.98 | 1 | 1 | 0.05 | 0.05 | 0.051 | 0.046 | 0.045 | 0.0066 | 0.044 | 0.044 | 0.0096 | -0.064 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.052 | 0.056 | 0.016 | 0.053 | 0.053 | 0.0073 | 0.027 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.1.179 | Beta-ketoacyl-[acyl-carrier-protein] synthase II. | 0.062 | 0.49 | 1 | 0.99 | 1 | 1 | 0.64 | 0.97 | 1 | 0.037 | 0.43 | 1 | 0.54 | 0.93 | 1 | 0.35 | 0.81 | 1 | 0.03 | 0.03 | 0.028 | 0.024 | 0.024 | 0.0042 | 0.024 | 0.023 | 0.008 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.036 | 0.03 | 0.025 | 0.032 | 0.03 | 0.008 | -0.17 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.16.- | 0.13 | 0.5 | 1 | 0.18 | 1 | 1 | 0.13 | 0.82 | 1 | 0.22 | 0.52 | 1 | 0.2 | 0.87 | 1 | 0.2 | 0.73 | 1 | 0.00027 | 0.00013 | 1.3e-05 | 0.00026 | 0.00011 | 0.00047 | 8.3e-05 | 0 | 0.00016 | -1.6 | 18 / 36 (50%) | 5 / 7 (71%) | 3 / 7 (43%) | 0.714 | 0.429 | 5.9e-05 | 0 | 7.3e-05 | 0.00016 | 0 | 0.00025 | 1.4 | 5 / 11 (45%) | 5 / 11 (45%) | 0.455 | 0.455 | |
| 4.1.99.18 | Transferred entry: 4.1.99.22 and 4.6.1.17. | 0.23 | 0.54 | 1 | 0.62 | 1 | 1 | 0.44 | 0.93 | 1 | 0.34 | 0.61 | 1 | 0.66 | 0.95 | 1 | 0.41 | 0.84 | 1 | 0.00013 | 6.1e-05 | 0 | 9.9e-05 | 2.8e-05 | 0.00014 | 8.5e-05 | 0 | 0.00017 | -0.22 | 17 / 36 (47%) | 4 / 7 (57%) | 3 / 7 (43%) | 0.571 | 0.429 | 3.7e-05 | 0 | 7.5e-05 | 4.5e-05 | 2.9e-05 | 4.9e-05 | 0.28 | 4 / 11 (36%) | 6 / 11 (55%) | 0.364 | 0.545 |
| 3.5.1.26 | N(4)-(beta-N-acetylglucosaminyl)-L-asparaginase. | 0.78 | 0.9 | 1 | 0.6 | 1 | 1 | 0.86 | 1 | 1 | 0.8 | 0.93 | 1 | 0.91 | 0.99 | 1 | 0.66 | 0.92 | 1 | 0.00025 | 2e-04 | 0.00013 | 0.00028 | 0.00014 | 0.00036 | 0.00016 | 0.00013 | 0.00014 | -0.81 | 28 / 36 (78%) | 5 / 7 (71%) | 6 / 7 (86%) | 0.714 | 0.857 | 0.00019 | 0.00013 | 0.00018 | 0.00018 | 0.00011 | 0.00021 | -0.078 | 9 / 11 (82%) | 8 / 11 (73%) | 0.818 | 0.727 |
| 2.4.99.13 | (Kdo)-lipid IV(A) 3-deoxy-D-manno-octulosonic acid transferase. | 0.078 | 0.49 | 1 | 0.075 | 1 | 1 | 0.16 | 0.85 | 1 | 0.07 | 0.43 | 1 | 0.13 | 0.87 | 1 | 0.27 | 0.77 | 1 | 0.00043 | 5.9e-05 | 0 | 0.00017 | 0 | 0.00035 | 0.00013 | 0 | 0.00033 | -0.39 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 3e-06 | 0 | 9.9e-06 | 2.7e-06 | 0 | 8.9e-06 | -0.15 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 1.1.1.67 | Mannitol 2-dehydrogenase. | 0.57 | 0.78 | 1 | 0.37 | 1 | 1 | 0.88 | 1 | 1 | 0.82 | 0.93 | 1 | 0.36 | 0.9 | 1 | 0.88 | 0.97 | 1 | 0.0017 | 0.0017 | 0.0018 | 0.0016 | 0.0018 | 0.00081 | 0.0021 | 0.0019 | 0.0015 | 0.39 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0014 | 0.0012 | 7e-04 | 0.0016 | 0.0018 | 0.00063 | 0.19 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.5.1.101 | N.N'-diacetyllegionaminate synthase. | 0.43 | 0.69 | 1 | 0.61 | 1 | 1 | 0.47 | 0.93 | 1 | 0.49 | 0.74 | 1 | 0.23 | 0.88 | 1 | 0.82 | 0.96 | 1 | 0.00079 | 0.00024 | 0 | 0.00027 | 0 | 0.00048 | 0.00024 | 0.00012 | 0.00037 | -0.17 | 11 / 36 (31%) | 2 / 7 (29%) | 4 / 7 (57%) | 0.286 | 0.571 | 9.5e-05 | 0 | 0.00028 | 0.00037 | 0 | 0.00081 | 2 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 3.1.11.1 | Exodeoxyribonuclease I. | 0.33 | 0.6 | 1 | 0.033 | 0.97 | 1 | 0.26 | 0.89 | 1 | 0.48 | 0.73 | 1 | 0.14 | 0.87 | 1 | 0.49 | 0.88 | 1 | 0.00056 | 0.00023 | 0 | 0.00046 | 4.8e-05 | 0.00077 | 0.00029 | 0 | 0.00061 | -0.67 | 15 / 36 (42%) | 4 / 7 (57%) | 2 / 7 (29%) | 0.571 | 0.286 | 0.00017 | 0 | 0.00036 | 0.00012 | 0 | 3e-04 | -0.5 | 5 / 11 (45%) | 4 / 11 (36%) | 0.455 | 0.364 |
| 1.17.1.4 | Xanthine dehydrogenase. | 0.47 | 0.71 | 1 | 0.61 | 1 | 1 | 0.5 | 0.95 | 1 | 0.36 | 0.63 | 1 | 0.94 | 0.99 | 1 | 0.23 | 0.75 | 1 | 0.016 | 0.016 | 0.016 | 0.016 | 0.015 | 0.005 | 0.015 | 0.019 | 0.0071 | -0.093 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.018 | 0.016 | 0.0052 | 0.016 | 0.015 | 0.0046 | -0.17 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 7.5.2.6 | ABC-type lipid A-core oligosaccharide transporter. | 0.1 | 0.49 | 1 | 0.094 | 1 | 1 | 0.057 | 0.71 | 1 | 0.085 | 0.44 | 1 | 0.15 | 0.87 | 1 | 0.11 | 0.66 | 1 | 0.00024 | 7.5e-05 | 0 | 0.00016 | 3.2e-05 | 0.00035 | 0.00013 | 0 | 3e-04 | -0.3 | 11 / 36 (31%) | 4 / 7 (57%) | 2 / 7 (29%) | 0.571 | 0.286 | 2e-05 | 0 | 5.1e-05 | 3.8e-05 | 0 | 8.3e-05 | 0.93 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 1.1.1.80 | Isopropanol dehydrogenase (NADP(+)). | 0.16 | 0.5 | 1 | 0.091 | 1 | 1 | 0.21 | 0.87 | 1 | 0.12 | 0.47 | 1 | 0.085 | 0.86 | 1 | 0.12 | 0.66 | 1 | 0.00013 | 4.5e-05 | 0 | 5e-05 | 0 | 8.9e-05 | 3.9e-06 | 0 | 1e-05 | -3.7 | 13 / 36 (36%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 0.00011 | 6.2e-05 | 0.00018 | 6.8e-06 | 0 | 2.3e-05 | -4 | 8 / 11 (73%) | 1 / 11 (9.1%) | 0.727 | 0.0909 |
| 5.3.1.22 | Hydroxypyruvate isomerase. | 0.77 | 0.89 | 1 | 0.63 | 1 | 1 | 0.81 | 1 | 1 | 0.98 | 0.99 | 1 | 0.82 | 0.98 | 1 | 0.8 | 0.96 | 1 | 0.00029 | 2e-04 | 1e-04 | 0.00021 | 8.5e-05 | 0.00033 | 0.00032 | 0.00011 | 0.00057 | 0.61 | 25 / 36 (69%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.00014 | 0.00011 | 0.00016 | 0.00018 | 9.9e-05 | 0.00024 | 0.36 | 8 / 11 (73%) | 7 / 11 (64%) | 0.727 | 0.636 |
| 2.7.1.83 | Pseudouridine kinase. | 0.58 | 0.79 | 1 | 0.34 | 1 | 1 | 0.26 | 0.89 | 1 | 0.97 | 0.99 | 1 | 0.37 | 0.9 | 1 | 0.36 | 0.81 | 1 | 0.0026 | 0.0026 | 0.002 | 0.0029 | 0.0019 | 0.0023 | 0.0026 | 0.0018 | 0.0024 | -0.16 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0024 | 0.0022 | 9e-04 | 0.0026 | 0.0021 | 0.0013 | 0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.7.59 | [Protein-PII] uridylyltransferase. | 0.13 | 0.5 | 1 | 0.42 | 1 | 1 | 0.46 | 0.93 | 1 | 0.13 | 0.47 | 1 | 0.42 | 0.91 | 1 | 0.75 | 0.95 | 1 | 0.0047 | 0.0042 | 0.0022 | 0.0017 | 0.00079 | 0.0019 | 0.0029 | 0.002 | 0.0034 | 0.77 | 32 / 36 (89%) | 6 / 7 (86%) | 5 / 7 (71%) | 0.857 | 0.714 | 0.0061 | 0.0027 | 0.011 | 0.0046 | 0.0032 | 0.0045 | -0.41 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 4.1.2.43 | 3-hexulose-6-phosphate synthase. | 0.7 | 0.86 | 1 | 0.18 | 1 | 1 | 0.57 | 0.96 | 1 | 0.5 | 0.74 | 1 | 0.21 | 0.87 | 1 | 0.75 | 0.95 | 1 | 0.00048 | 0.00044 | 0.00043 | 0.00037 | 0.00043 | 0.00032 | 0.00055 | 0.00047 | 0.00049 | 0.57 | 33 / 36 (92%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00041 | 4e-04 | 0.00029 | 0.00045 | 0.00043 | 0.00033 | 0.13 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 2.1.1.181 | 23S rRNA (adenine(1618)-N(6))-methyltransferase. | 0.08 | 0.49 | 1 | 0.94 | 1 | 1 | 0.33 | 0.89 | 1 | 0.14 | 0.47 | 1 | 0.78 | 0.97 | 1 | 0.59 | 0.91 | 1 | 0.00067 | 0.00064 | 0.00048 | 0.00081 | 0.00055 | 0.00065 | 0.00074 | 0.00065 | 4e-04 | -0.13 | 34 / 36 (94%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 4e-04 | 0.00032 | 0.00035 | 0.00069 | 0.00044 | 0.00057 | 0.79 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 3.4.11.6 | Aminopeptidase B. | 0.58 | 0.79 | 1 | 0.21 | 1 | 1 | 0.55 | 0.95 | 1 | 0.42 | 0.68 | 1 | 0.043 | 0.8 | 1 | 0.25 | 0.76 | 1 | 0.006 | 0.006 | 0.006 | 0.0055 | 0.0045 | 0.0031 | 0.0064 | 0.0067 | 0.0027 | 0.22 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0059 | 0.006 | 0.0024 | 0.0061 | 0.0058 | 0.0012 | 0.048 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.1.101 | Formylmethanofuran--tetrahydromethanopterin N-formyltransferase. | 0.32 | 0.6 | 1 | 0.18 | 1 | 1 | 0.11 | 0.82 | 1 | 0.61 | 0.82 | 1 | 0.5 | 0.93 | 1 | 0.25 | 0.76 | 1 | 0.00019 | 2.6e-05 | 0 | 7.3e-05 | 0 | 0.00019 | 8.8e-06 | 0 | 2.3e-05 | -3.1 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 9.5e-06 | 0 | 3.2e-05 | 2.3e-05 | 0 | 5.2e-05 | 1.3 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 3.2.1.78 | Mannan endo-1.4-beta-mannosidase. | 0.27 | 0.56 | 1 | 0.48 | 1 | 1 | 0.2 | 0.87 | 1 | 0.16 | 0.48 | 1 | 0.76 | 0.97 | 1 | 0.36 | 0.82 | 1 | 0.0059 | 0.0059 | 0.0039 | 0.0042 | 0.0036 | 0.0041 | 0.0028 | 0.0019 | 0.0025 | -0.58 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0064 | 0.0037 | 0.0055 | 0.0084 | 0.0064 | 0.0064 | 0.39 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.1.203 | UDP-N-acetylbacillosamine N-acetyltransferase. | 0.1 | 0.49 | 1 | 0.13 | 1 | 1 | 0.47 | 0.93 | 1 | 0.21 | 0.52 | 1 | 0.26 | 0.88 | 1 | 0.75 | 0.95 | 1 | 8.5e-05 | 9.5e-06 | 0 | 3.1e-05 | 0 | 5.4e-05 | 8.8e-06 | 0 | 2.3e-05 | -1.8 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 6e-06 | 0 | 2e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 3.5.1.78 | Glutathionylspermidine amidase. | 0.0031 | 0.49 | 1 | 0.006 | 0.7 | 1 | 0.0081 | 0.61 | 1 | 2e-04 | 0.23 | 0.45 | 0.017 | 0.76 | 1 | 0.017 | 0.55 | 1 | 0.00032 | 8.1e-05 | 0 | 0.00025 | 0.00014 | 0.00042 | 0.00015 | 0 | 0.00038 | -0.74 | 9 / 36 (25%) | 5 / 7 (71%) | 2 / 7 (29%) | 0.714 | 0.286 | 0 | 0 | 0 | 8.7e-06 | 0 | 2.1e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 5.3.3.14 | Trans-2-decenoyl-[acyl-carrier-protein] isomerase. | 0.012 | 0.49 | 1 | 0.0072 | 0.7 | 1 | 0.0017 | 0.51 | 1 | 0.007 | 0.37 | 1 | 0.0013 | 0.62 | 1 | 0.00019 | 0.44 | 0.44 | 0.00026 | 0.00013 | 2.4e-05 | 6.9e-06 | 0 | 1.8e-05 | 0.00016 | 0.00013 | 0.00018 | 4.5 | 18 / 36 (50%) | 1 / 7 (14%) | 5 / 7 (71%) | 0.143 | 0.714 | 0.00019 | 9.5e-05 | 0.00024 | 0.00012 | 0 | 0.00024 | -0.66 | 8 / 11 (73%) | 4 / 11 (36%) | 0.727 | 0.364 |
| 3.2.1.156 | Oligosaccharide reducing-end xylanase. | 0.062 | 0.49 | 1 | 0.69 | 1 | 1 | 1 | 1 | 1 | 0.061 | 0.43 | 1 | 0.99 | 1 | 1 | 0.85 | 0.96 | 1 | 0.0043 | 0.0043 | 0.0035 | 0.0025 | 0.0035 | 0.0014 | 0.0023 | 0.0013 | 0.0021 | -0.12 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.006 | 0.0037 | 0.0066 | 0.0049 | 0.0044 | 0.0031 | -0.29 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.3.99.23 | All-trans-retinol 13.14-reductase. | 0.64 | 0.83 | 1 | 0.76 | 1 | 1 | 0.63 | 0.96 | 1 | 0.6 | 0.81 | 1 | 0.73 | 0.96 | 1 | 0.85 | 0.96 | 1 | 7e-04 | 0.00041 | 9.7e-05 | 0.00021 | 0.00014 | 0.00034 | 0.00017 | 0 | 0.00029 | -0.3 | 21 / 36 (58%) | 4 / 7 (57%) | 3 / 7 (43%) | 0.571 | 0.429 | 0.00039 | 0.00011 | 0.00067 | 0.00071 | 7.5e-05 | 0.0015 | 0.86 | 7 / 11 (64%) | 7 / 11 (64%) | 0.636 | 0.636 |
| 3.1.26.- | 0.27 | 0.56 | 1 | 0.73 | 1 | 1 | 0.37 | 0.9 | 1 | 0.16 | 0.48 | 1 | 0.21 | 0.87 | 1 | 0.094 | 0.66 | 1 | 0.017 | 0.017 | 0.015 | 0.015 | 0.013 | 0.0044 | 0.016 | 0.017 | 0.0047 | 0.093 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.02 | 0.015 | 0.012 | 0.017 | 0.015 | 0.0048 | -0.23 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 1.6.5.3 | Transferred entry: 7.1.1.2. | 0.16 | 0.5 | 1 | 0.72 | 1 | 1 | 0.55 | 0.95 | 1 | 0.43 | 0.69 | 1 | 0.91 | 0.99 | 1 | 0.9 | 0.98 | 1 | 0.011 | 0.011 | 0.01 | 0.012 | 0.012 | 0.0029 | 0.012 | 0.01 | 0.0046 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0094 | 0.0099 | 0.0025 | 0.01 | 0.01 | 0.0058 | 0.089 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.3.2 | Ethanolamine-phosphate phospho-lyase. | 0.62 | 0.81 | 1 | 0.34 | 1 | 1 | 0.69 | 0.98 | 1 | 0.44 | 0.69 | 1 | 0.28 | 0.89 | 1 | 0.65 | 0.92 | 1 | 0.00072 | 0.00028 | 0 | 0.00028 | 0 | 0.00048 | 0.00012 | 0 | 0.00032 | -1.2 | 14 / 36 (39%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 4e-04 | 8e-05 | 0.00072 | 0.00027 | 0 | 0.00051 | -0.57 | 6 / 11 (55%) | 5 / 11 (45%) | 0.545 | 0.455 |
| 2.7.4.7 | Phosphooxymethylpyrimidine kinase. | 0.99 | 1 | 1 | 0.56 | 1 | 1 | 0.87 | 1 | 1 | 0.59 | 0.81 | 1 | 0.86 | 0.99 | 1 | 0.57 | 0.91 | 1 | 0.012 | 0.012 | 0.012 | 0.013 | 0.013 | 0.004 | 0.012 | 0.012 | 0.005 | -0.12 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.013 | 0.012 | 0.0043 | 0.012 | 0.011 | 0.0019 | -0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.6.1.83 | LL-diaminopimelate aminotransferase. | 0.11 | 0.5 | 1 | 0.86 | 1 | 1 | 0.89 | 1 | 1 | 0.16 | 0.48 | 1 | 0.15 | 0.87 | 1 | 0.17 | 0.71 | 1 | 0.02 | 0.02 | 0.02 | 0.018 | 0.018 | 0.0032 | 0.019 | 0.019 | 0.0037 | 0.078 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.021 | 0.021 | 0.0052 | 0.021 | 0.021 | 0.0032 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.4.8 | Guanylate kinase. | 0.056 | 0.49 | 1 | 0.4 | 1 | 1 | 0.27 | 0.89 | 1 | 0.089 | 0.45 | 1 | 0.078 | 0.86 | 1 | 0.039 | 0.63 | 1 | 0.016 | 0.016 | 0.016 | 0.014 | 0.014 | 0.003 | 0.015 | 0.014 | 0.0061 | 0.1 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.018 | 0.019 | 0.0038 | 0.017 | 0.016 | 0.0037 | -0.082 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.14.12.19 | 3-phenylpropanoate dioxygenase. | 0.025 | 0.49 | 1 | 0.042 | 0.97 | 1 | 0.05 | 0.71 | 1 | 0.076 | 0.44 | 1 | 0.062 | 0.86 | 1 | 0.066 | 0.66 | 1 | 0.00011 | 9.2e-06 | 0 | 3.9e-05 | 0 | 8.9e-05 | 0 | 0 | 0 | -Inf | 3 / 36 (8.3%) | 2 / 7 (29%) | 0 / 7 (0%) | 0.286 | 0 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.7.1.63 | Polyphosphate--glucose phosphotransferase. | 0.16 | 0.5 | 1 | 0.98 | 1 | 1 | 0.93 | 1 | 1 | 0.12 | 0.47 | 1 | 0.86 | 0.99 | 1 | 0.81 | 0.96 | 1 | 0.0016 | 0.0012 | 0.00059 | 0.00049 | 0.00011 | 0.00071 | 5e-04 | 0.00012 | 0.00074 | 0.029 | 27 / 36 (75%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.0019 | 0.00076 | 0.0039 | 0.0014 | 0.00098 | 0.0014 | -0.44 | 9 / 11 (82%) | 10 / 11 (91%) | 0.818 | 0.909 |
| 1.5.1.7 | Saccharopine dehydrogenase (NAD(+). L-lysine-forming). | 0.66 | 0.84 | 1 | 0.46 | 1 | 1 | 0.13 | 0.82 | 1 | 0.43 | 0.69 | 1 | 0.67 | 0.95 | 1 | 0.24 | 0.76 | 1 | 0.0092 | 0.0092 | 0.009 | 0.0083 | 0.0081 | 0.0017 | 0.0076 | 0.008 | 0.0029 | -0.13 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0091 | 0.0094 | 0.0028 | 0.011 | 0.0091 | 0.0043 | 0.27 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.3.5.8 | Transferred entry: 2.6.1.85. | 0.9 | 0.96 | 1 | 0.84 | 1 | 1 | 0.27 | 0.89 | 1 | 0.66 | 0.85 | 1 | 0.47 | 0.92 | 1 | 0.12 | 0.66 | 1 | 0.00057 | 0.00051 | 0.00038 | 0.00039 | 0.00036 | 0.00032 | 0.00045 | 0.00036 | 0.00041 | 0.21 | 32 / 36 (89%) | 7 / 7 (100%) | 5 / 7 (71%) | 1 | 0.714 | 0.00046 | 0.00039 | 5e-04 | 0.00067 | 6e-04 | 0.00046 | 0.54 | 9 / 11 (82%) | 11 / 11 (100%) | 0.818 | 1 |
| 6.3.4.2 | CTP synthase (glutamine hydrolyzing). | 0.11 | 0.5 | 1 | 0.32 | 1 | 1 | 0.95 | 1 | 1 | 0.15 | 0.47 | 1 | 0.55 | 0.93 | 1 | 0.14 | 0.68 | 1 | 0.027 | 0.027 | 0.027 | 0.026 | 0.024 | 0.0036 | 0.024 | 0.026 | 0.005 | -0.12 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.029 | 0.029 | 0.006 | 0.028 | 0.028 | 0.0031 | -0.051 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.6.1.13 | ADP-ribose diphosphatase. | 0.092 | 0.49 | 1 | 0.65 | 1 | 1 | 0.87 | 1 | 1 | 0.026 | 0.42 | 1 | 0.95 | 0.99 | 1 | 0.97 | 0.99 | 1 | 0.011 | 0.011 | 0.0091 | 0.0085 | 0.0079 | 0.004 | 0.0076 | 0.0072 | 0.0031 | -0.16 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.014 | 0.0098 | 0.012 | 0.012 | 0.012 | 0.0041 | -0.22 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.4.99.1 | Methylaspartate mutase. | 0.77 | 0.89 | 1 | 0.61 | 1 | 1 | 0.43 | 0.93 | 1 | 0.8 | 0.92 | 1 | 0.53 | 0.93 | 1 | 0.4 | 0.83 | 1 | 0.00031 | 0.00021 | 9.3e-05 | 0.00018 | 0.00012 | 0.00018 | 0.00011 | 0.00011 | 0.00011 | -0.71 | 24 / 36 (67%) | 6 / 7 (86%) | 5 / 7 (71%) | 0.857 | 0.714 | 0.00044 | 8.7e-05 | 0.0011 | 5.7e-05 | 0 | 9e-05 | -2.9 | 8 / 11 (73%) | 5 / 11 (45%) | 0.727 | 0.455 |
| 5.4.2.12 | Phosphoglycerate mutase (2.3-diphosphoglycerate-independent). | 0.67 | 0.85 | 1 | 0.55 | 1 | 1 | 0.99 | 1 | 1 | 0.46 | 0.71 | 1 | 0.58 | 0.93 | 1 | 0.27 | 0.77 | 1 | 0.023 | 0.023 | 0.022 | 0.023 | 0.022 | 0.0044 | 0.022 | 0.022 | 0.0044 | -0.064 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.024 | 0.023 | 0.0055 | 0.023 | 0.022 | 0.0028 | -0.061 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.1.88 | Biotin-independent malonate decarboxylase. | 1 | 1 | 1 | 0.27 | 1 | 1 | 0.97 | 1 | 1 | 0.82 | 0.93 | 1 | 0.2 | 0.87 | 1 | 0.9 | 0.98 | 1 | 0.00017 | 1.5e-05 | 0 | 0 | 0 | 0 | 3e-05 | 0 | 8.1e-05 | Inf | 3 / 36 (8.3%) | 0 / 7 (0%) | 1 / 7 (14%) | 0 | 0.143 | 0 | 0 | 0 | 2.8e-05 | 0 | 7.3e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 2.7.11.32 | [Pyruvate. phosphate dikinase] kinase. | 0.68 | 0.85 | 1 | 0.87 | 1 | 1 | 0.98 | 1 | 1 | 0.98 | 0.99 | 1 | 0.76 | 0.97 | 1 | 1 | 1 | 1 | 0.00046 | 0.00043 | 0.00036 | 0.00033 | 0.00036 | 2e-04 | 0.00035 | 0.00035 | 0.00019 | 0.085 | 34 / 36 (94%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00051 | 0.00022 | 0.00063 | 0.00047 | 0.00045 | 0.00033 | -0.12 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 4.2.1.99 | 2-methylisocitrate dehydratase. | 0.25 | 0.56 | 1 | 0.21 | 1 | 1 | 0.061 | 0.71 | 1 | 0.7 | 0.88 | 1 | 0.67 | 0.95 | 1 | 0.24 | 0.76 | 1 | 0.00051 | 0.00027 | 4.1e-05 | 0.00062 | 0 | 0.0014 | 0.00047 | 0 | 0.0011 | -0.4 | 19 / 36 (53%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 7.1e-05 | 2.8e-05 | 8.9e-05 | 0.00012 | 0.00011 | 0.00013 | 0.76 | 6 / 11 (55%) | 7 / 11 (64%) | 0.545 | 0.636 |
| 2.7.9.- | 0.82 | 0.92 | 1 | 0.51 | 1 | 1 | 0.96 | 1 | 1 | 0.96 | 0.99 | 1 | 0.83 | 0.98 | 1 | 0.75 | 0.95 | 1 | 0.0059 | 0.0059 | 0.0053 | 0.0064 | 0.0054 | 0.0032 | 0.0058 | 0.0061 | 0.0026 | -0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0062 | 0.0062 | 0.0036 | 0.0054 | 0.0051 | 0.003 | -0.2 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 4.1.1.18 | Lysine decarboxylase. | 0.46 | 0.71 | 1 | 0.44 | 1 | 1 | 0.86 | 1 | 1 | 0.27 | 0.56 | 1 | 0.27 | 0.88 | 1 | 0.93 | 0.98 | 1 | 0.0054 | 0.0054 | 0.0055 | 0.0052 | 0.0044 | 0.0033 | 0.0049 | 0.0057 | 0.003 | -0.086 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0059 | 0.0062 | 0.0025 | 0.0053 | 0.0052 | 0.0024 | -0.15 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.20 | Alpha-glucosidase. | 0.45 | 0.69 | 1 | 0.57 | 1 | 1 | 0.18 | 0.85 | 1 | 0.2 | 0.5 | 1 | 0.83 | 0.98 | 1 | 0.35 | 0.81 | 1 | 0.015 | 0.015 | 0.014 | 0.013 | 0.013 | 0.0048 | 0.012 | 0.014 | 0.0056 | -0.12 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.015 | 0.017 | 0.005 | 0.019 | 0.014 | 0.0094 | 0.34 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.4.1.1 | Alanine dehydrogenase. | 0.26 | 0.56 | 1 | 1 | 1 | 1 | 0.82 | 1 | 1 | 0.26 | 0.56 | 1 | 1 | 1 | 1 | 0.82 | 0.96 | 1 | 0.0016 | 0.0016 | 0.0012 | 0.0018 | 0.0016 | 0.00061 | 0.0019 | 0.0016 | 0.0014 | 0.078 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0013 | 0.0011 | 8e-04 | 0.0016 | 0.00085 | 0.0018 | 0.3 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.3.3.6 | Pyridoxal 5'-phosphate synthase (glutamine hydrolyzing). | 0.074 | 0.49 | 1 | 0.74 | 1 | 1 | 0.82 | 1 | 1 | 0.062 | 0.43 | 1 | 0.61 | 0.95 | 1 | 0.73 | 0.94 | 1 | 0.012 | 0.012 | 0.011 | 0.0088 | 0.0077 | 0.0036 | 0.0098 | 0.01 | 0.0051 | 0.16 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.013 | 0.012 | 0.007 | 0.013 | 0.011 | 0.0034 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.1.130 | Precorrin-2 C(20)-methyltransferase. | 0.74 | 0.88 | 1 | 0.3 | 1 | 1 | 0.78 | 1 | 1 | 0.86 | 0.95 | 1 | 0.62 | 0.95 | 1 | 0.72 | 0.94 | 1 | 0.0051 | 0.0051 | 0.0049 | 0.0055 | 0.0053 | 0.0019 | 0.0049 | 0.005 | 0.0021 | -0.17 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0052 | 0.0048 | 0.0017 | 0.0048 | 0.0047 | 0.0015 | -0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.1.49 | Phosphoenolpyruvate carboxykinase (ATP). | 0.4 | 0.66 | 1 | 0.36 | 1 | 1 | 0.055 | 0.71 | 1 | 0.99 | 1 | 1 | 0.93 | 0.99 | 1 | 0.23 | 0.75 | 1 | 0.022 | 0.022 | 0.022 | 0.023 | 0.021 | 0.0042 | 0.021 | 0.022 | 0.006 | -0.13 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.021 | 0.022 | 0.0046 | 0.024 | 0.022 | 0.0044 | 0.19 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.133 | Glucan 1.4-alpha-maltohydrolase. | 0.17 | 0.51 | 1 | 0.81 | 1 | 1 | 0.6 | 0.96 | 1 | 0.049 | 0.43 | 1 | 0.55 | 0.93 | 1 | 0.13 | 0.68 | 1 | 0.00039 | 6.6e-05 | 0 | 0 | 0 | 0 | 3.9e-06 | 0 | 1e-05 | Inf | 6 / 36 (17%) | 0 / 7 (0%) | 1 / 7 (14%) | 0 | 0.143 | 6.8e-05 | 0 | 9.7e-05 | 0.00014 | 0 | 0.00048 | 1 | 4 / 11 (36%) | 1 / 11 (9.1%) | 0.364 | 0.0909 |
| 2.1.1.183 | 18S rRNA (adenine(1779)-N(6)/adenine(1780)-N(6))-dimethyltransferase. | 0.61 | 0.81 | 1 | 0.7 | 1 | 1 | 0.83 | 1 | 1 | 0.93 | 0.98 | 1 | 0.54 | 0.93 | 1 | 0.68 | 0.93 | 1 | 0.00059 | 0.00051 | 0.00035 | 0.00058 | 0.00032 | 7e-04 | 0.00061 | 6e-04 | 0.00048 | 0.073 | 31 / 36 (86%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00036 | 0.00024 | 0.00034 | 0.00054 | 0.00037 | 0.00045 | 0.58 | 10 / 11 (91%) | 9 / 11 (82%) | 0.909 | 0.818 |
| 3.5.4.32 | 8-oxoguanine deaminase. | 0.74 | 0.88 | 1 | 0.66 | 1 | 1 | 0.94 | 1 | 1 | 0.76 | 0.91 | 1 | 0.92 | 0.99 | 1 | 0.86 | 0.96 | 1 | 0.00018 | 0.00011 | 6.4e-05 | 6.3e-05 | 5.7e-05 | 5.4e-05 | 0.00016 | 0 | 0.00025 | 1.3 | 23 / 36 (64%) | 5 / 7 (71%) | 3 / 7 (43%) | 0.714 | 0.429 | 9.7e-05 | 7.2e-05 | 0.00012 | 0.00013 | 0.00015 | 0.00014 | 0.42 | 8 / 11 (73%) | 7 / 11 (64%) | 0.727 | 0.636 |
| 4.2.2.6 | Oligogalacturonide lyase. | 0.89 | 0.95 | 1 | 0.4 | 1 | 1 | 0.036 | 0.71 | 1 | 0.69 | 0.87 | 1 | 0.39 | 0.9 | 1 | 0.088 | 0.66 | 1 | 0.00063 | 0.00035 | 6.5e-05 | 0.00021 | 0 | 0.00034 | 0.00012 | 0 | 0.00026 | -0.81 | 20 / 36 (56%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 0.00025 | 8.7e-05 | 0.00055 | 0.00069 | 2e-04 | 0.0011 | 1.5 | 7 / 11 (64%) | 8 / 11 (73%) | 0.636 | 0.727 |
| 3.1.31.1 | Micrococcal nuclease. | 0.86 | 0.94 | 1 | 0.0031 | 0.64 | 1 | 0.11 | 0.82 | 1 | 0.96 | 0.99 | 1 | 0.0016 | 0.62 | 1 | 0.068 | 0.66 | 1 | 0.00012 | 3.4e-05 | 0 | 2e-05 | 0 | 5.2e-05 | 7.1e-05 | 5.4e-05 | 8.3e-05 | 1.8 | 10 / 36 (28%) | 1 / 7 (14%) | 5 / 7 (71%) | 0.143 | 0.714 | 1.8e-05 | 0 | 5.9e-05 | 3.6e-05 | 0 | 6.5e-05 | 1 | 1 / 11 (9.1%) | 3 / 11 (27%) | 0.0909 | 0.273 |
| 2.1.1.176 | 16S rRNA (cytosine(967)-C(5))-methyltransferase. | 0.46 | 0.7 | 1 | 0.52 | 1 | 1 | 0.97 | 1 | 1 | 0.36 | 0.63 | 1 | 0.73 | 0.96 | 1 | 0.3 | 0.78 | 1 | 0.015 | 0.015 | 0.014 | 0.014 | 0.012 | 0.0037 | 0.013 | 0.013 | 0.004 | -0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.015 | 0.015 | 0.0036 | 0.015 | 0.014 | 0.0041 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.5 | Transferred entry: 1.1.1.303 and 1.1.1.304. | 0.24 | 0.55 | 1 | 0.029 | 0.97 | 1 | 0.022 | 0.71 | 1 | 0.27 | 0.56 | 1 | 0.0084 | 0.71 | 1 | 0.013 | 0.55 | 1 | 0.00016 | 4e-05 | 0 | 2.3e-06 | 0 | 6.1e-06 | 6.1e-05 | 8.1e-05 | 5.8e-05 | 4.7 | 9 / 36 (25%) | 1 / 7 (14%) | 4 / 7 (57%) | 0.143 | 0.571 | 7.3e-05 | 0 | 0.00017 | 1.7e-05 | 0 | 3.8e-05 | -2.1 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 3.4.13.9 | Xaa-Pro dipeptidase. | 0.62 | 0.82 | 1 | 0.61 | 1 | 1 | 0.36 | 0.9 | 1 | 0.34 | 0.61 | 1 | 0.17 | 0.87 | 1 | 0.1 | 0.66 | 1 | 0.01 | 0.01 | 0.0096 | 0.0098 | 0.012 | 0.0041 | 0.011 | 0.0091 | 0.0047 | 0.17 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.0081 | 0.0072 | 0.0096 | 0.0094 | 0.0027 | -0.2 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.58 | Glucan 1.3-beta-glucosidase. | 0.28 | 0.57 | 1 | 0.88 | 1 | 1 | 0.79 | 1 | 1 | 0.33 | 0.6 | 1 | 0.66 | 0.95 | 1 | 0.81 | 0.96 | 1 | 0.0029 | 0.0023 | 0.00096 | 0.0012 | 0.00073 | 0.0013 | 0.0014 | 0.00075 | 0.0017 | 0.22 | 29 / 36 (81%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.0034 | 0.0012 | 0.0062 | 0.0025 | 0.002 | 0.0025 | -0.44 | 9 / 11 (82%) | 10 / 11 (91%) | 0.818 | 0.909 |
| 2.7.7.61 | Citrate lyase holo-[acyl-carrier protein] synthase. | 0.71 | 0.86 | 1 | 0.71 | 1 | 1 | 0.58 | 0.96 | 1 | 0.6 | 0.81 | 1 | 0.8 | 0.98 | 1 | 0.92 | 0.98 | 1 | 0.00072 | 0.00066 | 0.00062 | 0.00064 | 0.00051 | 0.00041 | 0.00081 | 0.00095 | 0.00051 | 0.34 | 33 / 36 (92%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00063 | 0.00048 | 0.00055 | 6e-04 | 0.00023 | 0.00063 | -0.07 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 2.7.8.33 | UDP-N-acetylglucosamine--undecaprenyl-phosphate | 0.26 | 0.56 | 1 | 0.19 | 1 | 1 | 0.13 | 0.82 | 1 | 0.22 | 0.52 | 1 | 0.14 | 0.87 | 1 | 0.092 | 0.66 | 1 | 0.0049 | 0.0049 | 0.0036 | 0.0032 | 0.0031 | 0.0014 | 0.0052 | 0.0037 | 0.0051 | 0.7 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.006 | 0.0034 | 0.0072 | 0.0046 | 0.0039 | 0.0028 | -0.38 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.3.- | 0.43 | 0.69 | 1 | 0.18 | 1 | 1 | 0.13 | 0.82 | 1 | 0.36 | 0.63 | 1 | 0.11 | 0.87 | 1 | 0.082 | 0.66 | 1 | 8.9e-05 | 1.7e-05 | 0 | 4.1e-06 | 0 | 1.1e-05 | 2.9e-05 | 0 | 4.3e-05 | 2.8 | 7 / 36 (19%) | 1 / 7 (14%) | 3 / 7 (43%) | 0.143 | 0.429 | 2.6e-05 | 0 | 5.8e-05 | 9.9e-06 | 0 | 3.3e-05 | -1.4 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 | |
| 1.11.1.5 | Cytochrome-c peroxidase. | 0.42 | 0.68 | 1 | 0.17 | 1 | 1 | 0.098 | 0.81 | 1 | 0.28 | 0.57 | 1 | 0.16 | 0.87 | 1 | 0.046 | 0.65 | 1 | 0.00051 | 3e-04 | 6.2e-05 | 4e-04 | 0.00011 | 0.00069 | 0.00035 | 0 | 0.00086 | -0.19 | 21 / 36 (58%) | 5 / 7 (71%) | 3 / 7 (43%) | 0.714 | 0.429 | 0.00026 | 0 | 0.00057 | 0.00023 | 0.00011 | 0.00031 | -0.18 | 5 / 11 (45%) | 8 / 11 (73%) | 0.455 | 0.727 |
| 2.1.1.34 | tRNA (guanosine(18)-2'-O)-methyltransferase. | 0.25 | 0.56 | 1 | 0.79 | 1 | 1 | 0.96 | 1 | 1 | 0.18 | 0.5 | 1 | 0.73 | 0.96 | 1 | 1 | 1 | 1 | 0.0037 | 0.0037 | 0.0024 | 0.0023 | 0.0024 | 0.0015 | 0.0022 | 0.0021 | 0.0016 | -0.064 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0052 | 0.0022 | 0.0082 | 0.0041 | 0.0028 | 0.0037 | -0.34 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.3.7.8 | Benzoyl-CoA reductase. | 0.062 | 0.49 | 1 | 0.59 | 1 | 1 | 0.42 | 0.93 | 1 | 0.035 | 0.43 | 1 | 0.32 | 0.9 | 1 | 0.31 | 0.79 | 1 | 0.014 | 0.014 | 0.012 | 0.0085 | 0.0059 | 0.0044 | 0.011 | 0.01 | 0.007 | 0.37 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.019 | 0.011 | 0.022 | 0.015 | 0.013 | 0.0067 | -0.34 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.-.-.- | 0.38 | 0.65 | 1 | 0.77 | 1 | 1 | 0.55 | 0.95 | 1 | 0.18 | 0.5 | 1 | 0.33 | 0.9 | 1 | 0.89 | 0.97 | 1 | 0.011 | 0.011 | 0.0096 | 0.0094 | 0.0089 | 0.0028 | 0.0099 | 0.0088 | 0.004 | 0.075 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.0096 | 0.0031 | 0.012 | 0.011 | 0.0042 | 0.13 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 3.5.3.1 | Arginase. | 0.72 | 0.87 | 1 | 0.46 | 1 | 1 | 0.13 | 0.82 | 1 | 0.42 | 0.69 | 1 | 0.29 | 0.89 | 1 | 0.065 | 0.66 | 1 | 0.00056 | 0.00047 | 0.00016 | 0.00032 | 0.00024 | 0.00022 | 0.00025 | 0.00012 | 0.00035 | -0.36 | 30 / 36 (83%) | 6 / 7 (86%) | 5 / 7 (71%) | 0.857 | 0.714 | 0.00037 | 9.4e-05 | 0.00062 | 8e-04 | 0.00015 | 0.0015 | 1.1 | 8 / 11 (73%) | 11 / 11 (100%) | 0.727 | 1 |
| 2.1.1.206 | tRNA (cytidine(56)-2'-O)-methyltransferase. | 0.67 | 0.85 | 1 | 0.32 | 1 | 1 | 0.43 | 0.93 | 1 | 0.88 | 0.96 | 1 | 0.47 | 0.92 | 1 | 0.6 | 0.91 | 1 | 0.00011 | 9.2e-06 | 0 | 1.6e-05 | 0 | 4.3e-05 | 0 | 0 | 0 | -Inf | 3 / 36 (8.3%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 9.5e-06 | 0 | 3.2e-05 | 1e-05 | 0 | 3.4e-05 | 0.074 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.7.1.69 | Transferred entry: 2.7.1.191. 2.7.1.192. 2.7.1.193. 2.7.1.194. 2.7.1.195. | 0.55 | 0.77 | 1 | 0.28 | 1 | 1 | 0.31 | 0.89 | 1 | 0.23 | 0.53 | 1 | 0.19 | 0.87 | 1 | 0.22 | 0.74 | 1 | 0.047 | 0.047 | 0.048 | 0.044 | 0.049 | 0.02 | 0.052 | 0.054 | 0.026 | 0.24 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.048 | 0.047 | 0.016 | 0.046 | 0.046 | 0.0096 | -0.061 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.2.48 | tRNA-guanine(15) transglycosylase. | 0.46 | 0.71 | 1 | 0.48 | 1 | 1 | 0.25 | 0.89 | 1 | 0.71 | 0.89 | 1 | 0.97 | 0.99 | 1 | 0.51 | 0.89 | 1 | 0.00029 | 4e-05 | 0 | 8.1e-05 | 0 | 0.00021 | 4e-05 | 0 | 0.00011 | -1 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 1.9e-05 | 0 | 6.3e-05 | 3.4e-05 | 0 | 7.7e-05 | 0.84 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 4.2.1.103 | Cyclohexyl-isocyanide hydratase. | 0.07 | 0.49 | 1 | 1 | 1 | 1 | 0.4 | 0.92 | 1 | 0.056 | 0.43 | 1 | 0.83 | 0.98 | 1 | 0.36 | 0.82 | 1 | 0.00013 | 1.8e-05 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 5 / 36 (14%) | 0 / 7 (0%) | 0 / 7 (0%) | 0 | 0 | 4.8e-05 | 0 | 9.8e-05 | 1.2e-05 | 0 | 2.7e-05 | -2 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 4.1.3.16 | 4-hydroxy-2-oxoglutarate aldolase. | 0.0087 | 0.49 | 1 | 0.39 | 1 | 1 | 0.04 | 0.71 | 1 | 0.042 | 0.43 | 1 | 0.68 | 0.95 | 1 | 0.15 | 0.71 | 1 | 0.0039 | 0.0039 | 0.0036 | 0.0048 | 0.0044 | 0.0018 | 0.004 | 0.0044 | 0.0017 | -0.26 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0027 | 0.0029 | 0.0011 | 0.0043 | 0.0048 | 0.0019 | 0.67 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.23.49 | Omptin. | 0.045 | 0.49 | 1 | 0.048 | 0.97 | 1 | 0.038 | 0.71 | 1 | 0.02 | 0.42 | 1 | 0.13 | 0.87 | 1 | 0.093 | 0.66 | 1 | 0.00039 | 5.4e-05 | 0 | 0.00016 | 0 | 0.00035 | 0.00011 | 0 | 0.00028 | -0.54 | 5 / 36 (14%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 0 | 0 | 0 | 1e-05 | 0 | 3.4e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 5.3.1.24 | Phosphoribosylanthranilate isomerase. | 0.9 | 0.96 | 1 | 0.55 | 1 | 1 | 0.91 | 1 | 1 | 0.62 | 0.83 | 1 | 0.19 | 0.87 | 1 | 0.38 | 0.82 | 1 | 0.011 | 0.011 | 0.011 | 0.011 | 0.011 | 0.003 | 0.012 | 0.011 | 0.0043 | 0.13 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.01 | 0.0025 | 0.011 | 0.011 | 0.0023 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.3.99.n1 | Transferred entry: 5.3.99.11. | 0.53 | 0.75 | 1 | 0.97 | 1 | 1 | 0.52 | 0.95 | 1 | 0.58 | 0.8 | 1 | 0.97 | 0.99 | 1 | 0.5 | 0.88 | 1 | 0.00012 | 2.4e-05 | 0 | 1.8e-05 | 0 | 3.3e-05 | 4e-05 | 0 | 0.00011 | 1.2 | 7 / 36 (19%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 9.5e-06 | 0 | 3.2e-05 | 3.3e-05 | 0 | 5.7e-05 | 1.8 | 1 / 11 (9.1%) | 3 / 11 (27%) | 0.0909 | 0.273 |
| 3.5.1.108 | UDP-3-O-acyl-N-acetylglucosamine deacetylase. | 0.13 | 0.5 | 1 | 0.14 | 1 | 1 | 0.0087 | 0.61 | 1 | 0.034 | 0.43 | 1 | 0.15 | 0.87 | 1 | 0.0058 | 0.48 | 1 | 0.0016 | 0.0015 | 0.0012 | 0.0017 | 0.0018 | 0.00067 | 0.0012 | 0.001 | 0.0011 | -0.5 | 34 / 36 (94%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0011 | 0.00032 | 0.0016 | 0.0021 | 0.001 | 0.0023 | 0.93 | 9 / 11 (82%) | 11 / 11 (100%) | 0.818 | 1 |
| 2.7.1.51 | L-fuculokinase. | 0.24 | 0.55 | 1 | 0.8 | 1 | 1 | 0.55 | 0.95 | 1 | 0.22 | 0.52 | 1 | 0.74 | 0.96 | 1 | 0.53 | 0.9 | 1 | 0.0048 | 0.0048 | 0.0043 | 0.0043 | 0.0042 | 0.00091 | 0.0044 | 0.0039 | 0.0026 | 0.033 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0056 | 0.0056 | 0.0023 | 0.0047 | 0.0044 | 0.002 | -0.25 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.83 | Kappa-carrageenase. | 0.68 | 0.85 | 1 | 0.85 | 1 | 1 | 0.82 | 1 | 1 | 0.82 | 0.93 | 1 | 0.57 | 0.93 | 1 | 0.79 | 0.95 | 1 | 0.0099 | 0.0099 | 0.0095 | 0.011 | 0.0075 | 0.007 | 0.011 | 0.012 | 0.0058 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.009 | 0.0094 | 0.0031 | 0.0098 | 0.0098 | 0.0033 | 0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.341 | CDP-abequose synthase. | 0.51 | 0.74 | 1 | 0.91 | 1 | 1 | 0.68 | 0.98 | 1 | 0.34 | 0.61 | 1 | 0.98 | 0.99 | 1 | 0.93 | 0.98 | 1 | 0.00024 | 5.3e-05 | 0 | 9.2e-06 | 0 | 2.4e-05 | 3.9e-06 | 0 | 1e-05 | -1.2 | 8 / 36 (22%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 4.1e-05 | 0 | 8.2e-05 | 0.00012 | 0 | 0.00037 | 1.5 | 3 / 11 (27%) | 3 / 11 (27%) | 0.273 | 0.273 |
| 2.1.1.266 | 23S rRNA (adenine(2030)-N(6))-methyltransferase. | 0.14 | 0.5 | 1 | 0.37 | 1 | 1 | 0.11 | 0.82 | 1 | 0.18 | 0.49 | 1 | 0.53 | 0.93 | 1 | 0.19 | 0.72 | 1 | 0.00017 | 5e-05 | 0 | 1e-04 | 0 | 0.00022 | 7.2e-05 | 0 | 0.00018 | -0.47 | 11 / 36 (31%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 1.1e-05 | 0 | 3e-05 | 4.2e-05 | 0 | 6.1e-05 | 1.9 | 2 / 11 (18%) | 4 / 11 (36%) | 0.182 | 0.364 |
| 4.1.3.32 | 2.3-dimethylmalate lyase. | 0.93 | 0.97 | 1 | 0.36 | 1 | 1 | 0.85 | 1 | 1 | 0.96 | 0.99 | 1 | 0.45 | 0.92 | 1 | 1 | 1 | 1 | 0.00021 | 6.9e-05 | 0 | 6.7e-05 | 0 | 8.8e-05 | 3.3e-05 | 0 | 5.7e-05 | -1 | 12 / 36 (33%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 9.4e-05 | 0 | 0.00016 | 6.8e-05 | 0 | 0.00015 | -0.47 | 4 / 11 (36%) | 3 / 11 (27%) | 0.364 | 0.273 |
| 1.12.2.1 | Cytochrome-c3 hydrogenase. | 0.3 | 0.58 | 1 | 0.47 | 1 | 1 | 0.72 | 0.98 | 1 | 0.64 | 0.85 | 1 | 0.18 | 0.87 | 1 | 0.91 | 0.98 | 1 | 0.00027 | 8.4e-05 | 0 | 2e-04 | 0 | 0.00053 | 0.00016 | 0 | 0.00035 | -0.32 | 11 / 36 (31%) | 1 / 7 (14%) | 3 / 7 (43%) | 0.143 | 0.429 | 1.1e-05 | 0 | 2.7e-05 | 3.5e-05 | 0 | 4.6e-05 | 1.7 | 2 / 11 (18%) | 5 / 11 (45%) | 0.182 | 0.455 |
| 2.1.-.- | 0.6 | 0.8 | 1 | 0.17 | 1 | 1 | 0.15 | 0.84 | 1 | 0.96 | 0.99 | 1 | 0.061 | 0.86 | 1 | 0.11 | 0.66 | 1 | 4e-04 | 0.00026 | 8.5e-05 | 0.00045 | 1.6e-05 | 0.0011 | 0.00046 | 0.00025 | 7e-04 | 0.032 | 23 / 36 (64%) | 4 / 7 (57%) | 6 / 7 (86%) | 0.571 | 0.857 | 0.00018 | 7.2e-05 | 0.00034 | 8.1e-05 | 7.5e-05 | 9e-05 | -1.2 | 6 / 11 (55%) | 7 / 11 (64%) | 0.545 | 0.636 | |
| 3.4.22.70 | Sortase A. | 0.8 | 0.91 | 1 | 0.33 | 1 | 1 | 0.53 | 0.95 | 1 | 0.85 | 0.95 | 1 | 0.34 | 0.9 | 1 | 0.6 | 0.91 | 1 | 0.00015 | 6.4e-05 | 0 | 3e-05 | 0 | 4.7e-05 | 8.3e-05 | 0 | 0.00011 | 1.5 | 15 / 36 (42%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 2.9e-05 | 0 | 6.2e-05 | 0.00011 | 7.1e-05 | 0.00015 | 1.9 | 3 / 11 (27%) | 6 / 11 (55%) | 0.273 | 0.545 |
| 2.7.1.105 | 6-phosphofructo-2-kinase. | 0.34 | 0.61 | 1 | 0.75 | 1 | 1 | 0.95 | 1 | 1 | 0.38 | 0.65 | 1 | 1 | 1 | 1 | 0.93 | 0.98 | 1 | 0.0016 | 0.0013 | 0.00071 | 0.00063 | 0.00031 | 0.00077 | 0.00082 | 0.00072 | 0.001 | 0.38 | 30 / 36 (83%) | 6 / 7 (86%) | 5 / 7 (71%) | 0.857 | 0.714 | 0.0019 | 0.00061 | 0.0042 | 0.0016 | 0.0011 | 0.0018 | -0.25 | 10 / 11 (91%) | 9 / 11 (82%) | 0.909 | 0.818 |
| 1.8.1.- | 0.74 | 0.88 | 1 | 0.13 | 1 | 1 | 0.9 | 1 | 1 | 0.82 | 0.93 | 1 | 0.15 | 0.87 | 1 | 0.69 | 0.94 | 1 | 0.0032 | 0.0032 | 0.003 | 0.0027 | 0.0025 | 0.00069 | 0.0038 | 0.0037 | 0.00098 | 0.49 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0027 | 0.0024 | 0.0012 | 0.0037 | 0.0034 | 0.0022 | 0.45 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 5.3.1.28 | D-sedoheptulose 7-phosphate isomerase. | 0.9 | 0.96 | 1 | 0.18 | 1 | 1 | 0.25 | 0.89 | 1 | 0.52 | 0.76 | 1 | 0.07 | 0.86 | 1 | 0.16 | 0.71 | 1 | 0.00064 | 0.00062 | 0.00037 | 0.00068 | 0.00041 | 0.00092 | 0.00097 | 0.00034 | 0.001 | 0.51 | 35 / 36 (97%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.00052 | 0.00035 | 0.00047 | 0.00046 | 0.00038 | 0.00033 | -0.18 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.1.227 | 16S rRNA (cytidine(1409)-2'-O)-methyltransferase. | 0.075 | 0.49 | 1 | 0.36 | 1 | 1 | 0.32 | 0.89 | 1 | 0.076 | 0.44 | 1 | 0.17 | 0.87 | 1 | 0.35 | 0.81 | 1 | 0.0015 | 0.0011 | 0.00061 | 0.00036 | 4e-04 | 0.00036 | 0.00068 | 0.00032 | 0.00078 | 0.92 | 28 / 36 (78%) | 4 / 7 (57%) | 5 / 7 (71%) | 0.571 | 0.714 | 0.0018 | 0.00062 | 0.0032 | 0.0012 | 0.00071 | 0.0012 | -0.58 | 9 / 11 (82%) | 10 / 11 (91%) | 0.818 | 0.909 |
| 5.4.99.26 | tRNA pseudouridine(65) synthase. | 0.6 | 0.8 | 1 | 0.39 | 1 | 1 | 0.88 | 1 | 1 | 0.69 | 0.87 | 1 | 0.35 | 0.9 | 1 | 0.97 | 0.99 | 1 | 0.0013 | 0.0013 | 0.0011 | 0.0012 | 0.0011 | 0.00076 | 0.0016 | 0.0012 | 0.0011 | 0.42 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00099 | 0.00077 | 0.00069 | 0.0015 | 0.0011 | 0.0011 | 0.6 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.6 | Glycerol dehydrogenase. | 0.29 | 0.57 | 1 | 0.43 | 1 | 1 | 0.38 | 0.9 | 1 | 0.21 | 0.52 | 1 | 0.44 | 0.91 | 1 | 0.33 | 0.8 | 1 | 0.0091 | 0.0091 | 0.0094 | 0.0079 | 0.0066 | 0.0039 | 0.0095 | 0.01 | 0.006 | 0.27 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0096 | 0.0094 | 0.0027 | 0.0092 | 0.01 | 0.0029 | -0.061 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.298 | 3-hydroxypropionate dehydrogenase (NADP(+)). | 0.24 | 0.55 | 1 | 0.8 | 1 | 1 | 0.31 | 0.89 | 1 | 0.34 | 0.61 | 1 | 0.4 | 0.9 | 1 | 0.94 | 0.98 | 1 | 0.00073 | 8.1e-05 | 0 | 2e-04 | 0 | 0.00053 | 0.00019 | 0 | 0.00051 | -0.074 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3e-06 | 0 | 9.9e-06 | 1.1e-05 | 0 | 3.6e-05 | 1.9 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.7.1.12 | Cobalt-precorrin 5A hydrolase. | 0.049 | 0.49 | 1 | 0.069 | 1 | 1 | 0.37 | 0.9 | 1 | 0.035 | 0.43 | 1 | 0.049 | 0.81 | 1 | 0.33 | 0.8 | 1 | 0.00024 | 5.3e-05 | 0 | 1e-04 | 0 | 0.00021 | 0.00016 | 3.1e-05 | 0.00024 | 0.68 | 8 / 36 (22%) | 3 / 7 (43%) | 4 / 7 (57%) | 0.429 | 0.571 | 0 | 0 | 0 | 1e-05 | 0 | 3.4e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.1.1.86 | Tetrahydromethanopterin S-methyltransferase. | 0.88 | 0.95 | 1 | 0.1 | 1 | 1 | 0.29 | 0.89 | 1 | 0.85 | 0.95 | 1 | 0.45 | 0.92 | 1 | 0.86 | 0.96 | 1 | 0.00085 | 0.00014 | 0 | 0.00024 | 0 | 0.00064 | 6.2e-05 | 0 | 0.00016 | -2 | 6 / 36 (17%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0.00015 | 0 | 0.00036 | 0.00012 | 0 | 0.00031 | -0.32 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 4.1.2.2 | Ketotetrose-phosphate aldolase. | 0.064 | 0.49 | 1 | 0.41 | 1 | 1 | 0.15 | 0.84 | 1 | 0.072 | 0.43 | 1 | 0.82 | 0.98 | 1 | 0.22 | 0.74 | 1 | 4e-04 | 9.9e-05 | 0 | 0.00029 | 0 | 7e-04 | 0.00018 | 0 | 0.00034 | -0.69 | 9 / 36 (25%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 3e-06 | 0 | 9.9e-06 | 2.2e-05 | 0 | 4e-05 | 2.9 | 1 / 11 (9.1%) | 3 / 11 (27%) | 0.0909 | 0.273 |
| 4.2.1.85 | Dimethylmaleate hydratase. | 0.26 | 0.56 | 1 | 0.49 | 1 | 1 | 0.29 | 0.89 | 1 | 0.25 | 0.54 | 1 | 0.14 | 0.87 | 1 | 0.049 | 0.65 | 1 | 0.025 | 0.025 | 0.026 | 0.022 | 0.018 | 0.0084 | 0.025 | 0.03 | 0.012 | 0.18 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.027 | 0.03 | 0.0075 | 0.025 | 0.026 | 0.0068 | -0.11 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.7.18 | Nicotinate-nucleotide adenylyltransferase. | 0.02 | 0.49 | 1 | 0.34 | 1 | 1 | 0.95 | 1 | 1 | 0.02 | 0.42 | 1 | 0.79 | 0.97 | 1 | 0.42 | 0.85 | 1 | 0.011 | 0.011 | 0.01 | 0.0096 | 0.0097 | 0.00072 | 0.0088 | 0.0093 | 0.002 | -0.13 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.012 | 0.012 | 0.0033 | 0.011 | 0.011 | 0.0014 | -0.13 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.1.68 | L-fuconate dehydratase. | 0.68 | 0.85 | 1 | 0.52 | 1 | 1 | 0.15 | 0.84 | 1 | 0.58 | 0.8 | 1 | 0.38 | 0.9 | 1 | 0.24 | 0.76 | 1 | 0.00042 | 9.3e-05 | 0 | 1.6e-05 | 0 | 3.1e-05 | 0 | 0 | 0 | -Inf | 8 / 36 (22%) | 2 / 7 (29%) | 0 / 7 (0%) | 0.286 | 0 | 5.5e-05 | 0 | 0.00011 | 0.00024 | 0 | 0.00052 | 2.1 | 3 / 11 (27%) | 3 / 11 (27%) | 0.273 | 0.273 |
| 1.17.7.1 | (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase (ferredoxin). | 0.58 | 0.79 | 1 | 0.59 | 1 | 1 | 0.93 | 1 | 1 | 0.36 | 0.63 | 1 | 0.97 | 0.99 | 1 | 0.99 | 1 | 1 | 0.00057 | 0.00044 | 0.00023 | 0.00029 | 0.00022 | 0.00031 | 0.00049 | 0.00024 | 0.00066 | 0.76 | 28 / 36 (78%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 0.00044 | 0.00024 | 0.00075 | 0.00051 | 0.00023 | 0.00051 | 0.21 | 10 / 11 (91%) | 9 / 11 (82%) | 0.909 | 0.818 |
| 2.4.1.180 | Lipopolysaccharide N-acetylmannosaminouronosyltransferase. | 0.49 | 0.73 | 1 | 0.44 | 1 | 1 | 0.26 | 0.89 | 1 | 0.28 | 0.57 | 1 | 0.74 | 0.96 | 1 | 0.4 | 0.83 | 1 | 0.00021 | 8.1e-05 | 0 | 6.6e-05 | 8.1e-05 | 5.5e-05 | 0.00021 | 6.2e-05 | 0.00035 | 1.7 | 14 / 36 (39%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 6.9e-05 | 0 | 0.00012 | 2.3e-05 | 0 | 6.6e-05 | -1.6 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 2.5.1.79 | Thermospermine synthase. | 0.11 | 0.5 | 1 | 0.068 | 1 | 1 | 0.14 | 0.83 | 1 | 0.14 | 0.47 | 1 | 0.38 | 0.9 | 1 | 0.55 | 0.9 | 1 | 0.00031 | 4.3e-05 | 0 | 0.00015 | 0 | 4e-04 | 5.8e-05 | 0 | 0.00015 | -1.4 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 2.7e-06 | 0 | 8.9e-06 | 0.054 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.4.21.50 | Lysyl endopeptidase. | 0.14 | 0.5 | 1 | 0.51 | 1 | 1 | 0.79 | 1 | 1 | 0.11 | 0.47 | 1 | 0.61 | 0.95 | 1 | 0.77 | 0.95 | 1 | 0.00025 | 5.6e-05 | 0 | 9.2e-06 | 0 | 2.4e-05 | 0 | 0 | 0 | -Inf | 8 / 36 (22%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 0.00011 | 0 | 0.00021 | 7.3e-05 | 0 | 0.00018 | -0.59 | 4 / 11 (36%) | 3 / 11 (27%) | 0.364 | 0.273 |
| 1.7.1.7 | GMP reductase. | 0.9 | 0.96 | 1 | 0.89 | 1 | 1 | 0.38 | 0.91 | 1 | 0.71 | 0.89 | 1 | 0.55 | 0.93 | 1 | 0.25 | 0.76 | 1 | 0.00054 | 0.00049 | 0.00033 | 0.00049 | 0.00016 | 0.00088 | 5e-04 | 0.00021 | 0.00068 | 0.029 | 33 / 36 (92%) | 7 / 7 (100%) | 5 / 7 (71%) | 1 | 0.714 | 0.00044 | 0.00032 | 0.00049 | 0.00055 | 0.00043 | 0.00039 | 0.32 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 2.7.1.198 | Protein-N(pi)-phosphohistidine--D-sorbitol phosphotransferase. | 0.11 | 0.5 | 1 | 0.088 | 1 | 1 | 0.14 | 0.83 | 1 | 0.15 | 0.47 | 1 | 0.43 | 0.91 | 1 | 0.48 | 0.87 | 1 | 0.00028 | 3.8e-05 | 0 | 0.00014 | 0 | 0.00036 | 4.8e-05 | 0 | 0.00013 | -1.5 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 5.4e-06 | 0 | 1.8e-05 | 1.1 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.5.99.4 | Transferred entry: 3.5.4.42. | 0.16 | 0.5 | 1 | 0.56 | 1 | 1 | 0.49 | 0.95 | 1 | 0.14 | 0.47 | 1 | 0.49 | 0.92 | 1 | 0.57 | 0.9 | 1 | 0.00021 | 3.5e-05 | 0 | 7e-05 | 0 | 0.00018 | 9.1e-05 | 0 | 0.00023 | 0.38 | 6 / 36 (17%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0 | 0 | 0 | 1.3e-05 | 0 | 3.5e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 1.1.1.79 | Glyoxylate reductase (NADP(+)). | 0.14 | 0.5 | 1 | 0.071 | 1 | 1 | 0.39 | 0.91 | 1 | 0.031 | 0.43 | 1 | 0.093 | 0.86 | 1 | 0.35 | 0.81 | 1 | 0.0029 | 0.0029 | 0.0028 | 0.002 | 0.0017 | 0.00094 | 0.003 | 0.0031 | 0.0018 | 0.58 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0029 | 0.0024 | 0.0015 | 0.0033 | 0.0035 | 0.0011 | 0.19 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.8.36 | Undecaprenyl phosphate N.N'-diacetylbacillosamine 1-phosphate | 0.22 | 0.54 | 1 | 0.99 | 1 | 1 | 0.31 | 0.89 | 1 | 0.081 | 0.44 | 1 | 0.97 | 0.99 | 1 | 0.16 | 0.71 | 1 | 0.00082 | 0.00061 | 0.00036 | 0.00046 | 0.00022 | 0.00055 | 0.00051 | 0.00014 | 0.00072 | 0.15 | 27 / 36 (75%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.00073 | 0.00039 | 0.00068 | 0.00065 | 0.00034 | 9e-04 | -0.17 | 11 / 11 (100%) | 8 / 11 (73%) | 1 | 0.727 |
| 2.1.1.197 | Malonyl-[acyl-carrier protein] O-methyltransferase. | 0.48 | 0.72 | 1 | 0.31 | 1 | 1 | 0.98 | 1 | 1 | 0.95 | 0.98 | 1 | 0.94 | 0.99 | 1 | 0.42 | 0.85 | 1 | 0.0057 | 0.0057 | 0.0058 | 0.0064 | 0.0065 | 0.0023 | 0.0056 | 0.0052 | 0.0013 | -0.19 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0058 | 0.0063 | 0.0017 | 0.0051 | 0.0054 | 0.0013 | -0.19 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.6.3.20 | Transferred entry: 7.6.2.10. | 0.19 | 0.53 | 1 | 0.77 | 1 | 1 | 0.82 | 1 | 1 | 0.12 | 0.47 | 1 | 0.89 | 0.99 | 1 | 0.58 | 0.91 | 1 | 0.023 | 0.023 | 0.02 | 0.019 | 0.018 | 0.0077 | 0.018 | 0.019 | 0.0072 | -0.078 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.028 | 0.021 | 0.023 | 0.024 | 0.022 | 0.008 | -0.22 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.6.5.2 | NAD(P)H dehydrogenase (quinone). | 0.24 | 0.55 | 1 | 0.84 | 1 | 1 | 0.6 | 0.96 | 1 | 0.17 | 0.49 | 1 | 0.99 | 1 | 1 | 0.72 | 0.94 | 1 | 0.0017 | 0.0017 | 0.001 | 0.0011 | 0.001 | 0.00068 | 0.0013 | 0.00096 | 0.0012 | 0.24 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0022 | 0.001 | 0.003 | 0.0018 | 0.0016 | 0.0013 | -0.29 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.2.10 | Aminomethyltransferase. | 0.74 | 0.88 | 1 | 0.82 | 1 | 1 | 0.78 | 1 | 1 | 0.76 | 0.91 | 1 | 0.9 | 0.99 | 1 | 0.9 | 0.98 | 1 | 0.0021 | 0.0021 | 0.0018 | 0.0018 | 0.002 | 0.00063 | 0.0021 | 0.0017 | 0.0013 | 0.22 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0022 | 0.0018 | 0.0017 | 0.0021 | 0.0013 | 0.0017 | -0.067 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.197 | Protein-N(pi)-phosphohistidine--D-mannitol phosphotransferase. | 0.18 | 0.52 | 1 | 0.029 | 0.97 | 1 | 0.033 | 0.71 | 1 | 0.27 | 0.56 | 1 | 0.14 | 0.87 | 1 | 0.11 | 0.66 | 1 | 0.00057 | 0.00014 | 0 | 0.00038 | 0 | 0.00093 | 0.00024 | 0 | 0.00064 | -0.66 | 9 / 36 (25%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 3.4e-05 | 0 | 8.9e-05 | 4.1e-05 | 0 | 7.8e-05 | 0.27 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 3.4.19.13 | Glutathione gamma-glutamate hydrolase. | 0.16 | 0.5 | 1 | 0.26 | 1 | 1 | 0.14 | 0.83 | 1 | 0.2 | 0.5 | 1 | 0.72 | 0.95 | 1 | 0.4 | 0.83 | 1 | 0.00036 | 6e-05 | 0 | 0.00017 | 0 | 0.00044 | 0.00012 | 0 | 0.00031 | -0.5 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1.2e-05 | 0 | 2.7e-05 | 2.2 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 4.6.1.1 | Adenylate cyclase. | 0.95 | 0.98 | 1 | 0.37 | 1 | 1 | 0.61 | 0.96 | 1 | 0.79 | 0.92 | 1 | 0.66 | 0.95 | 1 | 0.97 | 0.99 | 1 | 0.0018 | 0.0018 | 0.0014 | 0.002 | 0.0011 | 0.002 | 0.0014 | 0.00064 | 0.0016 | -0.51 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.002 | 0.0014 | 0.0026 | 0.0017 | 0.0015 | 0.0012 | -0.23 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.3.3 | Creatinase. | 0.85 | 0.94 | 1 | 0.71 | 1 | 1 | 0.89 | 1 | 1 | 0.99 | 1 | 1 | 0.73 | 0.96 | 1 | 0.75 | 0.95 | 1 | 5e-04 | 4e-04 | 0.00026 | 0.00031 | 0.00018 | 0.00027 | 0.00037 | 0.00027 | 0.00029 | 0.26 | 29 / 36 (81%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00042 | 0.00012 | 5e-04 | 0.00047 | 0.00033 | 0.00048 | 0.16 | 9 / 11 (82%) | 8 / 11 (73%) | 0.818 | 0.727 |
| 1.3.98.1 | Dihydroorotate oxidase (fumarate). | 0.29 | 0.57 | 1 | 0.43 | 1 | 1 | 0.79 | 1 | 1 | 0.36 | 0.63 | 1 | 0.079 | 0.86 | 1 | 0.31 | 0.79 | 1 | 0.0033 | 0.003 | 0.002 | 0.0016 | 0.0019 | 0.0012 | 0.0021 | 0.0018 | 0.0014 | 0.39 | 33 / 36 (92%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.0039 | 0.0016 | 0.0068 | 0.0036 | 0.0025 | 0.0028 | -0.12 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 2.7.1.1 | Hexokinase. | 0.18 | 0.52 | 1 | 0.15 | 1 | 1 | 0.023 | 0.71 | 1 | 0.11 | 0.47 | 1 | 0.083 | 0.86 | 1 | 0.02 | 0.55 | 1 | 0.00036 | 9.9e-05 | 0 | 0.00013 | 3.2e-05 | 0.00022 | 9.4e-05 | 0 | 0.00024 | -0.47 | 10 / 36 (28%) | 4 / 7 (57%) | 2 / 7 (29%) | 0.571 | 0.286 | 3.6e-05 | 0 | 0.00012 | 0.00015 | 0 | 0.00039 | 2.1 | 1 / 11 (9.1%) | 3 / 11 (27%) | 0.0909 | 0.273 |
| 1.4.1.21 | Aspartate dehydrogenase. | 0.61 | 0.81 | 1 | 0.84 | 1 | 1 | 0.25 | 0.89 | 1 | 0.7 | 0.88 | 1 | 0.84 | 0.98 | 1 | 0.66 | 0.92 | 1 | 0.00078 | 0.00078 | 0.00075 | 0.00074 | 0.00082 | 4e-04 | 0.00066 | 0.00063 | 0.00027 | -0.17 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00065 | 0.00052 | 0.00045 | 0.001 | 0.00098 | 0.00084 | 0.62 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.2.1.12 | Glyceraldehyde-3-phosphate dehydrogenase (phosphorylating). | 0.2 | 0.53 | 1 | 1 | 1 | 1 | 0.83 | 1 | 1 | 0.19 | 0.5 | 1 | 0.47 | 0.92 | 1 | 0.7 | 0.94 | 1 | 0.011 | 0.011 | 0.011 | 0.0091 | 0.0084 | 0.0021 | 0.0093 | 0.0083 | 0.0033 | 0.031 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.011 | 0.0048 | 0.012 | 0.012 | 0.0027 | 0.13 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.97 | Endo-alpha-N-acetylgalactosaminidase. | 0.2 | 0.53 | 1 | 0.45 | 1 | 1 | 0.65 | 0.97 | 1 | 0.025 | 0.42 | 1 | 0.078 | 0.86 | 1 | 0.19 | 0.72 | 1 | 0.0045 | 0.0043 | 0.0031 | 0.0031 | 0.0027 | 0.0029 | 0.004 | 0.0038 | 0.0038 | 0.37 | 35 / 36 (97%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.0049 | 0.003 | 0.0056 | 0.0048 | 0.0034 | 0.0031 | -0.03 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.8.99.5 | Dissimilatory sulfite reductase. | 0.85 | 0.93 | 1 | 0.99 | 1 | 1 | 0.45 | 0.93 | 1 | 0.96 | 0.99 | 1 | 0.98 | 0.99 | 1 | 0.44 | 0.85 | 1 | 1e-04 | 1.4e-05 | 0 | 1.5e-05 | 0 | 2.6e-05 | 3e-05 | 0 | 8.1e-05 | 1 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 1.7e-05 | 0 | 3.8e-05 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 1.1.1.157 | 3-hydroxybutyryl-CoA dehydrogenase. | 0.37 | 0.65 | 1 | 0.98 | 1 | 1 | 0.57 | 0.96 | 1 | 0.25 | 0.54 | 1 | 0.39 | 0.9 | 1 | 0.62 | 0.92 | 1 | 0.0074 | 0.0074 | 0.007 | 0.0066 | 0.0067 | 0.0011 | 0.0066 | 0.0069 | 0.0016 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0075 | 0.0074 | 0.0021 | 0.0082 | 0.0072 | 0.0033 | 0.13 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.1.20 | Diaminopimelate decarboxylase. | 0.48 | 0.72 | 1 | 0.23 | 1 | 1 | 0.32 | 0.89 | 1 | 0.33 | 0.6 | 1 | 0.5 | 0.93 | 1 | 0.7 | 0.94 | 1 | 0.028 | 0.028 | 0.029 | 0.027 | 0.028 | 0.0053 | 0.025 | 0.029 | 0.0091 | -0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.03 | 0.031 | 0.0072 | 0.03 | 0.03 | 0.0044 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.3.83 | D-glycero-alpha-D-manno-heptose-1.7-bisphosphate 7-phosphatase. | 0.66 | 0.84 | 1 | 0.17 | 1 | 1 | 0.14 | 0.83 | 1 | 0.4 | 0.67 | 1 | 0.56 | 0.93 | 1 | 0.52 | 0.89 | 1 | 3.5e-05 | 4.8e-06 | 0 | 2.3e-06 | 0 | 6.1e-06 | 0 | 0 | 0 | -Inf | 5 / 36 (14%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 5.5e-06 | 0 | 1.2e-05 | 8.7e-06 | 0 | 2.1e-05 | 0.66 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 1.4.1.14 | Glutamate synthase (NADH). | 0.69 | 0.85 | 1 | 0.31 | 1 | 1 | 0.096 | 0.81 | 1 | 0.53 | 0.76 | 1 | 0.57 | 0.93 | 1 | 0.21 | 0.74 | 1 | 0.041 | 0.041 | 0.041 | 0.039 | 0.042 | 0.0082 | 0.036 | 0.036 | 0.014 | -0.12 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.042 | 0.047 | 0.014 | 0.046 | 0.048 | 0.014 | 0.13 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.14.13.240 | 2-polyprenylphenol 6-hydroxylase. | 0.2 | 0.53 | 1 | 0.94 | 1 | 1 | 0.38 | 0.9 | 1 | 0.23 | 0.53 | 1 | 0.71 | 0.95 | 1 | 0.72 | 0.94 | 1 | 0.00026 | 2.1e-05 | 0 | 5e-05 | 0 | 0.00013 | 4.8e-05 | 0 | 0.00013 | -0.059 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 6.8e-06 | 0 | 2.3e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.4.1.157 | Transferred entry: 2.4.1.336 and 2.4.1.337. | 0.027 | 0.49 | 1 | 0.29 | 1 | 1 | 0.13 | 0.82 | 1 | 0.019 | 0.42 | 1 | 0.16 | 0.87 | 1 | 0.099 | 0.66 | 1 | 0.00041 | 0.00027 | 0.00014 | 0.00012 | 0 | 0.00023 | 0.00022 | 0.00016 | 0.00025 | 0.87 | 24 / 36 (67%) | 2 / 7 (29%) | 5 / 7 (71%) | 0.286 | 0.714 | 0.00044 | 0.00029 | 0.00057 | 0.00024 | 9.9e-05 | 0.00027 | -0.87 | 9 / 11 (82%) | 8 / 11 (73%) | 0.818 | 0.727 |
| 1.8.1.4 | Dihydrolipoyl dehydrogenase. | 0.49 | 0.73 | 1 | 0.79 | 1 | 1 | 0.99 | 1 | 1 | 0.34 | 0.61 | 1 | 0.45 | 0.92 | 1 | 0.64 | 0.92 | 1 | 0.0075 | 0.0075 | 0.006 | 0.0061 | 0.0057 | 0.0017 | 0.0065 | 0.0058 | 0.0024 | 0.092 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0082 | 0.0068 | 0.0071 | 0.0082 | 0.0078 | 0.0048 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.64 | 2.6-beta-fructan 6-levanbiohydrolase. | 0.1 | 0.49 | 1 | 0.32 | 1 | 1 | 0.9 | 1 | 1 | 0.32 | 0.59 | 1 | 0.5 | 0.93 | 1 | 0.78 | 0.95 | 1 | 0.0038 | 0.0038 | 0.0039 | 0.0049 | 0.0046 | 0.0013 | 0.0044 | 0.0048 | 0.0022 | -0.16 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0036 | 0.0038 | 0.0013 | 0.0031 | 0.0035 | 0.0015 | -0.22 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.4.5 | Cytidine deaminase. | 0.3 | 0.58 | 1 | 0.51 | 1 | 1 | 0.23 | 0.88 | 1 | 0.87 | 0.96 | 1 | 0.95 | 0.99 | 1 | 0.63 | 0.92 | 1 | 0.0062 | 0.0062 | 0.0061 | 0.0065 | 0.0063 | 0.0012 | 0.0062 | 0.005 | 0.002 | -0.068 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0059 | 0.0063 | 0.0014 | 0.0064 | 0.0061 | 0.0012 | 0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.3.27 | Phosphatidylglycerophosphatase. | 0.39 | 0.66 | 1 | 0.56 | 1 | 1 | 0.74 | 0.99 | 1 | 0.6 | 0.81 | 1 | 0.78 | 0.97 | 1 | 0.9 | 0.98 | 1 | 0.00039 | 0.00033 | 0.00023 | 0.00033 | 0.00024 | 0.00025 | 0.00051 | 0.00024 | 0.00068 | 0.63 | 31 / 36 (86%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00018 | 0.00019 | 0.00016 | 0.00038 | 0.00033 | 4e-04 | 1.1 | 10 / 11 (91%) | 9 / 11 (82%) | 0.909 | 0.818 |
| 2.4.1.18 | 1.4-alpha-glucan branching enzyme. | 0.21 | 0.53 | 1 | 0.88 | 1 | 1 | 0.82 | 1 | 1 | 0.13 | 0.47 | 1 | 0.56 | 0.93 | 1 | 0.39 | 0.83 | 1 | 0.062 | 0.062 | 0.058 | 0.054 | 0.049 | 0.013 | 0.054 | 0.054 | 0.016 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.07 | 0.057 | 0.043 | 0.064 | 0.059 | 0.016 | -0.13 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.91 | Aryl-alcohol dehydrogenase (NADP(+)). | 0.32 | 0.59 | 1 | 0.8 | 1 | 1 | 0.26 | 0.89 | 1 | 0.27 | 0.56 | 1 | 0.98 | 0.99 | 1 | 0.2 | 0.73 | 1 | 0.00012 | 1.7e-05 | 0 | 1.7e-05 | 0 | 4.5e-05 | 7.7e-06 | 0 | 2e-05 | -1.1 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 4.1e-05 | 0 | 7e-05 | 0 | 0 | 0 | -Inf | 3 / 11 (27%) | 0 / 11 (0%) | 0.273 | 0 |
| 3.6.1.23 | dUTP diphosphatase. | 0.39 | 0.65 | 1 | 0.55 | 1 | 1 | 0.6 | 0.96 | 1 | 0.33 | 0.6 | 1 | 0.68 | 0.95 | 1 | 0.69 | 0.94 | 1 | 0.0097 | 0.0097 | 0.0094 | 0.009 | 0.0092 | 0.0018 | 0.0087 | 0.0087 | 0.0024 | -0.049 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.01 | 0.009 | 0.0037 | 0.01 | 0.0098 | 0.0022 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.1.1 | Amino-acid N-acetyltransferase. | 0.14 | 0.5 | 1 | 0.48 | 1 | 1 | 0.41 | 0.92 | 1 | 0.14 | 0.47 | 1 | 0.17 | 0.87 | 1 | 0.13 | 0.67 | 1 | 0.02 | 0.02 | 0.02 | 0.018 | 0.019 | 0.0026 | 0.02 | 0.022 | 0.0056 | 0.15 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.022 | 0.022 | 0.0051 | 0.021 | 0.02 | 0.0039 | -0.067 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.3.16 | Citrate synthase (unknown stereospecificity). | 0.1 | 0.49 | 1 | 0.93 | 1 | 1 | 0.9 | 1 | 1 | 0.093 | 0.45 | 1 | 0.7 | 0.95 | 1 | 0.7 | 0.94 | 1 | 0.00039 | 8.7e-05 | 0 | 0.00022 | 0 | 0.00053 | 2e-04 | 0 | 0.00045 | -0.14 | 8 / 36 (22%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 1e-05 | 0 | 3.4e-05 | 8.1e-06 | 0 | 2.7e-05 | -0.3 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.1.7.2 | Guanosine-3'.5'-bis(diphosphate) 3'-diphosphatase. | 0.88 | 0.95 | 1 | 0.28 | 1 | 1 | 0.84 | 1 | 1 | 0.65 | 0.85 | 1 | 0.16 | 0.87 | 1 | 0.38 | 0.83 | 1 | 0.0045 | 0.0045 | 0.0033 | 0.0037 | 0.003 | 0.0015 | 0.0048 | 0.0053 | 0.0024 | 0.38 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.004 | 0.0032 | 0.0023 | 0.0054 | 0.003 | 0.0056 | 0.43 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.17.7.3 | (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase (flavodoxin). | 0.22 | 0.54 | 1 | 0.95 | 1 | 1 | 0.93 | 1 | 1 | 0.21 | 0.52 | 1 | 0.22 | 0.88 | 1 | 0.18 | 0.71 | 1 | 0.017 | 0.017 | 0.017 | 0.016 | 0.016 | 0.0027 | 0.016 | 0.015 | 0.0036 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.018 | 0.016 | 0.0042 | 0.017 | 0.017 | 0.002 | -0.082 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.2.15 | Propionate kinase. | 0.11 | 0.5 | 1 | 0.36 | 1 | 1 | 0.29 | 0.89 | 1 | 0.053 | 0.43 | 1 | 0.26 | 0.88 | 1 | 0.22 | 0.74 | 1 | 0.001 | 0.00011 | 0 | 0.00027 | 0 | 0.00066 | 3e-04 | 0 | 0.00079 | 0.15 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 4.6.1.13 | Phosphatidylinositol diacylglycerol-lyase. | 0.43 | 0.69 | 1 | 0.9 | 1 | 1 | 0.1 | 0.82 | 1 | 0.16 | 0.49 | 1 | 0.48 | 0.92 | 1 | 0.13 | 0.67 | 1 | 0.0014 | 0.0013 | 0.00084 | 0.0015 | 0.00048 | 0.0019 | 0.0015 | 5e-04 | 0.0019 | 0 | 34 / 36 (94%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.0017 | 0.0011 | 0.0014 | 0.00081 | 0.00083 | 0.00037 | -1.1 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.3.3 | N-acetylneuraminate lyase. | 0.046 | 0.49 | 1 | 0.13 | 1 | 1 | 0.011 | 0.61 | 1 | 0.086 | 0.44 | 1 | 0.43 | 0.91 | 1 | 0.04 | 0.63 | 1 | 0.0032 | 0.0032 | 0.003 | 0.004 | 0.0035 | 0.0016 | 0.0031 | 0.003 | 0.0012 | -0.37 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0026 | 0.0022 | 0.0017 | 0.0035 | 0.0029 | 0.0019 | 0.43 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.- | 0.64 | 0.83 | 1 | 0.27 | 1 | 1 | 0.55 | 0.95 | 1 | 0.59 | 0.81 | 1 | 0.022 | 0.76 | 1 | 0.078 | 0.66 | 1 | 0.047 | 0.047 | 0.047 | 0.048 | 0.044 | 0.023 | 0.052 | 0.054 | 0.014 | 0.12 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.044 | 0.043 | 0.01 | 0.045 | 0.048 | 0.0076 | 0.032 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 1.1.1.192 | Long-chain-alcohol dehydrogenase. | 0.2 | 0.53 | 1 | 0.5 | 1 | 1 | 0.12 | 0.82 | 1 | 0.059 | 0.43 | 1 | 0.56 | 0.93 | 1 | 0.015 | 0.55 | 1 | 0.00077 | 0.00038 | 3.6e-05 | 0.00019 | 0 | 0.00033 | 0.00035 | 0 | 0.00059 | 0.88 | 18 / 36 (50%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0.00049 | 2e-04 | 0.00081 | 0.00043 | 0 | 0.00084 | -0.19 | 9 / 11 (82%) | 5 / 11 (45%) | 0.818 | 0.455 |
| 7.2.2.8 | P-type Cu(+) transporter. | 0.082 | 0.49 | 1 | 0.048 | 0.97 | 1 | 0.06 | 0.71 | 1 | 0.072 | 0.43 | 1 | 0.26 | 0.88 | 1 | 0.17 | 0.71 | 1 | 0.00042 | 4.7e-05 | 0 | 0.00017 | 0 | 0.00044 | 6.8e-05 | 0 | 0.00018 | -1.3 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.3.1.86 | Fatty-acyl-CoA synthase system. | 0.29 | 0.57 | 1 | 0.47 | 1 | 1 | 0.69 | 0.98 | 1 | 0.13 | 0.47 | 1 | 0.16 | 0.87 | 1 | 0.98 | 0.99 | 1 | 0.0039 | 0.0034 | 0.003 | 0.002 | 0.0023 | 0.0019 | 0.0028 | 0.0025 | 0.0029 | 0.49 | 32 / 36 (89%) | 5 / 7 (71%) | 6 / 7 (86%) | 0.714 | 0.857 | 0.0035 | 0.0021 | 0.0046 | 0.0046 | 0.0033 | 0.0034 | 0.39 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 2.7.7.64 | UTP-monosaccharide-1-phosphate uridylyltransferase. | 0.39 | 0.66 | 1 | 0.35 | 1 | 1 | 0.62 | 0.96 | 1 | 0.35 | 0.62 | 1 | 0.17 | 0.87 | 1 | 0.49 | 0.88 | 1 | 0.00024 | 2.6e-05 | 0 | 0 | 0 | 0 | 1.3e-05 | 0 | 3.5e-05 | Inf | 4 / 36 (11%) | 0 / 7 (0%) | 1 / 7 (14%) | 0 | 0.143 | 4.4e-05 | 0 | 0.00015 | 3.4e-05 | 0 | 7.7e-05 | -0.37 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 2.1.1.199 | 16S rRNA (cytosine(1402)-N(4))-methyltransferase. | 0.16 | 0.5 | 1 | 0.98 | 1 | 1 | 0.46 | 0.93 | 1 | 0.17 | 0.49 | 1 | 0.41 | 0.9 | 1 | 0.11 | 0.66 | 1 | 0.024 | 0.024 | 0.024 | 0.022 | 0.021 | 0.0045 | 0.022 | 0.024 | 0.0082 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.025 | 0.026 | 0.0039 | 0.024 | 0.024 | 0.0052 | -0.059 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.49 | Alpha-N-acetylgalactosaminidase. | 0.68 | 0.85 | 1 | 0.51 | 1 | 1 | 0.56 | 0.95 | 1 | 0.88 | 0.96 | 1 | 0.49 | 0.92 | 1 | 0.78 | 0.95 | 1 | 0.00044 | 0.00039 | 0.00033 | 0.00035 | 0.00023 | 0.00031 | 0.00044 | 0.00043 | 0.00033 | 0.33 | 32 / 36 (89%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00028 | 0.00013 | 0.00032 | 5e-04 | 0.00043 | 5e-04 | 0.84 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 2.3.1.169 | CO-methylating acetyl-CoA synthase. | 0.22 | 0.54 | 1 | 0.88 | 1 | 1 | 0.85 | 1 | 1 | 0.55 | 0.78 | 1 | 0.44 | 0.91 | 1 | 0.81 | 0.96 | 1 | 0.0075 | 0.0075 | 0.007 | 0.0089 | 0.009 | 0.0057 | 0.0089 | 0.0079 | 0.0045 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0065 | 0.0066 | 0.003 | 0.0068 | 0.007 | 0.0019 | 0.065 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.6.1.1 | Aspartate transaminase. | 0.22 | 0.54 | 1 | 0.12 | 1 | 1 | 0.13 | 0.82 | 1 | 0.076 | 0.44 | 1 | 0.0094 | 0.73 | 1 | 0.018 | 0.55 | 1 | 0.022 | 0.022 | 0.02 | 0.019 | 0.018 | 0.0053 | 0.023 | 0.024 | 0.0064 | 0.28 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.024 | 0.019 | 0.015 | 0.022 | 0.021 | 0.0066 | -0.13 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.5.1.29 | Geranylgeranyl diphosphate synthase. | 0.22 | 0.54 | 1 | 0.39 | 1 | 1 | 0.17 | 0.85 | 1 | 0.12 | 0.47 | 1 | 0.17 | 0.87 | 1 | 0.041 | 0.63 | 1 | 0.0031 | 0.0031 | 0.0024 | 0.0019 | 0.0014 | 0.0011 | 0.0015 | 0.0012 | 0.0011 | -0.34 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0039 | 0.0027 | 0.0057 | 0.0041 | 0.0038 | 0.0024 | 0.072 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.7.47 | Streptomycin 3''-adenylyltransferase. | 0.093 | 0.49 | 1 | 0.14 | 1 | 1 | 0.14 | 0.83 | 1 | 0.047 | 0.43 | 1 | 0.067 | 0.86 | 1 | 0.16 | 0.71 | 1 | 0.00039 | 0.00017 | 0 | 0.00025 | 0.00016 | 0.00027 | 0.00022 | 0 | 0.00035 | -0.18 | 16 / 36 (44%) | 6 / 7 (86%) | 3 / 7 (43%) | 0.857 | 0.429 | 8.6e-05 | 0 | 0.00016 | 0.00018 | 0 | 0.00044 | 1.1 | 4 / 11 (36%) | 3 / 11 (27%) | 0.364 | 0.273 |
| 2.7.13.- | 0.099 | 0.49 | 1 | 0.34 | 1 | 1 | 0.13 | 0.82 | 1 | 0.1 | 0.46 | 1 | 0.19 | 0.87 | 1 | 0.063 | 0.66 | 1 | 0.00035 | 0.00012 | 0 | 2e-05 | 0 | 5.4e-05 | 1e-04 | 0 | 2e-04 | 2.3 | 12 / 36 (33%) | 1 / 7 (14%) | 3 / 7 (43%) | 0.143 | 0.429 | 0.00022 | 0 | 0.00043 | 8.9e-05 | 0 | 0.00019 | -1.3 | 5 / 11 (45%) | 3 / 11 (27%) | 0.455 | 0.273 | |
| 3.1.4.58 | RNA 2'.3'-cyclic 3'-phosphodiesterase. | 0.7 | 0.86 | 1 | 0.48 | 1 | 1 | 0.58 | 0.96 | 1 | 0.76 | 0.91 | 1 | 0.36 | 0.9 | 1 | 0.58 | 0.91 | 1 | 0.00024 | 0.00017 | 8.8e-05 | 0.00017 | 0.00011 | 0.00019 | 0.00014 | 5.4e-05 | 0.00017 | -0.28 | 25 / 36 (69%) | 6 / 7 (86%) | 4 / 7 (57%) | 0.857 | 0.571 | 0.00024 | 0.00017 | 0.00027 | 0.00012 | 7.5e-05 | 0.00023 | -1 | 9 / 11 (82%) | 6 / 11 (55%) | 0.818 | 0.545 |
| 2.7.13.3 | Histidine kinase. | 0.65 | 0.84 | 1 | 0.62 | 1 | 1 | 0.63 | 0.96 | 1 | 0.66 | 0.86 | 1 | 0.018 | 0.76 | 1 | 0.22 | 0.74 | 1 | 0.53 | 0.53 | 0.51 | 0.53 | 0.55 | 0.11 | 0.55 | 0.63 | 0.13 | 0.053 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.51 | 0.49 | 0.064 | 0.55 | 0.52 | 0.11 | 0.11 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.1.19 | Imidazoleglycerol-phosphate dehydratase. | 0.087 | 0.49 | 1 | 0.39 | 1 | 1 | 0.99 | 1 | 1 | 0.065 | 0.43 | 1 | 0.046 | 0.81 | 1 | 0.22 | 0.74 | 1 | 0.0097 | 0.0097 | 0.0093 | 0.0084 | 0.0083 | 0.0011 | 0.0091 | 0.0091 | 0.0022 | 0.12 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.01 | 0.0089 | 0.0026 | 0.011 | 0.011 | 0.0019 | 0.14 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.60 | N-acylmannosamine kinase. | 0.13 | 0.5 | 1 | 0.45 | 1 | 1 | 0.18 | 0.86 | 1 | 0.065 | 0.43 | 1 | 0.16 | 0.87 | 1 | 0.056 | 0.66 | 1 | 0.0032 | 0.0032 | 0.0023 | 0.0021 | 0.0022 | 0.0013 | 0.0026 | 0.0025 | 0.0013 | 0.31 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0044 | 0.0027 | 0.0058 | 0.003 | 0.0022 | 0.0023 | -0.55 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.3.3.3 | Dethiobiotin synthase. | 0.15 | 0.5 | 1 | 0.68 | 1 | 1 | 0.37 | 0.9 | 1 | 0.56 | 0.79 | 1 | 0.27 | 0.88 | 1 | 0.091 | 0.66 | 1 | 0.0024 | 0.0024 | 0.0023 | 0.0029 | 0.0026 | 0.0012 | 0.003 | 0.0032 | 0.001 | 0.049 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0022 | 0.0019 | 0.00066 | 0.002 | 0.0018 | 0.00075 | -0.14 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.17.19 | Carboxypeptidase Taq. | 0.83 | 0.92 | 1 | 0.45 | 1 | 1 | 0.96 | 1 | 1 | 0.59 | 0.81 | 1 | 0.83 | 0.98 | 1 | 0.59 | 0.91 | 1 | 0.0044 | 0.0044 | 0.0045 | 0.0045 | 0.0041 | 0.002 | 0.004 | 0.0031 | 0.0017 | -0.17 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0047 | 0.0046 | 0.0016 | 0.0042 | 0.0034 | 0.0022 | -0.16 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.86 | Ketol-acid reductoisomerase (NADP(+)). | 0.16 | 0.5 | 1 | 0.95 | 1 | 1 | 0.87 | 1 | 1 | 0.088 | 0.45 | 1 | 0.38 | 0.9 | 1 | 0.68 | 0.93 | 1 | 0.019 | 0.019 | 0.018 | 0.016 | 0.015 | 0.0028 | 0.017 | 0.016 | 0.0046 | 0.087 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.021 | 0.019 | 0.012 | 0.021 | 0.021 | 0.0045 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.8.99.3 | Transferred entry: 1.8.99.5. | 0.95 | 0.98 | 1 | 0.64 | 1 | 1 | 0.61 | 0.96 | 1 | 0.88 | 0.96 | 1 | 0.69 | 0.95 | 1 | 0.74 | 0.94 | 1 | 0.0068 | 0.0068 | 0.0068 | 0.0067 | 0.0064 | 0.0017 | 0.0065 | 0.006 | 0.0033 | -0.044 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0068 | 0.0074 | 0.003 | 0.007 | 0.0072 | 0.0028 | 0.042 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.7.1.13 | PreQ(1) synthase. | 0.49 | 0.73 | 1 | 0.69 | 1 | 1 | 0.3 | 0.89 | 1 | 0.8 | 0.93 | 1 | 0.87 | 0.99 | 1 | 0.7 | 0.94 | 1 | 0.0021 | 0.0021 | 0.0022 | 0.0022 | 0.0017 | 0.00086 | 0.0021 | 0.0022 | 0.00073 | -0.067 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.002 | 0.0019 | 0.00083 | 0.0023 | 0.0026 | 0.001 | 0.2 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.99.2 | Aminopyrimidine aminohydrolase. | 0.94 | 0.97 | 1 | 0.47 | 1 | 1 | 0.59 | 0.96 | 1 | 0.96 | 0.99 | 1 | 0.5 | 0.93 | 1 | 0.37 | 0.82 | 1 | 0.0038 | 0.0038 | 0.0025 | 0.0036 | 0.0026 | 0.0024 | 0.0028 | 0.0017 | 0.0023 | -0.36 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0047 | 0.0018 | 0.0076 | 0.0036 | 0.0032 | 0.0024 | -0.38 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.4.3 | Guanine deaminase. | 0.33 | 0.6 | 1 | 0.3 | 1 | 1 | 0.81 | 1 | 1 | 0.65 | 0.85 | 1 | 0.41 | 0.9 | 1 | 0.69 | 0.94 | 1 | 0.0036 | 0.0036 | 0.0035 | 0.0044 | 0.0043 | 0.0018 | 0.0039 | 0.0042 | 0.0024 | -0.17 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0036 | 0.0037 | 0.0014 | 0.0027 | 0.0028 | 0.0011 | -0.42 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.68 | Isoamylase. | 0.14 | 0.5 | 1 | 0.64 | 1 | 1 | 0.73 | 0.99 | 1 | 0.053 | 0.43 | 1 | 0.18 | 0.87 | 1 | 0.39 | 0.83 | 1 | 0.0087 | 0.0082 | 0.0041 | 0.0033 | 0.0018 | 0.0035 | 0.0041 | 0.0038 | 0.004 | 0.31 | 34 / 36 (94%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.012 | 0.0043 | 0.025 | 0.0097 | 0.008 | 0.0097 | -0.31 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.251 | Galactitol-1-phosphate 5-dehydrogenase. | 0.91 | 0.96 | 1 | 0.47 | 1 | 1 | 0.49 | 0.94 | 1 | 0.77 | 0.91 | 1 | 0.42 | 0.91 | 1 | 0.43 | 0.85 | 1 | 0.0013 | 0.0013 | 0.0012 | 0.0012 | 0.001 | 0.00043 | 0.0015 | 0.0015 | 0.00074 | 0.32 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0013 | 0.0012 | 6e-04 | 0.0012 | 0.0011 | 0.00058 | -0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.99.- | 0.24 | 0.55 | 1 | 0.75 | 1 | 1 | 0.69 | 0.98 | 1 | 0.056 | 0.43 | 1 | 0.17 | 0.87 | 1 | 0.17 | 0.71 | 1 | 0.00052 | 0.00049 | 0.00043 | 0.00042 | 0.00041 | 0.00042 | 0.00035 | 0.00024 | 0.00025 | -0.26 | 34 / 36 (94%) | 5 / 7 (71%) | 7 / 7 (100%) | 0.714 | 1 | 0.00055 | 0.00044 | 0.00045 | 0.00056 | 5e-04 | 0.00046 | 0.026 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 3.4.13.22 | D-Ala-D-Ala dipeptidase. | 0.39 | 0.65 | 1 | 0.75 | 1 | 1 | 0.2 | 0.87 | 1 | 0.31 | 0.59 | 1 | 0.47 | 0.92 | 1 | 0.12 | 0.67 | 1 | 0.00077 | 0.00073 | 0.00048 | 0.00074 | 0.00055 | 5e-04 | 0.00079 | 0.00064 | 0.00067 | 0.094 | 34 / 36 (94%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 6e-04 | 0.00036 | 0.00085 | 0.00082 | 0.00045 | 0.00079 | 0.45 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 5.3.3.18 | 2-(1.2-epoxy-1.2-dihydrophenyl)acetyl-CoA isomerase. | 0.98 | 0.99 | 1 | 0.55 | 1 | 1 | 0.82 | 1 | 1 | 0.86 | 0.95 | 1 | 0.75 | 0.97 | 1 | 0.63 | 0.92 | 1 | 0.00011 | 2.9e-05 | 0 | 4e-05 | 0 | 7.6e-05 | 1.7e-05 | 0 | 3.5e-05 | -1.2 | 9 / 36 (25%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 4.1e-05 | 0 | 7.1e-05 | 1.6e-05 | 0 | 4.5e-05 | -1.4 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 4.2.1.119 | Enoyl-CoA hydratase 2. | 0.26 | 0.56 | 1 | 0.19 | 1 | 1 | 0.58 | 0.96 | 1 | 0.28 | 0.57 | 1 | 0.3 | 0.9 | 1 | 0.71 | 0.94 | 1 | 0.00017 | 2.3e-05 | 0 | 2.1e-05 | 0 | 4.5e-05 | 8.2e-05 | 0 | 0.00019 | 2 | 5 / 36 (14%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0 | 0 | 0 | 1e-05 | 0 | 3.4e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 3.1.1.72 | Acetylxylan esterase. | 0.67 | 0.85 | 1 | 0.088 | 1 | 1 | 0.012 | 0.61 | 1 | 0.54 | 0.77 | 1 | 0.0056 | 0.7 | 1 | 0.002 | 0.48 | 1 | 0.0012 | 0.0011 | 0.00033 | 0.00096 | 0.00067 | 0.0013 | 6e-04 | 0.00012 | 0.0013 | -0.68 | 33 / 36 (92%) | 7 / 7 (100%) | 5 / 7 (71%) | 1 | 0.714 | 8e-04 | 0.00031 | 0.0014 | 0.0019 | 4e-04 | 0.0033 | 1.2 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 3.1.3.23 | Sugar-phosphatase. | 0.12 | 0.5 | 1 | 0.99 | 1 | 1 | 0.45 | 0.93 | 1 | 0.1 | 0.46 | 1 | 0.74 | 0.96 | 1 | 0.37 | 0.82 | 1 | 0.0082 | 0.0082 | 0.007 | 0.0064 | 0.006 | 0.0019 | 0.0066 | 0.0062 | 0.0029 | 0.044 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.01 | 0.0082 | 0.0083 | 0.0082 | 0.008 | 0.0031 | -0.29 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.7.39 | Glycerol-3-phosphate cytidylyltransferase. | 0.32 | 0.6 | 1 | 0.68 | 1 | 1 | 0.09 | 0.79 | 1 | 0.66 | 0.86 | 1 | 0.33 | 0.9 | 1 | 0.017 | 0.55 | 1 | 0.0053 | 0.0053 | 0.0051 | 0.0062 | 0.0064 | 0.0025 | 0.0067 | 0.005 | 0.0036 | 0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0051 | 0.005 | 0.0019 | 0.0041 | 0.004 | 0.0014 | -0.31 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.6.3.40 | Transferred entry: 7.5.2.4. | 0.98 | 1 | 1 | 0.26 | 1 | 1 | 0.58 | 0.96 | 1 | 0.69 | 0.87 | 1 | 0.55 | 0.93 | 1 | 0.96 | 0.99 | 1 | 0.014 | 0.014 | 0.014 | 0.014 | 0.013 | 0.0032 | 0.013 | 0.011 | 0.0054 | -0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.014 | 0.015 | 0.0045 | 0.014 | 0.013 | 0.0032 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.8.4.8 | Phosphoadenylyl-sulfate reductase (thioredoxin). | 0.41 | 0.67 | 1 | 0.38 | 1 | 1 | 0.9 | 1 | 1 | 0.94 | 0.98 | 1 | 0.75 | 0.97 | 1 | 0.98 | 1 | 1 | 0.00051 | 0.00037 | 1e-04 | 0.00078 | 5.7e-05 | 0.0015 | 0.00052 | 0.00012 | 0.0011 | -0.58 | 26 / 36 (72%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 0.00027 | 9.4e-05 | 0.00041 | 0.00011 | 0.00011 | 8.6e-05 | -1.3 | 8 / 11 (73%) | 9 / 11 (82%) | 0.727 | 0.818 |
| 2.4.1.150 | N-acetyllactosaminide beta-1.6-N-acetylglucosaminyltransferase. | 0.95 | 0.98 | 1 | 0.84 | 1 | 1 | 0.56 | 0.96 | 1 | 0.76 | 0.91 | 1 | 0.53 | 0.93 | 1 | 0.34 | 0.8 | 1 | 0.00014 | 4e-05 | 0 | 5.4e-05 | 0 | 0.00014 | 3.6e-05 | 0 | 4.8e-05 | -0.58 | 10 / 36 (28%) | 2 / 7 (29%) | 3 / 7 (43%) | 0.286 | 0.429 | 5.1e-05 | 0 | 0.00012 | 2.3e-05 | 0 | 6.8e-05 | -1.1 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 3.1.4.44 | Glycerophosphoinositol glycerophosphodiesterase. | 0.2 | 0.53 | 1 | 0.41 | 1 | 1 | 0.31 | 0.89 | 1 | 0.16 | 0.48 | 1 | 0.26 | 0.88 | 1 | 0.24 | 0.76 | 1 | 0.00023 | 3.2e-05 | 0 | 0 | 0 | 0 | 2.6e-05 | 0 | 7e-05 | Inf | 5 / 36 (14%) | 0 / 7 (0%) | 1 / 7 (14%) | 0 | 0.143 | 6.2e-05 | 0 | 0.00015 | 2.7e-05 | 0 | 6.9e-05 | -1.2 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 1.2.1.- | 0.73 | 0.88 | 1 | 0.2 | 1 | 1 | 0.49 | 0.94 | 1 | 0.35 | 0.62 | 1 | 0.65 | 0.95 | 1 | 0.81 | 0.96 | 1 | 0.025 | 0.025 | 0.024 | 0.025 | 0.024 | 0.0032 | 0.022 | 0.023 | 0.0064 | -0.18 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.026 | 0.025 | 0.0045 | 0.025 | 0.024 | 0.0041 | -0.057 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 4.1.2.38 | Benzoin aldolase. | 0.78 | 0.9 | 1 | 0.85 | 1 | 1 | 0.56 | 0.95 | 1 | 0.57 | 0.79 | 1 | 0.7 | 0.95 | 1 | 0.4 | 0.83 | 1 | 0.0011 | 0.00047 | 0 | 0.00056 | 0 | 0.00096 | 0.00047 | 0 | 0.0011 | -0.25 | 15 / 36 (42%) | 2 / 7 (29%) | 3 / 7 (43%) | 0.286 | 0.429 | 0.00061 | 0 | 0.001 | 0.00028 | 0 | 0.00062 | -1.1 | 5 / 11 (45%) | 5 / 11 (45%) | 0.455 | 0.455 |
| 3.1.11.- | 0.13 | 0.5 | 1 | 0.87 | 1 | 1 | 0.18 | 0.86 | 1 | 0.16 | 0.48 | 1 | 0.78 | 0.97 | 1 | 0.17 | 0.71 | 1 | 0.00082 | 0.00075 | 0.00025 | 0.0013 | 0.00026 | 0.0026 | 0.0012 | 0.00025 | 0.0019 | -0.12 | 33 / 36 (92%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00029 | 0.00023 | 0.00025 | 0.00052 | 0.00034 | 0.00057 | 0.84 | 9 / 11 (82%) | 11 / 11 (100%) | 0.818 | 1 | |
| 3.1.4.- | 0.94 | 0.97 | 1 | 0.43 | 1 | 1 | 0.69 | 0.98 | 1 | 0.63 | 0.84 | 1 | 0.71 | 0.95 | 1 | 0.43 | 0.85 | 1 | 0.049 | 0.049 | 0.05 | 0.05 | 0.051 | 0.0068 | 0.047 | 0.048 | 0.012 | -0.089 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.05 | 0.053 | 0.012 | 0.049 | 0.049 | 0.0085 | -0.029 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 3.5.1.4 | Amidase. | 0.73 | 0.87 | 1 | 0.36 | 1 | 1 | 0.92 | 1 | 1 | 0.51 | 0.75 | 1 | 0.2 | 0.87 | 1 | 0.77 | 0.95 | 1 | 0.0023 | 0.0023 | 0.002 | 0.002 | 0.0018 | 0.00096 | 0.0025 | 0.0022 | 0.0013 | 0.32 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0022 | 0.0021 | 0.00088 | 0.0027 | 0.002 | 0.0014 | 0.3 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.99.19 | 2-iminoacetate synthase. | 0.59 | 0.8 | 1 | 0.24 | 1 | 1 | 0.72 | 0.98 | 1 | 0.94 | 0.98 | 1 | 0.065 | 0.86 | 1 | 0.35 | 0.81 | 1 | 0.019 | 0.019 | 0.019 | 0.019 | 0.017 | 0.0033 | 0.022 | 0.022 | 0.007 | 0.21 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.018 | 0.021 | 0.0067 | 0.019 | 0.019 | 0.0047 | 0.078 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.3.3.17 | Trans-2.3-dihydro-3-hydroxyanthranilate isomerase. | 0.39 | 0.65 | 1 | 0.7 | 1 | 1 | 0.93 | 1 | 1 | 0.34 | 0.61 | 1 | 0.91 | 0.99 | 1 | 0.9 | 0.98 | 1 | 0.00011 | 2.4e-05 | 0 | 1.2e-05 | 0 | 3.1e-05 | 3.9e-06 | 0 | 1e-05 | -1.6 | 8 / 36 (22%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3.9e-05 | 0 | 8.1e-05 | 3.1e-05 | 0 | 6.8e-05 | -0.33 | 3 / 11 (27%) | 3 / 11 (27%) | 0.273 | 0.273 |
| 5.3.99.- | 0.013 | 0.49 | 1 | 0.63 | 1 | 1 | 0.039 | 0.71 | 1 | 0.024 | 0.42 | 1 | 0.8 | 0.98 | 1 | 0.042 | 0.63 | 1 | 0.0013 | 0.0013 | 0.0014 | 0.0017 | 0.0015 | 0.00074 | 0.0016 | 0.0017 | 0.00091 | -0.087 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00089 | 0.00086 | 0.00059 | 0.0014 | 0.0014 | 0.00055 | 0.65 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 3.4.13.18 | Cytosol nonspecific dipeptidase. | 0.13 | 0.5 | 1 | 0.67 | 1 | 1 | 0.67 | 0.98 | 1 | 0.086 | 0.44 | 1 | 0.2 | 0.87 | 1 | 0.23 | 0.75 | 1 | 0.015 | 0.015 | 0.014 | 0.012 | 0.013 | 0.0041 | 0.013 | 0.014 | 0.0049 | 0.12 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.016 | 0.014 | 0.0064 | 0.016 | 0.014 | 0.0038 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.1.3.33 | 2-epi-5-epi-valiolone epimerase. | 0.021 | 0.49 | 1 | 0.014 | 0.89 | 1 | 0.047 | 0.71 | 1 | 0.016 | 0.42 | 1 | 0.039 | 0.8 | 1 | 0.12 | 0.66 | 1 | 0.00024 | 2e-05 | 0 | 8.4e-05 | 0 | 0.00018 | 1.9e-05 | 0 | 5.1e-05 | -2.1 | 3 / 36 (8.3%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 2.7.1.74 | Deoxycytidine kinase. | 0.82 | 0.92 | 1 | 0.091 | 1 | 1 | 0.092 | 0.79 | 1 | 0.95 | 0.98 | 1 | 0.16 | 0.87 | 1 | 0.21 | 0.74 | 1 | 0.00027 | 6.7e-05 | 0 | 7.9e-05 | 0 | 0.00015 | 3.9e-06 | 0 | 1e-05 | -4.3 | 9 / 36 (25%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 6.2e-05 | 0 | 0.00015 | 0.00011 | 0 | 0.00025 | 0.83 | 3 / 11 (27%) | 3 / 11 (27%) | 0.273 | 0.273 |
| 3.2.1.52 | Beta-N-acetylhexosaminidase. | 0.38 | 0.65 | 1 | 0.46 | 1 | 1 | 0.81 | 1 | 1 | 0.14 | 0.47 | 1 | 0.12 | 0.87 | 1 | 0.46 | 0.86 | 1 | 0.033 | 0.033 | 0.027 | 0.028 | 0.026 | 0.011 | 0.03 | 0.025 | 0.011 | 0.1 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.035 | 0.027 | 0.022 | 0.038 | 0.031 | 0.021 | 0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.3.99.4 | Choline trimethylamine-lyase. | 0.23 | 0.54 | 1 | 0.082 | 1 | 1 | 0.27 | 0.89 | 1 | 0.52 | 0.76 | 1 | 0.17 | 0.87 | 1 | 0.31 | 0.79 | 1 | 0.00046 | 0.00038 | 0.00019 | 0.00076 | 0.00028 | 0.0013 | 0.00057 | 0.00012 | 0.0012 | -0.42 | 30 / 36 (83%) | 6 / 7 (86%) | 5 / 7 (71%) | 0.857 | 0.714 | 0.00023 | 0.00019 | 0.00017 | 0.00018 | 0.00011 | 0.00015 | -0.35 | 9 / 11 (82%) | 10 / 11 (91%) | 0.818 | 0.909 |
| 2.3.1.82 | Aminoglycoside 6'-N-acetyltransferase. | 0.28 | 0.57 | 1 | 0.52 | 1 | 1 | 0.88 | 1 | 1 | 0.31 | 0.58 | 1 | 0.63 | 0.95 | 1 | 0.78 | 0.95 | 1 | 0.00015 | 2.5e-05 | 0 | 2.1e-05 | 0 | 4.5e-05 | 7.2e-05 | 0 | 0.00018 | 1.8 | 6 / 36 (17%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0 | 0 | 0 | 2.2e-05 | 0 | 5e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 4.2.99.9 | Transferred entry: 2.5.1.48. | 0.12 | 0.5 | 1 | 0.1 | 1 | 1 | 0.062 | 0.71 | 1 | 0.1 | 0.46 | 1 | 0.57 | 0.93 | 1 | 0.29 | 0.78 | 1 | 0.00078 | 8.7e-05 | 0 | 0.00025 | 0 | 0.00067 | 0.00018 | 0 | 0.00049 | -0.47 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.1.1.144 | Perillyl-alcohol dehydrogenase. | 0.56 | 0.78 | 1 | 0.62 | 1 | 1 | 0.6 | 0.96 | 1 | 0.72 | 0.89 | 1 | 0.51 | 0.93 | 1 | 0.74 | 0.94 | 1 | 7.8e-05 | 8.6e-06 | 0 | 4.1e-06 | 0 | 1.1e-05 | 1.5e-05 | 0 | 4e-05 | 1.9 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 1.6e-05 | 0 | 3.7e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 1.1.1.414 | L-galactonate 5-dehydrogenase. | 0.14 | 0.5 | 1 | 0.21 | 1 | 1 | 0.13 | 0.82 | 1 | 0.17 | 0.49 | 1 | 0.95 | 0.99 | 1 | 0.47 | 0.87 | 1 | 0.00043 | 3.6e-05 | 0 | 0.00012 | 0 | 0.00031 | 5.8e-05 | 0 | 0.00015 | -1 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.6.1.16 | Glutamine--fructose-6-phosphate transaminase (isomerizing). | 0.42 | 0.68 | 1 | 0.21 | 1 | 1 | 0.56 | 0.96 | 1 | 0.32 | 0.59 | 1 | 0.74 | 0.96 | 1 | 0.72 | 0.94 | 1 | 0.032 | 0.032 | 0.032 | 0.031 | 0.033 | 0.0074 | 0.028 | 0.029 | 0.0084 | -0.15 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.034 | 0.031 | 0.007 | 0.032 | 0.032 | 0.0033 | -0.087 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.8.98.1 | Dihydromethanophenazine:CoB--CoM heterodisulfide reductase. | 0.65 | 0.84 | 1 | 0.67 | 1 | 1 | 0.24 | 0.88 | 1 | 0.85 | 0.95 | 1 | 0.73 | 0.96 | 1 | 0.13 | 0.68 | 1 | 0.00098 | 0.00095 | 9e-04 | 0.0011 | 0.0012 | 0.00061 | 0.0011 | 0.00095 | 0.00094 | 0 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.001 | 0.00096 | 0.00056 | 0.00064 | 0.00052 | 0.00051 | -0.64 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 2.7.7.8 | Polyribonucleotide nucleotidyltransferase. | 0.058 | 0.49 | 1 | 0.48 | 1 | 1 | 0.96 | 1 | 1 | 0.074 | 0.43 | 1 | 0.74 | 0.96 | 1 | 0.34 | 0.8 | 1 | 0.035 | 0.035 | 0.034 | 0.032 | 0.031 | 0.0047 | 0.031 | 0.032 | 0.0073 | -0.046 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.039 | 0.037 | 0.009 | 0.037 | 0.037 | 0.0044 | -0.076 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.1.55 | 3-hydroxybutyryl-CoA dehydratase. | 0.44 | 0.69 | 1 | 0.83 | 1 | 1 | 0.25 | 0.89 | 1 | 0.74 | 0.9 | 1 | 0.71 | 0.95 | 1 | 0.54 | 0.9 | 1 | 0.005 | 0.005 | 0.0048 | 0.005 | 0.0047 | 0.001 | 0.0049 | 0.0049 | 0.0014 | -0.029 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0046 | 0.0045 | 0.0019 | 0.0054 | 0.0058 | 0.0021 | 0.23 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 7.3.2.1 | ABC-type phosphate transporter. | 0.099 | 0.49 | 1 | 0.22 | 1 | 1 | 0.056 | 0.71 | 1 | 0.098 | 0.46 | 1 | 0.047 | 0.81 | 1 | 0.0083 | 0.55 | 1 | 0.017 | 0.017 | 0.016 | 0.015 | 0.014 | 0.0029 | 0.017 | 0.015 | 0.0056 | 0.18 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.018 | 0.017 | 0.0044 | 0.016 | 0.016 | 0.0029 | -0.17 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.1.189 | 23S rRNA (uracil(747)-C(5))-methyltransferase. | 0.58 | 0.79 | 1 | 0.54 | 1 | 1 | 0.66 | 0.97 | 1 | 0.44 | 0.69 | 1 | 0.13 | 0.87 | 1 | 0.22 | 0.74 | 1 | 0.025 | 0.025 | 0.024 | 0.023 | 0.023 | 0.0055 | 0.025 | 0.026 | 0.0097 | 0.12 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.025 | 0.027 | 0.0083 | 0.025 | 0.025 | 0.0039 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.1.345 | Phosphatidyl-myo-inositol alpha-mannosyltransferase. | 0.95 | 0.98 | 1 | 0.86 | 1 | 1 | 0.43 | 0.93 | 1 | 0.92 | 0.98 | 1 | 0.98 | 0.99 | 1 | 0.54 | 0.9 | 1 | 0.00044 | 2e-04 | 0 | 0.00024 | 0 | 0.00035 | 0.00022 | 0 | 0.00035 | -0.13 | 16 / 36 (44%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 0.00029 | 0 | 0.00058 | 4.7e-05 | 0 | 6e-05 | -2.6 | 5 / 11 (45%) | 5 / 11 (45%) | 0.455 | 0.455 |
| 1.1.99.1 | Choline dehydrogenase. | 0.056 | 0.49 | 1 | 0.12 | 1 | 1 | 0.062 | 0.71 | 1 | 0.043 | 0.43 | 1 | 0.28 | 0.89 | 1 | 0.13 | 0.68 | 1 | 0.00043 | 6e-05 | 0 | 0.00017 | 0 | 0.00035 | 0.00012 | 0 | 0.00031 | -0.5 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 1.3e-05 | 0 | 3.5e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 2.7.7.7 | DNA-directed DNA polymerase. | 0.093 | 0.49 | 1 | 0.94 | 1 | 1 | 0.49 | 0.94 | 1 | 0.13 | 0.47 | 1 | 0.2 | 0.87 | 1 | 0.044 | 0.65 | 1 | 0.25 | 0.25 | 0.25 | 0.23 | 0.22 | 0.028 | 0.23 | 0.24 | 0.047 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.26 | 0.25 | 0.055 | 0.25 | 0.26 | 0.027 | -0.057 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.11.9 | Xaa-Pro aminopeptidase. | 0.066 | 0.49 | 1 | 0.99 | 1 | 1 | 0.55 | 0.95 | 1 | 0.06 | 0.43 | 1 | 0.84 | 0.98 | 1 | 0.62 | 0.92 | 1 | 0.016 | 0.016 | 0.015 | 0.012 | 0.0096 | 0.0059 | 0.012 | 0.0071 | 0.0088 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.018 | 0.019 | 0.0089 | 0.019 | 0.019 | 0.0054 | 0.078 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.205 | Protein-N(pi)-phosphohistidine--D-cellobiose phosphotransferase. | 0.091 | 0.49 | 1 | 0.64 | 1 | 1 | 0.52 | 0.95 | 1 | 0.13 | 0.47 | 1 | 0.39 | 0.9 | 1 | 0.3 | 0.78 | 1 | 0.00024 | 4e-05 | 0 | 1e-04 | 0 | 0.00022 | 9.4e-05 | 0 | 0.00014 | -0.089 | 6 / 36 (17%) | 2 / 7 (29%) | 3 / 7 (43%) | 0.286 | 0.429 | 6e-06 | 0 | 2e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.7.1.58 | 2-dehydro-3-deoxygalactonokinase. | 0.18 | 0.52 | 1 | 0.85 | 1 | 1 | 0.38 | 0.9 | 1 | 0.32 | 0.59 | 1 | 0.5 | 0.93 | 1 | 0.68 | 0.93 | 1 | 0.00029 | 9e-05 | 0 | 0.00026 | 0 | 0.00067 | 0.00013 | 0 | 0.00028 | -1 | 11 / 36 (31%) | 2 / 7 (29%) | 3 / 7 (43%) | 0.286 | 0.429 | 1.7e-05 | 0 | 5.8e-05 | 3.1e-05 | 0 | 4.1e-05 | 0.87 | 1 / 11 (9.1%) | 5 / 11 (45%) | 0.0909 | 0.455 |
| 6.2.1.25 | Benzoate--CoA ligase. | 0.86 | 0.94 | 1 | 0.77 | 1 | 1 | 0.31 | 0.89 | 1 | 0.96 | 0.99 | 1 | 0.92 | 0.99 | 1 | 0.54 | 0.9 | 1 | 0.0014 | 0.00027 | 0 | 0.00016 | 0 | 0.00043 | 4.6e-05 | 0 | 0.00012 | -1.8 | 7 / 36 (19%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 8.7e-05 | 0 | 0.00026 | 0.00065 | 0 | 0.0014 | 2.9 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 5.1.3.30 | D-psicose 3-epimerase. | 0.21 | 0.54 | 1 | 0.23 | 1 | 1 | 0.72 | 0.98 | 1 | 0.31 | 0.59 | 1 | 0.94 | 0.99 | 1 | 0.37 | 0.82 | 1 | 0.00032 | 2.7e-05 | 0 | 8.4e-05 | 0 | 0.00022 | 4.8e-05 | 0 | 0.00013 | -0.81 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3e-06 | 0 | 9.9e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 4.1.99.1 | Tryptophanase. | 0.94 | 0.97 | 1 | 0.93 | 1 | 1 | 0.58 | 0.96 | 1 | 0.99 | 1 | 1 | 0.91 | 0.99 | 1 | 0.38 | 0.82 | 1 | 0.00077 | 0.00067 | 0.00049 | 0.00059 | 0.00033 | 0.00062 | 0.00064 | 0.00063 | 6e-04 | 0.12 | 31 / 36 (86%) | 6 / 7 (86%) | 5 / 7 (71%) | 0.857 | 0.714 | 0.00069 | 0.00038 | 0.00092 | 0.00071 | 0.00055 | 0.00061 | 0.041 | 9 / 11 (82%) | 11 / 11 (100%) | 0.818 | 1 |
| 2.6.1.85 | Aminodeoxychorismate synthase. | 0.13 | 0.5 | 1 | 0.36 | 1 | 1 | 0.87 | 1 | 1 | 0.056 | 0.43 | 1 | 0.054 | 0.81 | 1 | 0.33 | 0.8 | 1 | 0.0074 | 0.0074 | 0.007 | 0.006 | 0.0056 | 0.0016 | 0.0067 | 0.0068 | 0.0014 | 0.16 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.008 | 0.007 | 0.0039 | 0.0083 | 0.0083 | 0.0021 | 0.053 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.7.3 | Pantetheine-phosphate adenylyltransferase. | 0.043 | 0.49 | 1 | 0.53 | 1 | 1 | 0.2 | 0.87 | 1 | 0.047 | 0.43 | 1 | 0.084 | 0.86 | 1 | 0.014 | 0.55 | 1 | 0.0092 | 0.0092 | 0.0091 | 0.0083 | 0.0079 | 0.0013 | 0.0089 | 0.0084 | 0.0023 | 0.1 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.01 | 0.0095 | 0.0025 | 0.009 | 0.0091 | 0.0013 | -0.15 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.1.129 | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase. | 0.26 | 0.56 | 1 | 0.4 | 1 | 1 | 0.057 | 0.71 | 1 | 0.06 | 0.43 | 1 | 0.41 | 0.91 | 1 | 0.038 | 0.63 | 1 | 0.002 | 0.0019 | 0.0012 | 0.0019 | 0.0016 | 8e-04 | 0.0016 | 0.00071 | 0.0017 | -0.25 | 34 / 36 (94%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0014 | 0.00064 | 0.0017 | 0.0026 | 0.0013 | 0.003 | 0.89 | 9 / 11 (82%) | 11 / 11 (100%) | 0.818 | 1 |
| 1.3.99.- | 0.22 | 0.54 | 1 | 0.81 | 1 | 1 | 0.9 | 1 | 1 | 0.14 | 0.47 | 1 | 0.74 | 0.96 | 1 | 0.87 | 0.97 | 1 | 0.0081 | 0.0081 | 0.0043 | 0.0047 | 0.0033 | 0.0033 | 0.0044 | 0.0034 | 0.0039 | -0.095 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.012 | 0.0051 | 0.021 | 0.0088 | 0.0085 | 0.0077 | -0.45 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 1.1.1.52 | 3-alpha-hydroxycholanate dehydrogenase (NAD(+)). | 0.32 | 0.59 | 1 | 0.92 | 1 | 1 | 0.71 | 0.98 | 1 | 0.45 | 0.7 | 1 | 0.92 | 0.99 | 1 | 0.55 | 0.9 | 1 | 6.6e-05 | 1.8e-05 | 0 | 3.2e-05 | 0 | 4.8e-05 | 2.9e-05 | 0 | 4.5e-05 | -0.14 | 10 / 36 (28%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 1.4e-05 | 0 | 2.9e-05 | 6.8e-06 | 0 | 2.3e-05 | -1 | 3 / 11 (27%) | 1 / 11 (9.1%) | 0.273 | 0.0909 |
| 5.5.1.6 | Chalcone isomerase. | 0.73 | 0.87 | 1 | 0.98 | 1 | 1 | 0.63 | 0.96 | 1 | 0.9 | 0.97 | 1 | 0.88 | 0.99 | 1 | 0.8 | 0.96 | 1 | 0.00012 | 1.6e-05 | 0 | 1.6e-05 | 0 | 4.2e-05 | 1.5e-05 | 0 | 4e-05 | -0.093 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 8.7e-06 | 0 | 2.9e-05 | 2.4e-05 | 0 | 5.4e-05 | 1.5 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 2.7.8.31 | Undecaprenyl-phosphate glucose phosphotransferase. | 0.086 | 0.49 | 1 | 0.86 | 1 | 1 | 0.45 | 0.93 | 1 | 0.24 | 0.54 | 1 | 0.55 | 0.93 | 1 | 0.79 | 0.96 | 1 | 0.0092 | 0.0092 | 0.0086 | 0.01 | 0.0099 | 0.0024 | 0.01 | 0.01 | 0.0025 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0075 | 0.0076 | 0.0025 | 0.0096 | 0.0077 | 0.0049 | 0.36 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.77 | Lactaldehyde reductase. | 0.96 | 0.98 | 1 | 0.44 | 1 | 1 | 0.27 | 0.89 | 1 | 0.57 | 0.79 | 1 | 0.92 | 0.99 | 1 | 0.73 | 0.94 | 1 | 0.016 | 0.016 | 0.015 | 0.016 | 0.016 | 0.0051 | 0.015 | 0.015 | 0.0048 | -0.093 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.016 | 0.014 | 0.0052 | 0.017 | 0.016 | 0.0035 | 0.087 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.26 | Riboflavin kinase. | 0.19 | 0.53 | 1 | 0.63 | 1 | 1 | 0.83 | 1 | 1 | 0.19 | 0.5 | 1 | 0.066 | 0.86 | 1 | 0.1 | 0.66 | 1 | 0.013 | 0.013 | 0.013 | 0.012 | 0.012 | 0.002 | 0.012 | 0.014 | 0.0027 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.013 | 0.013 | 0.0023 | 0.013 | 0.013 | 0.0015 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.2.13 | UDP-4-amino-4-deoxy-L-arabinose formyltransferase. | 0.31 | 0.59 | 1 | 0.17 | 1 | 1 | 0.52 | 0.95 | 1 | 0.49 | 0.73 | 1 | 0.48 | 0.92 | 1 | 0.19 | 0.72 | 1 | 0.00035 | 3.9e-05 | 0 | 1e-04 | 0 | 0.00027 | 7.7e-05 | 0 | 2e-04 | -0.38 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 8.9e-06 | 0 | 3e-05 | 5.4e-06 | 0 | 1.8e-05 | -0.72 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.5.1.42 | Geranylgeranylglycerol-phosphate geranylgeranyltransferase. | 0.89 | 0.95 | 1 | 0.2 | 1 | 1 | 0.51 | 0.95 | 1 | 0.83 | 0.94 | 1 | 0.31 | 0.9 | 1 | 0.76 | 0.95 | 1 | 0.00022 | 2.5e-05 | 0 | 4.9e-05 | 0 | 0.00013 | 0 | 0 | 0 | -Inf | 4 / 36 (11%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 3e-05 | 0 | 7e-05 | 2.1e-05 | 0 | 6.8e-05 | -0.51 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 3.5.2.3 | Dihydroorotase. | 0.021 | 0.49 | 1 | 0.96 | 1 | 1 | 0.79 | 1 | 1 | 0.073 | 0.43 | 1 | 0.17 | 0.87 | 1 | 0.23 | 0.75 | 1 | 0.022 | 0.022 | 0.021 | 0.019 | 0.02 | 0.0034 | 0.019 | 0.02 | 0.004 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.023 | 0.022 | 0.0046 | 0.024 | 0.024 | 0.0036 | 0.061 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.7.48 | RNA-directed RNA polymerase. | 0.95 | 0.98 | 1 | 0.92 | 1 | 1 | 0.41 | 0.92 | 1 | 0.55 | 0.78 | 1 | 0.95 | 0.99 | 1 | 0.73 | 0.94 | 1 | 0.00056 | 0.00023 | 0 | 0.00021 | 0 | 0.00039 | 0.00023 | 0 | 0.00057 | 0.13 | 15 / 36 (42%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0.00013 | 6.5e-05 | 0.00017 | 0.00035 | 0 | 0.00049 | 1.4 | 6 / 11 (55%) | 5 / 11 (45%) | 0.545 | 0.455 |
| 2.1.1.98 | Diphthine synthase. | 0.52 | 0.75 | 1 | 0.41 | 1 | 1 | 0.26 | 0.89 | 1 | 0.71 | 0.89 | 1 | 0.54 | 0.93 | 1 | 0.37 | 0.82 | 1 | 0.00019 | 4.1e-05 | 0 | 6.4e-05 | 0 | 0.00015 | 1.5e-05 | 0 | 4.1e-05 | -2.1 | 8 / 36 (22%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.4e-05 | 0 | 5.5e-05 | 6.1e-05 | 0 | 0.00013 | 1.3 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 6.2.1.12 | 4-coumarate--CoA ligase. | 0.92 | 0.96 | 1 | 0.27 | 1 | 1 | 0.24 | 0.88 | 1 | 0.44 | 0.69 | 1 | 0.73 | 0.96 | 1 | 0.44 | 0.85 | 1 | 0.0011 | 0.00053 | 0 | 0.00079 | 0 | 0.0016 | 0.00027 | 0 | 0.00058 | -1.5 | 17 / 36 (47%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0.00052 | 0.00019 | 0.00083 | 0.00053 | 0.00046 | 0.00065 | 0.027 | 6 / 11 (55%) | 7 / 11 (64%) | 0.545 | 0.636 |
| 1.1.1.30 | 3-hydroxybutyrate dehydrogenase. | 0.63 | 0.83 | 1 | 0.82 | 1 | 1 | 0.23 | 0.88 | 1 | 0.37 | 0.64 | 1 | 0.9 | 0.99 | 1 | 0.39 | 0.83 | 1 | 0.0012 | 0.0012 | 0.00094 | 9e-04 | 0.00096 | 0.00051 | 0.00087 | 0.00073 | 0.00064 | -0.049 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.001 | 0.00085 | 0.00061 | 0.0016 | 0.0016 | 0.00095 | 0.68 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.2.50 | 6-carboxytetrahydropterin synthase. | 0.26 | 0.56 | 1 | 0.18 | 1 | 1 | 0.64 | 0.97 | 1 | 0.31 | 0.59 | 1 | 0.11 | 0.87 | 1 | 0.46 | 0.86 | 1 | 0.00018 | 2e-05 | 0 | 3.4e-05 | 0 | 8.9e-05 | 5.4e-05 | 0 | 1e-04 | 0.67 | 4 / 36 (11%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 0 | 0 | 0 | 9e-06 | 0 | 3e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.5.1.141 | Heme o synthase. | 0.44 | 0.69 | 1 | 0.28 | 1 | 1 | 0.16 | 0.85 | 1 | 0.8 | 0.92 | 1 | 0.18 | 0.87 | 1 | 0.1 | 0.66 | 1 | 0.00032 | 5.3e-05 | 0 | 0.00012 | 0 | 0.00031 | 0.00012 | 0 | 0.00028 | 0 | 6 / 36 (17%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 1.5e-05 | 0 | 3.3e-05 | 5.4e-06 | 0 | 1.8e-05 | -1.5 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 3.4.22.8 | Clostripain. | 0.47 | 0.72 | 1 | 0.78 | 1 | 1 | 0.29 | 0.89 | 1 | 0.3 | 0.58 | 1 | 0.48 | 0.92 | 1 | 0.15 | 0.7 | 1 | 0.0012 | 0.0011 | 0.00086 | 0.001 | 0.00079 | 0.00075 | 0.0011 | 0.0013 | 0.00088 | 0.14 | 33 / 36 (92%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00085 | 8e-04 | 0.00072 | 0.0013 | 9e-04 | 0.0013 | 0.61 | 9 / 11 (82%) | 11 / 11 (100%) | 0.818 | 1 |
| 2.3.2.13 | Protein-glutamine gamma-glutamyltransferase. | 0.63 | 0.83 | 1 | 0.6 | 1 | 1 | 0.97 | 1 | 1 | 0.79 | 0.92 | 1 | 0.23 | 0.88 | 1 | 0.85 | 0.96 | 1 | 0.0017 | 0.0012 | 0.00063 | 0.00093 | 0.00045 | 0.0012 | 0.001 | 0.00021 | 0.0016 | 0.1 | 26 / 36 (72%) | 6 / 7 (86%) | 4 / 7 (57%) | 0.857 | 0.571 | 0.0014 | 0.0012 | 0.0016 | 0.0013 | 0.00082 | 0.0017 | -0.11 | 9 / 11 (82%) | 7 / 11 (64%) | 0.818 | 0.636 |
| 1.2.1.91 | 3-oxo-5.6-dehydrosuberyl-CoA semialdehyde dehydrogenase. | 0.83 | 0.92 | 1 | 0.48 | 1 | 1 | 0.91 | 1 | 1 | 0.87 | 0.96 | 1 | 0.51 | 0.93 | 1 | 0.85 | 0.96 | 1 | 0.00022 | 1.9e-05 | 0 | 1.7e-05 | 0 | 4.5e-05 | 0 | 0 | 0 | -Inf | 3 / 36 (8.3%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 4.4e-05 | 0 | 0.00015 | 6.5e-06 | 0 | 2.1e-05 | -2.8 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 5.4.99.18 | 5-(carboxyamino)imidazole ribonucleotide mutase. | 0.73 | 0.87 | 1 | 0.65 | 1 | 1 | 0.42 | 0.93 | 1 | 0.44 | 0.69 | 1 | 0.18 | 0.87 | 1 | 0.086 | 0.66 | 1 | 0.0096 | 0.0096 | 0.0095 | 0.0095 | 0.0097 | 0.0024 | 0.0099 | 0.011 | 0.0028 | 0.06 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0097 | 0.0096 | 0.0019 | 0.0092 | 0.0092 | 0.0014 | -0.076 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.1.2.3 | 3-hydroxybutyryl-CoA epimerase. | 0.17 | 0.5 | 1 | 0.64 | 1 | 1 | 0.97 | 1 | 1 | 0.17 | 0.49 | 1 | 0.69 | 0.95 | 1 | 0.82 | 0.96 | 1 | 8e-04 | 0.00018 | 0 | 0.00042 | 0 | 0.0011 | 0.00044 | 0 | 0.0011 | 0.067 | 8 / 36 (22%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 1.4e-05 | 0 | 3.3e-05 | 1.6e-05 | 0 | 5.4e-05 | 0.19 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 1.1.1.60 | 2-hydroxy-3-oxopropionate reductase. | 0.85 | 0.93 | 1 | 0.45 | 1 | 1 | 0.37 | 0.9 | 1 | 0.53 | 0.76 | 1 | 0.87 | 0.99 | 1 | 0.29 | 0.78 | 1 | 0.0024 | 0.0024 | 0.0022 | 0.0027 | 0.0015 | 0.0021 | 0.0024 | 0.0022 | 0.0015 | -0.17 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0027 | 0.0026 | 0.0012 | 0.0019 | 0.0017 | 0.00081 | -0.51 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.3.2.14 | Enterobactin synthase. | 0.32 | 0.59 | 1 | 0.51 | 1 | 1 | 0.43 | 0.93 | 1 | 0.58 | 0.8 | 1 | 0.45 | 0.92 | 1 | 0.37 | 0.82 | 1 | 0.00069 | 0.00017 | 0 | 0.00039 | 0 | 0.001 | 0.00042 | 0 | 0.0011 | 0.11 | 9 / 36 (25%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 2.7e-05 | 0 | 7.2e-05 | 2.1e-05 | 0 | 4.7e-05 | -0.36 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 2.5.1.75 | tRNA dimethylallyltransferase. | 0.12 | 0.5 | 1 | 0.32 | 1 | 1 | 0.65 | 0.97 | 1 | 0.068 | 0.43 | 1 | 0.95 | 0.99 | 1 | 0.59 | 0.91 | 1 | 0.014 | 0.014 | 0.014 | 0.013 | 0.013 | 0.0018 | 0.012 | 0.011 | 0.0027 | -0.12 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.016 | 0.014 | 0.0045 | 0.015 | 0.014 | 0.0021 | -0.093 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.1.66 | Transferred entry: 2.1.1.230. | 0.38 | 0.65 | 1 | 0.41 | 1 | 1 | 0.13 | 0.82 | 1 | 0.25 | 0.54 | 1 | 0.16 | 0.87 | 1 | 0.065 | 0.66 | 1 | 0.00044 | 0.00021 | 0 | 0.00019 | 0 | 0.00041 | 0.00021 | 6.8e-05 | 0.00031 | 0.14 | 17 / 36 (47%) | 2 / 7 (29%) | 4 / 7 (57%) | 0.286 | 0.571 | 0.00028 | 9.5e-05 | 0.00037 | 0.00014 | 0 | 0.00027 | -1 | 6 / 11 (55%) | 5 / 11 (45%) | 0.545 | 0.455 |
| 3.6.1.29 | Bis(5'-adenosyl)-triphosphatase. | 0.87 | 0.95 | 1 | 0.58 | 1 | 1 | 0.8 | 1 | 1 | 0.97 | 0.99 | 1 | 0.63 | 0.95 | 1 | 0.96 | 0.99 | 1 | 0.00016 | 7.1e-05 | 0 | 7.2e-05 | 0 | 0.00013 | 0.00012 | 0 | 0.00021 | 0.74 | 16 / 36 (44%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 6.2e-05 | 0 | 1e-04 | 5.2e-05 | 3e-05 | 7.2e-05 | -0.25 | 4 / 11 (36%) | 6 / 11 (55%) | 0.364 | 0.545 |
| 3.1.11.5 | Exodeoxyribonuclease V. | 0.52 | 0.75 | 1 | 0.92 | 1 | 1 | 0.74 | 0.99 | 1 | 0.84 | 0.94 | 1 | 0.62 | 0.95 | 1 | 0.91 | 0.98 | 1 | 0.0044 | 0.0044 | 0.0038 | 0.0048 | 0.0048 | 0.0038 | 0.0048 | 0.0044 | 0.0032 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0037 | 0.0039 | 0.0018 | 0.0045 | 0.0037 | 0.0033 | 0.28 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.7.81 | Pseudaminic acid cytidylyltransferase. | 0.21 | 0.54 | 1 | 0.57 | 1 | 1 | 0.96 | 1 | 1 | 0.39 | 0.66 | 1 | 0.51 | 0.93 | 1 | 0.99 | 1 | 1 | 0.00033 | 0.00023 | 9.4e-05 | 0.00039 | 0.00011 | 0.00054 | 0.00034 | 0.00024 | 0.00042 | -0.2 | 25 / 36 (69%) | 6 / 7 (86%) | 4 / 7 (57%) | 0.857 | 0.571 | 0.00015 | 9.5e-05 | 0.00019 | 0.00012 | 5.9e-05 | 0.00021 | -0.32 | 8 / 11 (73%) | 7 / 11 (64%) | 0.727 | 0.636 |
| 2.7.1.24 | Dephospho-CoA kinase. | 0.0039 | 0.49 | 1 | 0.16 | 1 | 1 | 0.056 | 0.71 | 1 | 0.016 | 0.42 | 1 | 0.013 | 0.76 | 1 | 0.0044 | 0.48 | 1 | 0.0097 | 0.0097 | 0.0094 | 0.0081 | 0.0084 | 0.0015 | 0.0091 | 0.0098 | 0.0018 | 0.17 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.0099 | 0.0027 | 0.01 | 0.0095 | 0.0016 | -0.14 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.196 | Limit dextrin alpha-1.6-maltotetraose-hydrolase. | 0.031 | 0.49 | 1 | 0.014 | 0.89 | 1 | 0.019 | 0.71 | 1 | 0.018 | 0.42 | 1 | 0.081 | 0.86 | 1 | 0.07 | 0.66 | 1 | 0.00051 | 7.1e-05 | 0 | 0.00026 | 0 | 0.00062 | 8.7e-05 | 0 | 0.00023 | -1.6 | 5 / 36 (14%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 0 | 0 | 0 | 1e-05 | 0 | 3.4e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 3.4.21.110 | C5a peptidase. | 0.36 | 0.63 | 1 | 0.65 | 1 | 1 | 0.062 | 0.71 | 1 | 0.4 | 0.67 | 1 | 0.77 | 0.97 | 1 | 0.074 | 0.66 | 1 | 0.00063 | 0.00052 | 0.00036 | 0.00047 | 4e-04 | 0.00041 | 0.00068 | 0.00046 | 0.00077 | 0.53 | 30 / 36 (83%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 7e-04 | 0.00077 | 0.00046 | 0.00028 | 7.1e-05 | 0.00051 | -1.3 | 10 / 11 (91%) | 8 / 11 (73%) | 0.909 | 0.727 |
| 6.3.5.- | 0.13 | 0.5 | 1 | 0.93 | 1 | 1 | 0.57 | 0.96 | 1 | 0.15 | 0.47 | 1 | 0.47 | 0.92 | 1 | 0.22 | 0.74 | 1 | 0.026 | 0.026 | 0.027 | 0.023 | 0.022 | 0.0049 | 0.023 | 0.026 | 0.0068 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.029 | 0.029 | 0.0097 | 0.026 | 0.026 | 0.0078 | -0.16 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 4.3.2.- | 0.081 | 0.49 | 1 | 0.072 | 1 | 1 | 0.05 | 0.71 | 1 | 0.066 | 0.43 | 1 | 0.38 | 0.9 | 1 | 0.23 | 0.75 | 1 | 0.00015 | 1.7e-05 | 0 | 5.3e-05 | 0 | 0.00013 | 2.9e-05 | 0 | 7.7e-05 | -0.87 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 | |
| 1.2.1.58 | Phenylglyoxylate dehydrogenase (acylating). | 0.024 | 0.49 | 1 | 0.23 | 1 | 1 | 0.071 | 0.73 | 1 | 0.025 | 0.42 | 1 | 0.31 | 0.9 | 1 | 0.11 | 0.66 | 1 | 0.00016 | 3e-05 | 0 | 6.5e-05 | 0 | 8.5e-05 | 2e-05 | 0 | 4e-05 | -1.7 | 7 / 36 (19%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 0 | 0 | 0 | 4.6e-05 | 0 | 1e-04 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 3.6.3.21 | Transferred entry: 7.4.2.1. | 0.16 | 0.5 | 1 | 0.38 | 1 | 1 | 0.2 | 0.87 | 1 | 0.073 | 0.43 | 1 | 0.54 | 0.93 | 1 | 0.29 | 0.78 | 1 | 0.0065 | 0.0065 | 0.0067 | 0.0056 | 0.0056 | 0.0012 | 0.0051 | 0.0044 | 0.0023 | -0.13 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.007 | 0.0067 | 0.0027 | 0.0076 | 0.0078 | 0.0015 | 0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.5.1.94 | Adenosyl-chloride synthase. | 0.85 | 0.93 | 1 | 0.088 | 1 | 1 | 0.068 | 0.73 | 1 | 0.8 | 0.92 | 1 | 0.16 | 0.87 | 1 | 0.18 | 0.72 | 1 | 0.00033 | 0.00024 | 9.1e-05 | 0.00022 | 0.00011 | 0.00033 | 0.00052 | 0.00036 | 0.00053 | 1.2 | 26 / 36 (72%) | 5 / 7 (71%) | 6 / 7 (86%) | 0.714 | 0.857 | 0.00023 | 7.2e-05 | 0.00039 | 8.7e-05 | 7.5e-05 | 9.8e-05 | -1.4 | 7 / 11 (64%) | 8 / 11 (73%) | 0.636 | 0.727 |
| 1.1.1.122 | D-threo-aldose 1-dehydrogenase. | 0.12 | 0.5 | 1 | 0.36 | 1 | 1 | 0.19 | 0.87 | 1 | 0.046 | 0.43 | 1 | 0.22 | 0.88 | 1 | 0.26 | 0.76 | 1 | 0.00062 | 0.00038 | 0.00012 | 0.00054 | 0.00014 | 0.00075 | 0.00051 | 0.00012 | 0.00078 | -0.082 | 22 / 36 (61%) | 7 / 7 (100%) | 5 / 7 (71%) | 1 | 0.714 | 0.00018 | 0 | 0.00034 | 0.00039 | 0 | 0.00099 | 1.1 | 5 / 11 (45%) | 5 / 11 (45%) | 0.455 | 0.455 |
| 2.3.1.39 | [Acyl-carrier-protein] S-malonyltransferase. | 0.27 | 0.56 | 1 | 0.64 | 1 | 1 | 0.82 | 1 | 1 | 0.09 | 0.45 | 1 | 0.16 | 0.87 | 1 | 0.43 | 0.85 | 1 | 0.017 | 0.017 | 0.017 | 0.015 | 0.015 | 0.0029 | 0.016 | 0.018 | 0.0043 | 0.093 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.017 | 0.017 | 0.0026 | 0.018 | 0.017 | 0.0041 | 0.082 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.1.247 | Beta-D-galactosyl-(1->4)-L-rhamnose phosphorylase. | 0.82 | 0.92 | 1 | 0.98 | 1 | 1 | 0.41 | 0.92 | 1 | 0.85 | 0.95 | 1 | 0.31 | 0.9 | 1 | 0.12 | 0.67 | 1 | 0.00088 | 0.00085 | 0.00079 | 7e-04 | 0.00079 | 0.00038 | 0.00091 | 5e-04 | 0.001 | 0.38 | 35 / 36 (97%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00079 | 0.00078 | 0.00049 | 0.00097 | 0.001 | 0.00047 | 0.3 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.4.1.2 | Transferred entry: 5.4.99.61. | 0.63 | 0.83 | 1 | 0.83 | 1 | 1 | 0.52 | 0.95 | 1 | 0.95 | 0.98 | 1 | 0.79 | 0.97 | 1 | 0.93 | 0.98 | 1 | 0.0018 | 0.0018 | 0.0019 | 0.0019 | 0.0019 | 0.00068 | 0.0018 | 0.0018 | 8e-04 | -0.078 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0017 | 0.0018 | 0.00079 | 0.0019 | 0.002 | 0.00071 | 0.16 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.6.1.87 | UDP-4-amino-4-deoxy-L-arabinose aminotransferase. | 0.25 | 0.56 | 1 | 0.44 | 1 | 1 | 0.61 | 0.96 | 1 | 0.74 | 0.9 | 1 | 0.095 | 0.86 | 1 | 0.19 | 0.72 | 1 | 0.011 | 0.011 | 0.011 | 0.011 | 0.011 | 0.0012 | 0.012 | 0.013 | 0.0029 | 0.13 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.01 | 0.009 | 0.0035 | 0.01 | 0.01 | 0.002 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.1.21 | Phosphoribosylaminoimidazole carboxylase. | 0.61 | 0.81 | 1 | 0.65 | 1 | 1 | 0.45 | 0.93 | 1 | 0.7 | 0.88 | 1 | 0.58 | 0.93 | 1 | 0.51 | 0.89 | 1 | 0.00078 | 0.00078 | 0.00078 | 0.00073 | 6e-04 | 0.00054 | 0.00087 | 0.00081 | 0.00075 | 0.25 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00061 | 0.00049 | 0.00041 | 0.00092 | 0.00085 | 0.00061 | 0.59 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.3.1.20 | Lipoate--protein ligase. | 0.24 | 0.55 | 1 | 0.63 | 1 | 1 | 0.026 | 0.71 | 1 | 0.17 | 0.49 | 1 | 0.55 | 0.93 | 1 | 0.024 | 0.57 | 1 | 0.0011 | 0.0011 | 0.00098 | 0.0011 | 0.0012 | 0.00052 | 0.0013 | 0.0011 | 0.00078 | 0.24 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0015 | 0.0012 | 0.00084 | 0.00075 | 0.00075 | 0.00027 | -1 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.1.41 | Cephalosporin-C deacetylase. | 0.43 | 0.69 | 1 | 0.52 | 1 | 1 | 0.66 | 0.97 | 1 | 0.48 | 0.72 | 1 | 0.46 | 0.92 | 1 | 0.73 | 0.94 | 1 | 0.0016 | 0.0015 | 0.0011 | 0.0011 | 0.001 | 0.00061 | 0.0014 | 0.0012 | 0.0013 | 0.35 | 34 / 36 (94%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.0016 | 0.0011 | 0.0013 | 0.0017 | 9e-04 | 0.0018 | 0.087 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 2.4.2.17 | ATP phosphoribosyltransferase. | 0.32 | 0.6 | 1 | 0.54 | 1 | 1 | 0.48 | 0.94 | 1 | 0.23 | 0.53 | 1 | 0.76 | 0.97 | 1 | 0.84 | 0.96 | 1 | 0.012 | 0.012 | 0.012 | 0.011 | 0.011 | 0.0015 | 0.011 | 0.011 | 0.0023 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.012 | 0.012 | 0.0036 | 0.013 | 0.012 | 0.0024 | 0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.168 | D-glycero-alpha-D-manno-heptose-7-phosphate kinase. | 0.67 | 0.85 | 1 | 0.36 | 1 | 1 | 0.37 | 0.9 | 1 | 0.55 | 0.78 | 1 | 0.1 | 0.86 | 1 | 0.32 | 0.79 | 1 | 0.0013 | 0.001 | 0.00042 | 0.00079 | 0.00034 | 0.00097 | 8e-04 | 6.8e-05 | 0.0013 | 0.018 | 29 / 36 (81%) | 6 / 7 (86%) | 4 / 7 (57%) | 0.857 | 0.571 | 0.001 | 0.00061 | 0.0012 | 0.0013 | 5e-04 | 0.002 | 0.38 | 10 / 11 (91%) | 9 / 11 (82%) | 0.909 | 0.818 |
| 2.1.2.9 | Methionyl-tRNA formyltransferase. | 0.008 | 0.49 | 1 | 0.72 | 1 | 1 | 0.8 | 1 | 1 | 0.026 | 0.42 | 1 | 0.65 | 0.95 | 1 | 0.3 | 0.78 | 1 | 0.014 | 0.014 | 0.014 | 0.012 | 0.012 | 0.0016 | 0.012 | 0.012 | 0.0033 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.016 | 0.015 | 0.0038 | 0.015 | 0.015 | 0.0019 | -0.093 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.3.1.15 | Diaminopropionate ammonia-lyase. | 0.9 | 0.96 | 1 | 0.66 | 1 | 1 | 0.071 | 0.73 | 1 | 0.56 | 0.79 | 1 | 0.53 | 0.93 | 1 | 0.027 | 0.61 | 1 | 0.0025 | 0.0025 | 0.0025 | 0.0026 | 0.0028 | 0.00097 | 0.003 | 0.0028 | 0.002 | 0.21 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0027 | 0.0027 | 0.00068 | 0.0019 | 0.0017 | 0.00086 | -0.51 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.5.5.- | 0.083 | 0.49 | 1 | 0.023 | 0.92 | 1 | 0.062 | 0.71 | 1 | 0.055 | 0.43 | 1 | 0.027 | 0.76 | 1 | 0.079 | 0.66 | 1 | 4e-04 | 0.00012 | 0 | 3e-04 | 4.8e-05 | 0.00065 | 0.00027 | 0 | 0.00072 | -0.15 | 11 / 36 (31%) | 4 / 7 (57%) | 1 / 7 (14%) | 0.571 | 0.143 | 1.7e-05 | 0 | 3.3e-05 | 1.9e-05 | 0 | 3.7e-05 | 0.16 | 3 / 11 (27%) | 3 / 11 (27%) | 0.273 | 0.273 | |
| 4.3.1.19 | Threonine ammonia-lyase. | 0.2 | 0.53 | 1 | 0.85 | 1 | 1 | 0.74 | 0.99 | 1 | 0.18 | 0.5 | 1 | 0.47 | 0.92 | 1 | 0.64 | 0.92 | 1 | 0.018 | 0.018 | 0.018 | 0.016 | 0.016 | 0.0032 | 0.016 | 0.017 | 0.0046 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.019 | 0.018 | 0.0069 | 0.019 | 0.02 | 0.0034 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.6.1.57 | Aromatic-amino-acid transaminase. | 0.59 | 0.79 | 1 | 0.94 | 1 | 1 | 0.66 | 0.97 | 1 | 0.31 | 0.59 | 1 | 0.75 | 0.97 | 1 | 0.61 | 0.92 | 1 | 0.00068 | 0.00056 | 0.00039 | 0.00056 | 0.00028 | 0.00073 | 0.00055 | 0.00047 | 0.00064 | -0.026 | 30 / 36 (83%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.00064 | 0.00056 | 0.00075 | 0.00051 | 0.00023 | 0.00048 | -0.33 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 3.4.23.- | 1 | 1 | 1 | 0.61 | 1 | 1 | 0.57 | 0.96 | 1 | 0.89 | 0.97 | 1 | 0.39 | 0.9 | 1 | 0.67 | 0.93 | 1 | 0.00083 | 0.00078 | 0.00067 | 0.00088 | 0.00064 | 0.00077 | 0.00087 | 0.0012 | 0.00071 | -0.016 | 34 / 36 (94%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00089 | 0.00094 | 0.00053 | 0.00057 | 5e-04 | 0.00031 | -0.64 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 | |
| 1.2.7.5 | Aldehyde ferredoxin oxidoreductase. | 0.097 | 0.49 | 1 | 0.47 | 1 | 1 | 0.75 | 0.99 | 1 | 0.26 | 0.56 | 1 | 0.64 | 0.95 | 1 | 0.53 | 0.9 | 1 | 0.00033 | 0.00028 | 0.00016 | 0.00052 | 3e-04 | 0.00069 | 0.00042 | 0.00036 | 0.00041 | -0.31 | 31 / 36 (86%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 2e-04 | 0.00013 | 0.00016 | 0.00013 | 0.00011 | 0.00011 | -0.62 | 10 / 11 (91%) | 8 / 11 (73%) | 0.909 | 0.727 |
| 3.6.3.- | 0.2 | 0.53 | 1 | 0.98 | 1 | 1 | 0.77 | 0.99 | 1 | 0.14 | 0.47 | 1 | 0.41 | 0.9 | 1 | 0.28 | 0.78 | 1 | 0.72 | 0.72 | 0.73 | 0.66 | 0.64 | 0.15 | 0.66 | 0.73 | 0.21 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.78 | 0.77 | 0.28 | 0.74 | 0.76 | 0.11 | -0.076 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 6.3.2.7 | UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--L-lysine ligase. | 0.041 | 0.49 | 1 | 0.35 | 1 | 1 | 0.23 | 0.88 | 1 | 0.054 | 0.43 | 1 | 0.067 | 0.86 | 1 | 0.083 | 0.66 | 1 | 0.0048 | 0.0046 | 0.0029 | 0.0023 | 0.0021 | 0.0021 | 0.0033 | 0.0026 | 0.0038 | 0.52 | 34 / 36 (94%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.0065 | 0.0043 | 0.0084 | 0.0048 | 0.0043 | 0.003 | -0.44 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.n2 | Blood group B linear chain alpha-1.3-galactosidase. | 0.81 | 0.91 | 1 | 0.69 | 1 | 1 | 0.44 | 0.93 | 1 | 0.65 | 0.85 | 1 | 0.88 | 0.99 | 1 | 0.66 | 0.92 | 1 | 0.0011 | 0.00086 | 0.00034 | 0.00074 | 0.00069 | 0.00072 | 0.00052 | 0.00032 | 0.00055 | -0.51 | 28 / 36 (78%) | 5 / 7 (71%) | 6 / 7 (86%) | 0.714 | 0.857 | 9e-04 | 0.00011 | 0.0017 | 0.0011 | 0.00057 | 0.0017 | 0.29 | 8 / 11 (73%) | 9 / 11 (82%) | 0.727 | 0.818 |
| 1.3.1.- | 0.097 | 0.49 | 1 | 0.38 | 1 | 1 | 0.17 | 0.85 | 1 | 0.14 | 0.47 | 1 | 0.013 | 0.76 | 1 | 0.0026 | 0.48 | 1 | 0.034 | 0.034 | 0.034 | 0.031 | 0.031 | 0.0042 | 0.033 | 0.038 | 0.009 | 0.09 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.036 | 0.036 | 0.0047 | 0.034 | 0.032 | 0.0036 | -0.082 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 1.3.1.74 | 2-alkenal reductase (NAD(P)(+)). | 0.35 | 0.63 | 1 | 0.62 | 1 | 1 | 0.79 | 1 | 1 | 0.1 | 0.46 | 1 | 0.22 | 0.88 | 1 | 0.57 | 0.9 | 1 | 0.0054 | 0.0054 | 0.0051 | 0.0046 | 0.0048 | 0.0021 | 0.0049 | 0.0055 | 0.0025 | 0.091 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0056 | 0.0052 | 0.0024 | 0.0062 | 0.0051 | 0.0029 | 0.15 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.6.99.5 | Transferred entry: 1.6.5.11. | 0.13 | 0.5 | 1 | 0.55 | 1 | 1 | 0.96 | 1 | 1 | 0.19 | 0.5 | 1 | 0.82 | 0.98 | 1 | 0.24 | 0.76 | 1 | 0.0022 | 0.0022 | 0.0016 | 0.0034 | 0.002 | 0.0039 | 0.0029 | 0.0023 | 0.0025 | -0.23 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0017 | 0.0015 | 0.0016 | 0.0016 | 0.00085 | 0.0016 | -0.087 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.1.222 | Phosphate propanoyltransferase. | 0.98 | 0.99 | 1 | 0.27 | 1 | 1 | 0.2 | 0.87 | 1 | 0.81 | 0.93 | 1 | 0.16 | 0.87 | 1 | 0.11 | 0.66 | 1 | 0.0038 | 0.0038 | 0.004 | 0.0037 | 0.0038 | 0.0011 | 0.0043 | 0.0043 | 0.0016 | 0.22 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0038 | 0.0041 | 0.0012 | 0.0035 | 0.0037 | 0.0013 | -0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.21.7 | Deoxyribonuclease V. | 0.71 | 0.86 | 1 | 0.048 | 0.97 | 1 | 0.3 | 0.89 | 1 | 0.91 | 0.97 | 1 | 0.081 | 0.86 | 1 | 0.45 | 0.86 | 1 | 0.00022 | 9.3e-05 | 0 | 0.00017 | 0 | 0.00022 | 3.4e-05 | 0 | 9e-05 | -2.3 | 15 / 36 (42%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 0.00011 | 6.2e-05 | 0.00013 | 6.6e-05 | 0 | 0.00012 | -0.74 | 6 / 11 (55%) | 5 / 11 (45%) | 0.545 | 0.455 |
| 7.5.2.11 | ABC-type D-galactose transporter. | 0.23 | 0.54 | 1 | 0.094 | 1 | 1 | 0.75 | 0.99 | 1 | 0.42 | 0.68 | 1 | 0.28 | 0.88 | 1 | 0.66 | 0.92 | 1 | 0.00036 | 6e-05 | 0 | 0.00019 | 0 | 0.00044 | 7.7e-05 | 0 | 2e-04 | -1.3 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.7e-05 | 0 | 7.2e-05 | 0 | 0 | 0 | -Inf | 3 / 11 (27%) | 0 / 11 (0%) | 0.273 | 0 |
| 3.5.4.33 | tRNA(adenine(34)) deaminase. | 0.1 | 0.49 | 1 | 0.57 | 1 | 1 | 0.73 | 0.99 | 1 | 0.093 | 0.45 | 1 | 0.98 | 0.99 | 1 | 0.32 | 0.8 | 1 | 0.0097 | 0.0097 | 0.01 | 0.0091 | 0.0095 | 0.0021 | 0.0087 | 0.01 | 0.0029 | -0.065 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.011 | 0.0023 | 0.0098 | 0.0098 | 0.0015 | -0.17 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.1.47 | N-acetyldiaminopimelate deacetylase. | 0.21 | 0.54 | 1 | 0.76 | 1 | 1 | 0.19 | 0.87 | 1 | 0.19 | 0.5 | 1 | 0.73 | 0.96 | 1 | 0.24 | 0.76 | 1 | 0.0014 | 0.0014 | 0.0013 | 0.0013 | 0.00071 | 0.0011 | 0.0014 | 0.0013 | 0.0012 | 0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0018 | 0.0015 | 0.001 | 0.0011 | 0.00086 | 0.00053 | -0.71 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.2.8 | Ribosylpyrimidine nucleosidase. | 0.052 | 0.49 | 1 | 0.4 | 1 | 1 | 0.26 | 0.89 | 1 | 0.0056 | 0.36 | 1 | 0.023 | 0.76 | 1 | 0.031 | 0.62 | 1 | 0.0035 | 0.0035 | 0.0017 | 0.0013 | 0.00059 | 0.0016 | 0.0019 | 8e-04 | 0.0021 | 0.55 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0057 | 0.0022 | 0.011 | 0.0037 | 0.0028 | 0.0035 | -0.62 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.1.42 | Nicotinamide-nucleotide amidase. | 0.74 | 0.88 | 1 | 0.14 | 1 | 1 | 0.58 | 0.96 | 1 | 0.6 | 0.81 | 1 | 0.2 | 0.87 | 1 | 0.84 | 0.96 | 1 | 0.00079 | 0.00075 | 0.00072 | 0.00078 | 0.00079 | 0.00053 | 0.00053 | 0.00047 | 0.00046 | -0.56 | 34 / 36 (94%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00087 | 0.00064 | 0.00064 | 0.00075 | 0.00075 | 6e-04 | -0.21 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 2.5.1.55 | 3-deoxy-8-phosphooctulonate synthase. | 0.49 | 0.73 | 1 | 0.87 | 1 | 1 | 0.36 | 0.9 | 1 | 0.3 | 0.58 | 1 | 0.79 | 0.97 | 1 | 0.55 | 0.9 | 1 | 0.0012 | 0.0012 | 0.00078 | 0.0011 | 0.00081 | 0.00063 | 0.0012 | 0.00053 | 0.0012 | 0.13 | 34 / 36 (94%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00096 | 0.00056 | 0.0012 | 0.0014 | 0.0011 | 0.0015 | 0.54 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 5.1.3.- | 0.9 | 0.96 | 1 | 0.6 | 1 | 1 | 0.47 | 0.93 | 1 | 0.42 | 0.69 | 1 | 0.94 | 0.99 | 1 | 0.87 | 0.97 | 1 | 0.006 | 0.006 | 0.0055 | 0.0059 | 0.0058 | 0.0024 | 0.0053 | 0.005 | 0.002 | -0.15 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0059 | 0.0054 | 0.0015 | 0.0066 | 0.006 | 0.0044 | 0.16 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 4.2.3.12 | 6-pyruvoyltetrahydropterin synthase. | 0.81 | 0.91 | 1 | 0.77 | 1 | 1 | 0.96 | 1 | 1 | 0.79 | 0.92 | 1 | 0.65 | 0.95 | 1 | 0.81 | 0.96 | 1 | 0.00012 | 4.7e-05 | 0 | 2.9e-05 | 0 | 4.4e-05 | 3.8e-05 | 0 | 5e-05 | 0.39 | 14 / 36 (39%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 4.7e-05 | 0 | 7.7e-05 | 6.5e-05 | 0 | 0.00011 | 0.47 | 4 / 11 (36%) | 4 / 11 (36%) | 0.364 | 0.364 |
| 3.1.5.1 | dGTPase. | 0.33 | 0.6 | 1 | 0.74 | 1 | 1 | 0.63 | 0.96 | 1 | 0.25 | 0.54 | 1 | 0.9 | 0.99 | 1 | 0.72 | 0.94 | 1 | 0.0071 | 0.0071 | 0.0061 | 0.0057 | 0.0064 | 0.0013 | 0.0055 | 0.0057 | 0.0025 | -0.052 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.008 | 0.0059 | 0.0066 | 0.008 | 0.0067 | 0.0036 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.1.81 | Threonine-phosphate decarboxylase. | 0.79 | 0.9 | 1 | 0.52 | 1 | 1 | 0.56 | 0.96 | 1 | 0.7 | 0.88 | 1 | 0.44 | 0.91 | 1 | 0.48 | 0.87 | 1 | 0.0089 | 0.0089 | 0.0085 | 0.0084 | 0.0077 | 0.0025 | 0.0083 | 0.0078 | 0.0051 | -0.017 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0093 | 0.0084 | 0.0044 | 0.0091 | 0.0086 | 0.0032 | -0.031 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.10.- | 0.51 | 0.74 | 1 | 0.72 | 1 | 1 | 0.63 | 0.96 | 1 | 0.43 | 0.69 | 1 | 0.97 | 0.99 | 1 | 0.55 | 0.9 | 1 | 0.0052 | 0.0052 | 0.0028 | 0.0047 | 0.0035 | 0.0039 | 0.0069 | 0.0024 | 0.011 | 0.55 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0036 | 0.0025 | 0.0051 | 0.0061 | 0.003 | 0.0092 | 0.76 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 1.21.4.3 | Sarcosine reductase. | 0.36 | 0.63 | 1 | 0.24 | 1 | 1 | 0.0018 | 0.51 | 1 | 0.14 | 0.47 | 1 | 0.25 | 0.88 | 1 | 0.0019 | 0.48 | 1 | 0.00061 | 5e-04 | 0.00025 | 0.00054 | 0.00014 | 0.00072 | 0.00069 | 0.00049 | 0.00081 | 0.35 | 30 / 36 (83%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.00061 | 0.00079 | 0.00046 | 0.00026 | 0.00015 | 0.00025 | -1.2 | 11 / 11 (100%) | 9 / 11 (82%) | 1 | 0.818 |
| 2.6.1.88 | Methionine transaminase. | 0.67 | 0.84 | 1 | 1 | 1 | 1 | 0.59 | 0.96 | 1 | 0.84 | 0.94 | 1 | 0.64 | 0.95 | 1 | 0.96 | 0.99 | 1 | 0.00033 | 0.00012 | 0 | 2e-04 | 0 | 0.00053 | 0.00014 | 0 | 0.00036 | -0.51 | 13 / 36 (36%) | 2 / 7 (29%) | 3 / 7 (43%) | 0.286 | 0.429 | 5.1e-05 | 0 | 9.4e-05 | 0.00012 | 0 | 0.00022 | 1.2 | 4 / 11 (36%) | 4 / 11 (36%) | 0.364 | 0.364 |
| 3.2.1.24 | Alpha-mannosidase. | 0.25 | 0.56 | 1 | 0.54 | 1 | 1 | 0.67 | 0.98 | 1 | 0.05 | 0.43 | 1 | 0.071 | 0.86 | 1 | 0.074 | 0.66 | 1 | 0.0028 | 0.0027 | 0.0016 | 0.0015 | 0.0011 | 0.0014 | 0.0016 | 0.0019 | 0.0014 | 0.093 | 35 / 36 (97%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.0034 | 0.0015 | 0.0057 | 0.0035 | 0.002 | 0.0053 | 0.042 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.1.337 | 1.2-diacylglycerol 3-alpha-glucosyltransferase. | 0.72 | 0.87 | 1 | 1 | 1 | 1 | 0.34 | 0.89 | 1 | 0.49 | 0.74 | 1 | 0.85 | 0.98 | 1 | 0.48 | 0.87 | 1 | 0.00021 | 2.4e-05 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 4 / 36 (11%) | 0 / 7 (0%) | 0 / 7 (0%) | 0 | 0 | 6.6e-06 | 0 | 2.2e-05 | 7.2e-05 | 0 | 2e-04 | 3.4 | 1 / 11 (9.1%) | 3 / 11 (27%) | 0.0909 | 0.273 |
| 3.2.2.31 | Adenine glycosylase. | 0.039 | 0.49 | 1 | 0.98 | 1 | 1 | 0.44 | 0.93 | 1 | 0.29 | 0.58 | 1 | 0.49 | 0.92 | 1 | 0.89 | 0.98 | 1 | 0.0093 | 0.0093 | 0.0093 | 0.01 | 0.011 | 0.0027 | 0.01 | 0.011 | 0.0022 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0081 | 0.0092 | 0.0022 | 0.0091 | 0.009 | 0.0022 | 0.17 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.5.1.- | 0.13 | 0.5 | 1 | 0.5 | 1 | 1 | 0.3 | 0.89 | 1 | 0.31 | 0.59 | 1 | 0.67 | 0.95 | 1 | 0.36 | 0.81 | 1 | 0.0024 | 0.0024 | 0.0022 | 0.0028 | 0.003 | 0.0011 | 0.0027 | 0.0018 | 0.0018 | -0.052 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.002 | 0.0021 | 0.001 | 0.0023 | 0.0022 | 0.00079 | 0.2 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 3.6.1.61 | Diadenosine hexaphosphate hydrolase (ATP-forming). | 0.62 | 0.82 | 1 | 0.034 | 0.97 | 1 | 0.13 | 0.82 | 1 | 0.63 | 0.83 | 1 | 0.02 | 0.76 | 1 | 0.083 | 0.66 | 1 | 0.00015 | 7.3e-05 | 8e-06 | 3.1e-05 | 0 | 4.9e-05 | 9.6e-05 | 0.00011 | 8.6e-05 | 1.6 | 18 / 36 (50%) | 3 / 7 (43%) | 6 / 7 (86%) | 0.429 | 0.857 | 7.4e-05 | 0 | 0.00013 | 8.5e-05 | 0 | 0.00015 | 0.2 | 4 / 11 (36%) | 5 / 11 (45%) | 0.364 | 0.455 |
| 1.4.-.- | 0.44 | 0.69 | 1 | 0.88 | 1 | 1 | 0.27 | 0.89 | 1 | 0.11 | 0.47 | 1 | 0.73 | 0.96 | 1 | 0.098 | 0.66 | 1 | 0.0013 | 0.0011 | 0.00042 | 0.00074 | 0.00043 | 0.00067 | 0.00098 | 0.0011 | 0.00088 | 0.41 | 31 / 36 (86%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00054 | 0.00036 | 0.00065 | 0.0019 | 0.00041 | 0.0035 | 1.8 | 7 / 11 (64%) | 11 / 11 (100%) | 0.636 | 1 | |
| 2.7.1.5 | Rhamnulokinase. | 0.068 | 0.49 | 1 | 0.81 | 1 | 1 | 0.46 | 0.93 | 1 | 0.18 | 0.5 | 1 | 0.52 | 0.93 | 1 | 0.82 | 0.96 | 1 | 0.0079 | 0.0079 | 0.0075 | 0.0088 | 0.0084 | 0.0028 | 0.0092 | 0.01 | 0.0036 | 0.064 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0064 | 0.0067 | 0.0017 | 0.0078 | 0.0075 | 0.0026 | 0.29 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.7.6 | DNA-directed RNA polymerase. | 0.0057 | 0.49 | 1 | 0.32 | 1 | 1 | 0.96 | 1 | 1 | 0.062 | 0.43 | 1 | 0.47 | 0.92 | 1 | 0.1 | 0.66 | 1 | 0.15 | 0.15 | 0.16 | 0.14 | 0.14 | 0.015 | 0.13 | 0.14 | 0.024 | -0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.17 | 0.17 | 0.026 | 0.16 | 0.16 | 0.017 | -0.087 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.1.129 | Squalene--hopanol cyclase. | 0.67 | 0.84 | 1 | 0.7 | 1 | 1 | 0.14 | 0.83 | 1 | 0.74 | 0.9 | 1 | 0.75 | 0.97 | 1 | 0.17 | 0.71 | 1 | 0.00022 | 1.9e-05 | 0 | 4.6e-06 | 0 | 1.2e-05 | 0 | 0 | 0 | -Inf | 3 / 36 (8.3%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 0 | 0 | 0 | 5.8e-05 | 0 | 0.00013 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 3.2.1.25 | Beta-mannosidase. | 0.028 | 0.49 | 1 | 0.8 | 1 | 1 | 0.97 | 1 | 1 | 0.0033 | 0.36 | 1 | 0.33 | 0.9 | 1 | 0.33 | 0.8 | 1 | 0.005 | 0.0047 | 0.003 | 0.002 | 0.0017 | 0.0021 | 0.0018 | 0.00053 | 0.0031 | -0.15 | 34 / 36 (94%) | 5 / 7 (71%) | 7 / 7 (100%) | 0.714 | 1 | 0.0069 | 0.0032 | 0.0098 | 0.0062 | 0.0038 | 0.0064 | -0.15 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.1.19 | Arginine decarboxylase. | 0.45 | 0.7 | 1 | 0.15 | 1 | 1 | 0.11 | 0.82 | 1 | 0.93 | 0.98 | 1 | 0.54 | 0.93 | 1 | 0.37 | 0.82 | 1 | 0.028 | 0.028 | 0.028 | 0.03 | 0.031 | 0.0055 | 0.026 | 0.028 | 0.0068 | -0.21 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.028 | 0.03 | 0.008 | 0.029 | 0.027 | 0.0062 | 0.051 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.131 | Xylan alpha-1.2-glucuronosidase. | 0.71 | 0.86 | 1 | 0.83 | 1 | 1 | 0.46 | 0.93 | 1 | 0.66 | 0.85 | 1 | 0.84 | 0.98 | 1 | 0.73 | 0.94 | 1 | 0.00087 | 0.00073 | 0.00021 | 0.00061 | 0.00028 | 0.00074 | 0.00056 | 0.00021 | 0.00065 | -0.12 | 30 / 36 (83%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00051 | 0.00016 | 0.00082 | 0.0011 | 0.00022 | 0.0024 | 1.1 | 9 / 11 (82%) | 9 / 11 (82%) | 0.818 | 0.818 |
| 4.3.1.2 | Methylaspartate ammonia-lyase. | 0.056 | 0.49 | 1 | 0.7 | 1 | 1 | 0.9 | 1 | 1 | 0.095 | 0.45 | 1 | 0.63 | 0.95 | 1 | 0.83 | 0.96 | 1 | 0.00018 | 4.5e-05 | 0 | 9.5e-05 | 0 | 0.00016 | 0.00011 | 0 | 0.00016 | 0.21 | 9 / 36 (25%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 6e-06 | 0 | 2e-05 | 9.5e-06 | 0 | 2.3e-05 | 0.66 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 2.7.14.1 | Protein arginine kinase. | 0.66 | 0.84 | 1 | 0.84 | 1 | 1 | 0.39 | 0.91 | 1 | 0.88 | 0.96 | 1 | 0.97 | 0.99 | 1 | 0.4 | 0.83 | 1 | 0.0018 | 0.0018 | 0.0017 | 0.0015 | 0.0014 | 0.00086 | 0.0015 | 0.0016 | 0.00095 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0019 | 0.0019 | 0.0011 | 0.0021 | 0.002 | 0.00087 | 0.14 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.7.68 | Phosphoenolpyruvate guanylyltransferase. | 0.053 | 0.49 | 1 | 0.15 | 1 | 1 | 0.078 | 0.74 | 1 | 0.09 | 0.45 | 1 | 0.25 | 0.88 | 1 | 0.14 | 0.69 | 1 | 0.00019 | 2.1e-05 | 0 | 5.8e-05 | 0 | 0.00011 | 1.8e-05 | 0 | 4.7e-05 | -1.7 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.1e-05 | 0 | 6.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 4.1.2.4 | Deoxyribose-phosphate aldolase. | 0.95 | 0.98 | 1 | 0.82 | 1 | 1 | 0.44 | 0.93 | 1 | 0.59 | 0.81 | 1 | 0.35 | 0.9 | 1 | 0.95 | 0.99 | 1 | 0.011 | 0.011 | 0.011 | 0.011 | 0.011 | 0.0026 | 0.011 | 0.013 | 0.0032 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.011 | 0.0024 | 0.012 | 0.012 | 0.0028 | 0.13 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.1.100 | (R)-amidase. | 0.29 | 0.58 | 1 | 0.68 | 1 | 1 | 0.57 | 0.96 | 1 | 0.56 | 0.79 | 1 | 0.48 | 0.92 | 1 | 0.91 | 0.98 | 1 | 0.0041 | 0.0041 | 0.0037 | 0.0043 | 0.0047 | 0.0018 | 0.0047 | 0.0053 | 0.0022 | 0.13 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0033 | 0.0035 | 0.0013 | 0.0044 | 0.0039 | 0.0024 | 0.42 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.2.7.7 | 3-methyl-2-oxobutanoate dehydrogenase (ferredoxin). | 0.099 | 0.49 | 1 | 0.69 | 1 | 1 | 0.13 | 0.82 | 1 | 0.091 | 0.45 | 1 | 0.85 | 0.98 | 1 | 0.6 | 0.91 | 1 | 0.0012 | 0.0011 | 0.00074 | 0.0013 | 0.001 | 0.00061 | 0.0012 | 0.00088 | 0.0011 | -0.12 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 7e-04 | 0.00026 | 0.00089 | 0.0014 | 0.00071 | 0.0017 | 1 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 2.1.1.223 | tRNA(1)(Val) (adenine(37)-N(6))-methyltransferase. | 0.51 | 0.74 | 1 | 0.79 | 1 | 1 | 0.51 | 0.95 | 1 | 0.97 | 0.99 | 1 | 0.93 | 0.99 | 1 | 0.69 | 0.94 | 1 | 0.0063 | 0.0063 | 0.0068 | 0.0067 | 0.0071 | 0.003 | 0.0067 | 0.0059 | 0.0039 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0059 | 0.0068 | 0.0017 | 0.0063 | 0.0067 | 0.0012 | 0.095 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.1.217 | tRNA (adenine(22)-N(1))-methyltransferase. | 0.41 | 0.67 | 1 | 0.74 | 1 | 1 | 0.7 | 0.98 | 1 | 0.37 | 0.64 | 1 | 0.34 | 0.9 | 1 | 0.91 | 0.98 | 1 | 0.0061 | 0.0061 | 0.0059 | 0.0053 | 0.0051 | 0.002 | 0.0055 | 0.006 | 0.0022 | 0.053 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0063 | 0.0052 | 0.0029 | 0.0068 | 0.0065 | 0.002 | 0.11 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.1.85 | 3-dehydro-L-gulonate-6-phosphate decarboxylase. | 0.61 | 0.81 | 1 | 0.92 | 1 | 1 | 0.3 | 0.89 | 1 | 0.98 | 0.99 | 1 | 0.93 | 0.99 | 1 | 0.3 | 0.78 | 1 | 0.00027 | 9.1e-05 | 0 | 0.00017 | 0 | 0.00035 | 0.00017 | 0 | 0.00031 | 0 | 12 / 36 (33%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 6.5e-05 | 0 | 8.7e-05 | 1.5e-05 | 0 | 3.3e-05 | -2.1 | 5 / 11 (45%) | 2 / 11 (18%) | 0.455 | 0.182 |
| 1.1.1.271 | GDP-L-fucose synthase. | 0.15 | 0.5 | 1 | 0.81 | 1 | 1 | 0.37 | 0.9 | 1 | 0.16 | 0.48 | 1 | 0.84 | 0.98 | 1 | 0.36 | 0.82 | 1 | 0.0029 | 0.0029 | 0.0027 | 0.003 | 0.0028 | 0.00056 | 0.0034 | 0.0035 | 0.002 | 0.18 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0022 | 0.0021 | 0.0015 | 0.0032 | 0.0027 | 0.0017 | 0.54 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.1.1.12 | Aspartate--tRNA ligase. | 0.25 | 0.56 | 1 | 0.38 | 1 | 1 | 0.061 | 0.71 | 1 | 0.32 | 0.59 | 1 | 0.79 | 0.97 | 1 | 0.15 | 0.71 | 1 | 0.021 | 0.021 | 0.02 | 0.018 | 0.018 | 0.005 | 0.016 | 0.014 | 0.0062 | -0.17 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.022 | 0.021 | 0.0087 | 0.025 | 0.024 | 0.0071 | 0.18 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.152 | Mannosylglycoprotein endo-beta-mannosidase. | 0.015 | 0.49 | 1 | 0.91 | 1 | 1 | 0.33 | 0.89 | 1 | 0.00037 | 0.28 | 0.84 | 0.75 | 0.97 | 1 | 0.98 | 1 | 1 | 0.0031 | 0.0019 | 0.00011 | 2.3e-05 | 0 | 4.3e-05 | 1.5e-05 | 0 | 4e-05 | -0.62 | 22 / 36 (61%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0.0046 | 0.00085 | 0.011 | 0.0016 | 0.00056 | 0.0028 | -1.5 | 9 / 11 (82%) | 10 / 11 (91%) | 0.818 | 0.909 |
| 1.17.1.2 | Transferred entry: 1.17.7.4. | 0.14 | 0.5 | 1 | 0.39 | 1 | 1 | 0.71 | 0.98 | 1 | 0.075 | 0.43 | 1 | 0.11 | 0.87 | 1 | 0.33 | 0.8 | 1 | 0.0026 | 0.0022 | 0.0011 | 0.00077 | 0.00017 | 0.00097 | 0.0014 | 0.00084 | 0.0018 | 0.86 | 30 / 36 (83%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.0029 | 0.0012 | 0.0058 | 0.0028 | 0.0012 | 0.0028 | -0.051 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 3.1.2.27 | Choloyl-CoA hydrolase. | 0.12 | 0.5 | 1 | 0.68 | 1 | 1 | 0.38 | 0.9 | 1 | 0.091 | 0.45 | 1 | 0.38 | 0.9 | 1 | 0.096 | 0.66 | 1 | 0.0024 | 0.002 | 0.0012 | 9e-04 | 4e-04 | 0.0011 | 0.0011 | 0.00084 | 0.0013 | 0.29 | 30 / 36 (83%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.0031 | 0.0015 | 0.0056 | 0.0022 | 0.002 | 0.0022 | -0.49 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 2.1.1.79 | Cyclopropane-fatty-acyl-phospholipid synthase. | 0.66 | 0.84 | 1 | 0.42 | 1 | 1 | 0.15 | 0.83 | 1 | 0.91 | 0.97 | 1 | 0.23 | 0.88 | 1 | 0.046 | 0.65 | 1 | 0.0038 | 0.0038 | 0.0033 | 0.0042 | 0.0037 | 0.0024 | 0.0047 | 0.0047 | 0.0024 | 0.16 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0037 | 0.003 | 0.0015 | 0.003 | 0.0029 | 0.0013 | -0.3 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.93 | Tartrate dehydrogenase. | 0.11 | 0.5 | 1 | 0.36 | 1 | 1 | 0.65 | 0.97 | 1 | 0.13 | 0.47 | 1 | 0.27 | 0.88 | 1 | 0.71 | 0.94 | 1 | 5e-04 | 0.00035 | 0.00022 | 0.00014 | 0 | 0.00023 | 0.00022 | 0.00011 | 0.00034 | 0.65 | 25 / 36 (69%) | 3 / 7 (43%) | 4 / 7 (57%) | 0.429 | 0.571 | 4e-04 | 0.00019 | 0.00044 | 0.00051 | 5e-04 | 0.00032 | 0.35 | 8 / 11 (73%) | 10 / 11 (91%) | 0.727 | 0.909 |
| 3.5.3.18 | Dimethylargininase. | 0.69 | 0.85 | 1 | 0.66 | 1 | 1 | 0.67 | 0.98 | 1 | 0.53 | 0.77 | 1 | 0.89 | 0.99 | 1 | 0.73 | 0.94 | 1 | 0.00044 | 0.00032 | 0.00013 | 0.00026 | 0.00014 | 0.00034 | 0.00016 | 0.00012 | 0.00016 | -0.7 | 26 / 36 (72%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 3e-04 | 0.00014 | 0.00029 | 0.00049 | 8.8e-05 | 0.00074 | 0.71 | 9 / 11 (82%) | 7 / 11 (64%) | 0.818 | 0.636 |
| 3.2.1.65 | Levanase. | 0.13 | 0.5 | 1 | 0.61 | 1 | 1 | 0.41 | 0.92 | 1 | 0.048 | 0.43 | 1 | 0.85 | 0.98 | 1 | 0.22 | 0.74 | 1 | 0.00061 | 0.00049 | 0.00021 | 0.00047 | 0.00033 | 0.00042 | 0.00086 | 0.00021 | 0.0013 | 0.87 | 29 / 36 (81%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00021 | 7.2e-05 | 0.00028 | 0.00055 | 0.00021 | 0.00064 | 1.4 | 7 / 11 (64%) | 9 / 11 (82%) | 0.636 | 0.818 |
| 3.1.21.5 | Type III site-specific deoxyribonuclease. | 0.057 | 0.49 | 1 | 0.5 | 1 | 1 | 0.054 | 0.71 | 1 | 0.084 | 0.44 | 1 | 0.31 | 0.9 | 1 | 0.041 | 0.63 | 1 | 0.00048 | 0.00025 | 2.9e-05 | 6.4e-05 | 0 | 1e-04 | 0.00012 | 9.3e-05 | 0.00014 | 0.91 | 19 / 36 (53%) | 3 / 7 (43%) | 4 / 7 (57%) | 0.429 | 0.571 | 0.00053 | 0.00022 | 8e-04 | 0.00018 | 0 | 0.00044 | -1.6 | 7 / 11 (64%) | 5 / 11 (45%) | 0.636 | 0.455 |
| 2.8.3.17 | 3-(aryl)acryloyl-CoA:(R)-3-(aryl)lactate CoA-transferase. | 0.76 | 0.89 | 1 | 0.15 | 1 | 1 | 0.13 | 0.82 | 1 | 0.63 | 0.84 | 1 | 0.1 | 0.86 | 1 | 0.088 | 0.66 | 1 | 6.9e-05 | 1.3e-05 | 0 | 8.1e-06 | 0 | 2.1e-05 | 3e-05 | 0 | 4.6e-05 | 1.9 | 7 / 36 (19%) | 1 / 7 (14%) | 3 / 7 (43%) | 0.143 | 0.429 | 1.5e-05 | 0 | 3.9e-05 | 5.4e-06 | 0 | 1.8e-05 | -1.5 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 5.4.99.6 | Transferred entry: 5.4.4.2. | 0.42 | 0.68 | 1 | 0.17 | 1 | 1 | 0.12 | 0.82 | 1 | 0.46 | 0.71 | 1 | 0.21 | 0.88 | 1 | 0.081 | 0.66 | 1 | 0.00027 | 2.2e-05 | 0 | 3.4e-05 | 0 | 8.9e-05 | 7.7e-05 | 0 | 2e-04 | 1.2 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 3.1.4.52 | Cyclic-guanylate-specific phosphodiesterase. | 0.46 | 0.71 | 1 | 0.33 | 1 | 1 | 0.3 | 0.89 | 1 | 0.88 | 0.96 | 1 | 0.62 | 0.95 | 1 | 0.61 | 0.92 | 1 | 0.068 | 0.068 | 0.064 | 0.073 | 0.064 | 0.021 | 0.068 | 0.082 | 0.031 | -0.1 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.065 | 0.063 | 0.022 | 0.067 | 0.072 | 0.019 | 0.044 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.4 | Cellulase. | 0.9 | 0.96 | 1 | 0.86 | 1 | 1 | 0.59 | 0.96 | 1 | 0.75 | 0.9 | 1 | 0.47 | 0.92 | 1 | 0.92 | 0.98 | 1 | 0.03 | 0.03 | 0.025 | 0.027 | 0.024 | 0.012 | 0.028 | 0.025 | 0.014 | 0.052 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.029 | 0.02 | 0.015 | 0.033 | 0.03 | 0.017 | 0.19 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.1.1.19 | Arginine--tRNA ligase. | 0.043 | 0.49 | 1 | 0.5 | 1 | 1 | 0.89 | 1 | 1 | 0.092 | 0.45 | 1 | 0.65 | 0.95 | 1 | 0.19 | 0.72 | 1 | 0.029 | 0.029 | 0.03 | 0.027 | 0.027 | 0.0037 | 0.026 | 0.029 | 0.0067 | -0.054 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.032 | 0.031 | 0.005 | 0.03 | 0.03 | 0.0035 | -0.093 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.27 | L-lactate dehydrogenase. | 0.016 | 0.49 | 1 | 0.5 | 1 | 1 | 0.12 | 0.82 | 1 | 0.029 | 0.43 | 1 | 0.2 | 0.87 | 1 | 0.04 | 0.63 | 1 | 0.015 | 0.015 | 0.015 | 0.012 | 0.012 | 0.0041 | 0.014 | 0.014 | 0.0051 | 0.22 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.018 | 0.017 | 0.0054 | 0.015 | 0.016 | 0.0026 | -0.26 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.4.1.8 | Acetophenone carboxylase. | 0.82 | 0.92 | 1 | 0.56 | 1 | 1 | 0.36 | 0.9 | 1 | 0.75 | 0.9 | 1 | 0.78 | 0.97 | 1 | 0.22 | 0.74 | 1 | 0.00042 | 0.00039 | 3e-04 | 0.00048 | 0.00042 | 0.00037 | 0.00042 | 0.00059 | 0.00029 | -0.19 | 33 / 36 (92%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00042 | 0.00029 | 0.00043 | 0.00026 | 2e-04 | 0.00024 | -0.69 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 1.1.1.325 | Sepiapterin reductase (L-threo-7.8-dihydrobiopterin forming). | 0.7 | 0.86 | 1 | 0.93 | 1 | 1 | 0.91 | 1 | 1 | 0.66 | 0.85 | 1 | 0.69 | 0.95 | 1 | 0.72 | 0.94 | 1 | 0.00044 | 0.00012 | 0 | 6.9e-05 | 0 | 0.00017 | 6.1e-05 | 0 | 0.00012 | -0.18 | 10 / 36 (28%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0.00013 | 0 | 0.00029 | 0.00019 | 0 | 0.00051 | 0.55 | 3 / 11 (27%) | 3 / 11 (27%) | 0.273 | 0.273 |
| 4.1.2.56 | 2-amino-4.5-dihydroxy-6-oxo-7-(phosphonooxy)heptanoate synthase. | 0.12 | 0.5 | 1 | 0.5 | 1 | 1 | 0.8 | 1 | 1 | 0.083 | 0.44 | 1 | 0.43 | 0.91 | 1 | 0.83 | 0.96 | 1 | 0.00019 | 2.6e-05 | 0 | 4.5e-05 | 0 | 8.9e-05 | 8.6e-05 | 0 | 2e-04 | 0.93 | 5 / 36 (14%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0 | 0 | 0 | 3e-06 | 0 | 1e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 3.5.4.4 | Adenosine deaminase. | 0.16 | 0.5 | 1 | 0.38 | 1 | 1 | 0.93 | 1 | 1 | 0.2 | 0.51 | 1 | 0.62 | 0.95 | 1 | 0.59 | 0.91 | 1 | 0.0036 | 0.0036 | 0.0035 | 0.0032 | 0.0037 | 0.00084 | 0.0029 | 0.0028 | 0.0013 | -0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0042 | 0.0047 | 0.0018 | 0.0037 | 0.0035 | 0.0012 | -0.18 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.103 | L-threonine 3-dehydrogenase. | 0.94 | 0.97 | 1 | 0.72 | 1 | 1 | 0.75 | 0.99 | 1 | 0.72 | 0.89 | 1 | 0.97 | 0.99 | 1 | 0.46 | 0.86 | 1 | 0.011 | 0.011 | 0.011 | 0.011 | 0.012 | 0.0024 | 0.011 | 0.0099 | 0.0051 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.012 | 0.0033 | 0.01 | 0.01 | 0.0027 | -0.14 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.1.1.1 | Tyrosine--tRNA ligase. | 0.14 | 0.5 | 1 | 0.59 | 1 | 1 | 0.62 | 0.96 | 1 | 0.18 | 0.5 | 1 | 0.54 | 0.93 | 1 | 0.15 | 0.71 | 1 | 0.021 | 0.021 | 0.021 | 0.02 | 0.019 | 0.0022 | 0.02 | 0.02 | 0.004 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.023 | 0.023 | 0.0044 | 0.021 | 0.021 | 0.0022 | -0.13 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.305 | UDP-glucuronic acid oxidase (UDP-4-keto-hexauronic acid decarboxylating). | 0.31 | 0.59 | 1 | 0.17 | 1 | 1 | 0.52 | 0.95 | 1 | 0.49 | 0.73 | 1 | 0.48 | 0.92 | 1 | 0.19 | 0.72 | 1 | 0.00035 | 3.9e-05 | 0 | 1e-04 | 0 | 0.00027 | 7.7e-05 | 0 | 2e-04 | -0.38 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 8.9e-06 | 0 | 3e-05 | 5.4e-06 | 0 | 1.8e-05 | -0.72 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 4.1.1.71 | 2-oxoglutarate decarboxylase. | 0.14 | 0.5 | 1 | 0.71 | 1 | 1 | 0.75 | 0.99 | 1 | 0.21 | 0.52 | 1 | 0.94 | 0.99 | 1 | 0.51 | 0.89 | 1 | 0.00019 | 7.4e-05 | 0 | 0.00017 | 4.8e-05 | 0.00034 | 0.00012 | 5.4e-05 | 0.00019 | -0.5 | 14 / 36 (39%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 4.5e-05 | 0 | 0.00011 | 1.4e-05 | 0 | 3e-05 | -1.7 | 4 / 11 (36%) | 2 / 11 (18%) | 0.364 | 0.182 |
| 6.5.1.2 | DNA ligase (NAD(+)). | 0.074 | 0.49 | 1 | 0.64 | 1 | 1 | 0.77 | 1 | 1 | 0.098 | 0.46 | 1 | 0.64 | 0.95 | 1 | 0.19 | 0.72 | 1 | 0.032 | 0.032 | 0.031 | 0.03 | 0.029 | 0.0044 | 0.029 | 0.029 | 0.0068 | -0.049 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.035 | 0.036 | 0.008 | 0.033 | 0.033 | 0.0047 | -0.085 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.19.11 | Gamma-D-glutamyl-meso-diaminopimelate peptidase. | 0.54 | 0.76 | 1 | 0.22 | 1 | 1 | 0.62 | 0.96 | 1 | 0.89 | 0.97 | 1 | 0.34 | 0.9 | 1 | 0.87 | 0.97 | 1 | 0.013 | 0.013 | 0.011 | 0.015 | 0.015 | 0.0069 | 0.012 | 0.012 | 0.0072 | -0.32 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.013 | 0.0097 | 0.0059 | 0.011 | 0.01 | 0.0046 | -0.24 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.4.1.1 | Pyruvate carboxylase. | 0.28 | 0.56 | 1 | 0.41 | 1 | 1 | 0.64 | 0.97 | 1 | 0.45 | 0.7 | 1 | 0.32 | 0.9 | 1 | 0.91 | 0.98 | 1 | 0.0082 | 0.0082 | 0.0073 | 0.0086 | 0.0077 | 0.0041 | 0.01 | 0.0062 | 0.007 | 0.22 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0065 | 0.0066 | 0.0031 | 0.0084 | 0.0081 | 0.0048 | 0.37 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.1.82 | Phosphonopyruvate decarboxylase. | 0.35 | 0.62 | 1 | 0.9 | 1 | 1 | 0.55 | 0.95 | 1 | 0.73 | 0.9 | 1 | 0.96 | 0.99 | 1 | 0.49 | 0.88 | 1 | 4e-04 | 0.00027 | 9.7e-05 | 0.00046 | 0.00014 | 0.00057 | 0.00044 | 0.00012 | 0.00052 | -0.064 | 24 / 36 (67%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 2e-04 | 0.00014 | 2e-04 | 1e-04 | 7.1e-05 | 0.00017 | -1 | 8 / 11 (73%) | 6 / 11 (55%) | 0.727 | 0.545 |
| 2.5.1.145 | Phosphatidylglycerol--prolipoprotein diacylglyceryl transferase. | 0.033 | 0.49 | 1 | 0.55 | 1 | 1 | 0.87 | 1 | 1 | 0.047 | 0.43 | 1 | 0.66 | 0.95 | 1 | 0.35 | 0.81 | 1 | 0.016 | 0.016 | 0.016 | 0.014 | 0.013 | 0.0031 | 0.014 | 0.013 | 0.0038 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.018 | 0.017 | 0.0031 | 0.017 | 0.017 | 0.003 | -0.082 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.1.3.2 | UDP-glucose 4-epimerase. | 0.052 | 0.49 | 1 | 0.35 | 1 | 1 | 0.33 | 0.89 | 1 | 0.042 | 0.43 | 1 | 0.63 | 0.95 | 1 | 0.69 | 0.93 | 1 | 0.038 | 0.038 | 0.035 | 0.034 | 0.033 | 0.0057 | 0.032 | 0.032 | 0.0023 | -0.087 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.041 | 0.04 | 0.012 | 0.042 | 0.042 | 0.0079 | 0.035 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.6.2.2 | Cytochrome-b5 reductase. | 0.069 | 0.49 | 1 | 0.67 | 1 | 1 | 0.3 | 0.89 | 1 | 0.043 | 0.43 | 1 | 0.9 | 0.99 | 1 | 0.12 | 0.66 | 1 | 0.00039 | 0.00017 | 0 | 0.00012 | 0 | 0.00025 | 5.1e-05 | 0 | 9.3e-05 | -1.2 | 16 / 36 (44%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 3e-04 | 0.00024 | 0.00032 | 0.00016 | 0 | 0.00028 | -0.91 | 8 / 11 (73%) | 4 / 11 (36%) | 0.727 | 0.364 |
| 5.3.1.9 | Glucose-6-phosphate isomerase. | 0.21 | 0.54 | 1 | 0.16 | 1 | 1 | 0.054 | 0.71 | 1 | 0.13 | 0.47 | 1 | 0.011 | 0.75 | 1 | 0.0032 | 0.48 | 1 | 0.026 | 0.026 | 0.027 | 0.025 | 0.023 | 0.0048 | 0.026 | 0.027 | 0.006 | 0.057 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.027 | 0.027 | 0.0033 | 0.026 | 0.025 | 0.0039 | -0.054 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.13.12.4 | Lactate 2-monooxygenase. | 0.6 | 0.8 | 1 | 0.85 | 1 | 1 | 0.27 | 0.89 | 1 | 0.46 | 0.71 | 1 | 0.72 | 0.95 | 1 | 0.2 | 0.73 | 1 | 0.00014 | 3.4e-05 | 0 | 3.8e-05 | 0 | 8.8e-05 | 4.2e-05 | 0 | 7.6e-05 | 0.14 | 9 / 36 (25%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 5.1e-05 | 0 | 8.1e-05 | 9.9e-06 | 0 | 3.3e-05 | -2.4 | 4 / 11 (36%) | 1 / 11 (9.1%) | 0.364 | 0.0909 |
| 1.1.1.274 | 2.5-didehydrogluconate reductase (2-dehydro-D-gluconate-forming). | 0.35 | 0.62 | 1 | 0.83 | 1 | 1 | 0.88 | 1 | 1 | 0.18 | 0.5 | 1 | 0.39 | 0.9 | 1 | 0.72 | 0.94 | 1 | 0.0041 | 0.0041 | 0.0031 | 0.003 | 0.003 | 0.0021 | 0.0031 | 0.003 | 0.0019 | 0.047 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0047 | 0.0028 | 0.0057 | 0.0048 | 0.0039 | 0.003 | 0.03 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.194 | Protein-N(pi)-phosphohistidine--L-ascorbate phosphotransferase. | 0.42 | 0.68 | 1 | 0.54 | 1 | 1 | 0.94 | 1 | 1 | 0.86 | 0.95 | 1 | 0.85 | 0.98 | 1 | 0.56 | 0.9 | 1 | 7e-04 | 0.00012 | 0 | 0.00029 | 0 | 0.00076 | 0.00022 | 0 | 0.00059 | -0.4 | 6 / 36 (17%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3.5e-05 | 0 | 8.1e-05 | 2.3e-05 | 0 | 6.8e-05 | -0.61 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 1.3.5.3 | Protoporphyrinogen IX dehydrogenase (menaquinone). | 0.059 | 0.49 | 1 | 0.48 | 1 | 1 | 0.32 | 0.89 | 1 | 0.056 | 0.43 | 1 | 0.77 | 0.97 | 1 | 0.59 | 0.91 | 1 | 0.00017 | 4.8e-05 | 0 | 0.00012 | 0 | 0.00022 | 9.5e-05 | 0 | 2e-04 | -0.34 | 10 / 36 (28%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 5.5e-06 | 0 | 1.2e-05 | 1.8e-05 | 0 | 4e-05 | 1.7 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 2.7.7.- | 0.67 | 0.85 | 1 | 0.96 | 1 | 1 | 0.63 | 0.96 | 1 | 0.24 | 0.54 | 1 | 0.16 | 0.87 | 1 | 0.047 | 0.65 | 1 | 0.066 | 0.066 | 0.064 | 0.065 | 0.063 | 0.007 | 0.065 | 0.064 | 0.0071 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.067 | 0.064 | 0.01 | 0.065 | 0.066 | 0.013 | -0.044 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 4.4.1.5 | Lactoylglutathione lyase. | 0.52 | 0.75 | 1 | 0.72 | 1 | 1 | 0.97 | 1 | 1 | 1 | 1 | 1 | 0.28 | 0.88 | 1 | 0.5 | 0.88 | 1 | 0.0042 | 0.0042 | 0.004 | 0.0043 | 0.0041 | 0.0015 | 0.0046 | 0.0044 | 0.002 | 0.097 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0039 | 0.0036 | 0.0012 | 0.0041 | 0.004 | 0.00091 | 0.072 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.20.4.1 | Arsenate reductase (glutaredoxin). | 0.34 | 0.62 | 1 | 0.044 | 0.97 | 1 | 0.063 | 0.71 | 1 | 0.08 | 0.44 | 1 | 0.027 | 0.76 | 1 | 0.017 | 0.55 | 1 | 0.0014 | 0.0014 | 0.0013 | 0.0011 | 0.0013 | 0.00075 | 0.0017 | 0.0019 | 0.0013 | 0.63 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0013 | 0.0012 | 2e-04 | 0.0014 | 0.0014 | 0.00091 | 0.11 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.7 | Mannokinase. | 0.44 | 0.69 | 1 | 0.89 | 1 | 1 | 0.58 | 0.96 | 1 | 0.72 | 0.89 | 1 | 0.88 | 0.99 | 1 | 0.87 | 0.97 | 1 | 0.00031 | 5.1e-05 | 0 | 1e-04 | 0 | 0.00027 | 8.7e-05 | 0 | 0.00023 | -0.2 | 6 / 36 (17%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 9.9e-06 | 0 | 2.5e-05 | 3.7e-05 | 0 | 1e-04 | 1.9 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 3.5.1.53 | N-carbamoylputrescine amidase. | 0.65 | 0.84 | 1 | 0.45 | 1 | 1 | 0.65 | 0.97 | 1 | 0.91 | 0.97 | 1 | 0.19 | 0.87 | 1 | 0.37 | 0.82 | 1 | 0.0044 | 0.0044 | 0.0042 | 0.0045 | 0.0044 | 0.0018 | 0.005 | 0.006 | 0.0019 | 0.15 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0042 | 0.0046 | 0.0017 | 0.0042 | 0.0037 | 0.0019 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.3.1.15 | D-lyxose ketol-isomerase. | 0.99 | 1 | 1 | 0.17 | 1 | 1 | 0.048 | 0.71 | 1 | 0.79 | 0.92 | 1 | 0.14 | 0.87 | 1 | 0.035 | 0.63 | 1 | 1e-04 | 1.4e-05 | 0 | 1.6e-05 | 0 | 4.3e-05 | 3.8e-05 | 0 | 7.2e-05 | 1.2 | 5 / 36 (14%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 1.3e-05 | 0 | 2.8e-05 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 1.4.4.2 | Glycine dehydrogenase (aminomethyl-transferring). | 0.4 | 0.66 | 1 | 0.65 | 1 | 1 | 0.84 | 1 | 1 | 0.36 | 0.63 | 1 | 0.83 | 0.98 | 1 | 0.67 | 0.93 | 1 | 0.0046 | 0.0046 | 0.0035 | 0.0052 | 0.0054 | 0.0016 | 0.005 | 0.0058 | 0.0024 | -0.057 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0046 | 0.0034 | 0.0046 | 0.0039 | 0.0026 | 0.0036 | -0.24 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.2.1 | Glycine hydroxymethyltransferase. | 0.055 | 0.49 | 1 | 1 | 1 | 1 | 0.18 | 0.86 | 1 | 0.13 | 0.47 | 1 | 0.21 | 0.87 | 1 | 0.016 | 0.55 | 1 | 0.016 | 0.016 | 0.016 | 0.015 | 0.015 | 0.0033 | 0.015 | 0.015 | 0.0038 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.018 | 0.018 | 0.0036 | 0.016 | 0.014 | 0.0029 | -0.17 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.2.14 | 2-dehydro-3-deoxy-phosphogluconate aldolase. | 0.0066 | 0.49 | 1 | 0.38 | 1 | 1 | 0.031 | 0.71 | 1 | 0.035 | 0.43 | 1 | 0.67 | 0.95 | 1 | 0.13 | 0.67 | 1 | 0.004 | 0.004 | 0.0039 | 0.0049 | 0.0049 | 0.0018 | 0.0042 | 0.0046 | 0.0016 | -0.22 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0028 | 0.0031 | 0.0011 | 0.0045 | 0.0049 | 0.0018 | 0.68 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.6.1.21 | D-amino-acid transaminase. | 0.26 | 0.56 | 1 | 0.33 | 1 | 1 | 0.14 | 0.83 | 1 | 0.54 | 0.77 | 1 | 0.61 | 0.95 | 1 | 0.3 | 0.78 | 1 | 0.00048 | 0.00041 | 0.00035 | 0.00054 | 0.00036 | 0.00043 | 0.00039 | 0.00034 | 0.00041 | -0.47 | 31 / 36 (86%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00032 | 0.00026 | 0.00027 | 0.00044 | 0.00045 | 0.00029 | 0.46 | 9 / 11 (82%) | 10 / 11 (91%) | 0.818 | 0.909 |
| 1.14.11.17 | Taurine dioxygenase. | 0.13 | 0.5 | 1 | 0.16 | 1 | 1 | 0.12 | 0.82 | 1 | 0.15 | 0.47 | 1 | 0.92 | 0.99 | 1 | 0.5 | 0.88 | 1 | 0.00036 | 3e-05 | 0 | 1e-04 | 0 | 0.00027 | 4.8e-05 | 0 | 0.00013 | -1.1 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.1.1.64 | 3-demethylubiquinol 3-O-methyltransferase. | 0.15 | 0.5 | 1 | 0.082 | 1 | 1 | 0.042 | 0.71 | 1 | 0.28 | 0.57 | 1 | 0.16 | 0.87 | 1 | 0.1 | 0.66 | 1 | 0.0027 | 0.0027 | 0.0025 | 0.0032 | 0.003 | 0.00092 | 0.0021 | 0.0023 | 0.00066 | -0.61 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0024 | 0.0023 | 0.0012 | 0.003 | 0.0028 | 0.0015 | 0.32 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.8.- | 0.46 | 0.71 | 1 | 0.6 | 1 | 1 | 0.42 | 0.93 | 1 | 0.24 | 0.53 | 1 | 0.43 | 0.91 | 1 | 0.55 | 0.9 | 1 | 0.069 | 0.069 | 0.066 | 0.065 | 0.066 | 0.013 | 0.063 | 0.065 | 0.015 | -0.045 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.07 | 0.066 | 0.014 | 0.073 | 0.07 | 0.018 | 0.061 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 2.3.2.2 | Gamma-glutamyltransferase. | 0.044 | 0.49 | 1 | 0.23 | 1 | 1 | 0.16 | 0.85 | 1 | 0.057 | 0.43 | 1 | 0.2 | 0.87 | 1 | 0.14 | 0.69 | 1 | 0.00041 | 0.00035 | 0.00025 | 0.00055 | 0.00028 | 0.00055 | 4e-04 | 0.00028 | 4e-04 | -0.46 | 30 / 36 (83%) | 7 / 7 (100%) | 5 / 7 (71%) | 1 | 0.714 | 0.00023 | 0.00014 | 0.00021 | 3e-04 | 0.00022 | 0.00031 | 0.38 | 8 / 11 (73%) | 10 / 11 (91%) | 0.727 | 0.909 |
| 3.5.4.12 | dCMP deaminase. | 0.94 | 0.97 | 1 | 0.55 | 1 | 1 | 0.029 | 0.71 | 1 | 0.76 | 0.91 | 1 | 0.64 | 0.95 | 1 | 0.064 | 0.66 | 1 | 0.0037 | 0.0037 | 0.0037 | 0.0032 | 0.0028 | 0.00073 | 0.003 | 0.002 | 0.0017 | -0.093 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0033 | 0.003 | 0.0014 | 0.0047 | 0.0044 | 0.0013 | 0.51 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.1.99.6 | NAD(P)H-hydrate epimerase. | 0.062 | 0.49 | 1 | 0.55 | 1 | 1 | 0.18 | 0.85 | 1 | 0.098 | 0.46 | 1 | 0.072 | 0.86 | 1 | 0.015 | 0.55 | 1 | 0.016 | 0.016 | 0.016 | 0.015 | 0.016 | 0.0039 | 0.016 | 0.015 | 0.0041 | 0.093 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.018 | 0.017 | 0.0049 | 0.016 | 0.016 | 0.0012 | -0.17 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.399 | 2-oxoglutarate reductase. | 0.11 | 0.5 | 1 | 0.74 | 1 | 1 | 0.95 | 1 | 1 | 0.06 | 0.43 | 1 | 0.34 | 0.9 | 1 | 0.97 | 0.99 | 1 | 0.0023 | 0.0021 | 0.001 | 0.00078 | 6e-04 | 0.00093 | 0.00094 | 0.00032 | 0.0011 | 0.27 | 33 / 36 (92%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.0032 | 0.0012 | 0.0063 | 0.0027 | 0.0019 | 0.0026 | -0.25 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 3.6.1.55 | 8-oxo-dGTP diphosphatase. | 0.28 | 0.57 | 1 | 0.97 | 1 | 1 | 0.91 | 1 | 1 | 0.22 | 0.52 | 1 | 0.68 | 0.95 | 1 | 0.81 | 0.96 | 1 | 0.012 | 0.012 | 0.011 | 0.01 | 0.0086 | 0.0044 | 0.01 | 0.011 | 0.006 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.015 | 0.011 | 0.014 | 0.013 | 0.014 | 0.0047 | -0.21 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.1.287 | Rhamnopyranosyl-N-acetylglucosaminyl-diphospho-decaprenol beta-1.4/1.5- | 0.48 | 0.72 | 1 | 0.48 | 1 | 1 | 0.27 | 0.89 | 1 | 0.42 | 0.68 | 1 | 0.68 | 0.95 | 1 | 0.62 | 0.92 | 1 | 0.00047 | 0.00021 | 0 | 0.00021 | 0.00011 | 0.00031 | 9.7e-05 | 5.4e-05 | 0.00014 | -1.1 | 16 / 36 (44%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.00013 | 0 | 0.00027 | 0.00035 | 0 | 0.00072 | 1.4 | 4 / 11 (36%) | 4 / 11 (36%) | 0.364 | 0.364 |
| 2.7.1.29 | Glycerone kinase. | 0.72 | 0.87 | 1 | 0.085 | 1 | 1 | 0.45 | 0.93 | 1 | 0.44 | 0.69 | 1 | 0.026 | 0.76 | 1 | 0.19 | 0.72 | 1 | 0.0024 | 0.0024 | 0.0024 | 0.0021 | 0.0024 | 0.00085 | 0.0027 | 0.0025 | 0.00098 | 0.36 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0022 | 0.0022 | 0.00097 | 0.0025 | 0.0025 | 0.0011 | 0.18 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.62 | 17-beta-estradiol 17-dehydrogenase. | 0.0085 | 0.49 | 1 | 0.52 | 1 | 1 | 0.13 | 0.82 | 1 | 0.0018 | 0.31 | 1 | 0.085 | 0.86 | 1 | 0.029 | 0.61 | 1 | 0.0015 | 0.00087 | 0.00014 | 7e-05 | 0 | 0.00018 | 0.00015 | 0 | 0.00027 | 1.1 | 21 / 36 (58%) | 1 / 7 (14%) | 3 / 7 (43%) | 0.143 | 0.429 | 0.0019 | 0.00087 | 0.0036 | 0.00085 | 0.00023 | 0.0011 | -1.2 | 9 / 11 (82%) | 8 / 11 (73%) | 0.818 | 0.727 |
| 2.7.11.5 | [Isocitrate dehydrogenase (NADP(+))] kinase. | 0.2 | 0.53 | 1 | 0.6 | 1 | 1 | 0.27 | 0.89 | 1 | 0.24 | 0.54 | 1 | 0.73 | 0.96 | 1 | 0.4 | 0.83 | 1 | 0.00041 | 6.9e-05 | 0 | 0.00016 | 0 | 4e-04 | 0.00015 | 0 | 0.00038 | -0.093 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 5.1e-06 | 0 | 1.7e-05 | 2.9e-05 | 0 | 6.8e-05 | 2.5 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 1.1.1.73 | Octanol dehydrogenase. | 0.74 | 0.88 | 1 | 0.59 | 1 | 1 | 0.51 | 0.95 | 1 | 0.93 | 0.98 | 1 | 0.93 | 0.99 | 1 | 0.47 | 0.87 | 1 | 0.00066 | 0.00022 | 0 | 0.00031 | 0 | 0.00056 | 0.00022 | 0 | 0.00045 | -0.49 | 12 / 36 (33%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0.00021 | 0 | 4e-04 | 0.00017 | 0 | 0.00028 | -0.3 | 3 / 11 (27%) | 5 / 11 (45%) | 0.273 | 0.455 |
| 2.3.1.50 | Serine C-palmitoyltransferase. | 0.46 | 0.71 | 1 | 0.75 | 1 | 1 | 0.32 | 0.89 | 1 | 0.61 | 0.82 | 1 | 0.83 | 0.98 | 1 | 0.83 | 0.96 | 1 | 0.0012 | 0.001 | 0.00038 | 0.00098 | 0.0011 | 0.00085 | 0.00092 | 0.00083 | 0.0011 | -0.091 | 31 / 36 (86%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00059 | 0.00024 | 9e-04 | 0.0015 | 0.00033 | 0.0028 | 1.3 | 10 / 11 (91%) | 9 / 11 (82%) | 0.909 | 0.818 |
| 1.18.1.1 | Rubredoxin--NAD(+) reductase. | 0.18 | 0.52 | 1 | 0.1 | 1 | 1 | 0.26 | 0.89 | 1 | 0.37 | 0.64 | 1 | 0.21 | 0.87 | 1 | 0.41 | 0.84 | 1 | 3e-04 | 0.00021 | 9.4e-05 | 0.00046 | 0.00016 | 0.00089 | 0.00027 | 2.7e-05 | 0.00063 | -0.77 | 25 / 36 (69%) | 6 / 7 (86%) | 4 / 7 (57%) | 0.857 | 0.571 | 0.00012 | 9.5e-05 | 0.00012 | 1e-04 | 0.00011 | 0.00011 | -0.26 | 8 / 11 (73%) | 7 / 11 (64%) | 0.727 | 0.636 |
| 3.1.8.1 | Aryldialkylphosphatase. | 0.74 | 0.88 | 1 | 0.87 | 1 | 1 | 0.58 | 0.96 | 1 | 0.83 | 0.93 | 1 | 0.64 | 0.95 | 1 | 0.67 | 0.93 | 1 | 0.00029 | 0.00013 | 0 | 0.00019 | 0 | 0.00039 | 0.00027 | 0 | 0.00066 | 0.51 | 16 / 36 (44%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 7.6e-05 | 3.3e-05 | 0.00011 | 5.2e-05 | 0 | 8.3e-05 | -0.55 | 7 / 11 (64%) | 4 / 11 (36%) | 0.636 | 0.364 |
| 3.2.1.28 | Alpha.alpha-trehalase. | 0.7 | 0.86 | 1 | 0.38 | 1 | 1 | 0.25 | 0.89 | 1 | 0.29 | 0.58 | 1 | 0.39 | 0.9 | 1 | 0.58 | 0.91 | 1 | 0.0018 | 0.0017 | 0.00063 | 0.0012 | 0.00098 | 0.00082 | 8e-04 | 0.00063 | 8e-04 | -0.58 | 33 / 36 (92%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0015 | 0.00048 | 0.0026 | 0.0027 | 5e-04 | 0.0054 | 0.85 | 10 / 11 (91%) | 9 / 11 (82%) | 0.909 | 0.818 |
| 2.6.1.9 | Histidinol-phosphate transaminase. | 0.0091 | 0.49 | 1 | 0.21 | 1 | 1 | 0.14 | 0.83 | 1 | 0.025 | 0.42 | 1 | 0.0095 | 0.73 | 1 | 0.0052 | 0.48 | 1 | 0.019 | 0.019 | 0.019 | 0.015 | 0.014 | 0.0031 | 0.017 | 0.018 | 0.0053 | 0.18 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.021 | 0.021 | 0.0044 | 0.019 | 0.02 | 0.0027 | -0.14 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.12.98.4 | Sulfhydrogenase. | 0.054 | 0.49 | 1 | 0.86 | 1 | 1 | 0.55 | 0.95 | 1 | 0.047 | 0.43 | 1 | 0.81 | 0.98 | 1 | 0.44 | 0.86 | 1 | 0.0013 | 0.00081 | 0.00031 | 0.00019 | 0 | 0.00027 | 2e-04 | 0 | 0.00044 | 0.074 | 22 / 36 (61%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 0.0015 | 7e-04 | 0.003 | 9e-04 | 7e-04 | 0.0011 | -0.74 | 9 / 11 (82%) | 7 / 11 (64%) | 0.818 | 0.636 |
| 4.2.2.n2 | Peptidoglycan lytic endotransglycosylase. | 0.18 | 0.52 | 1 | 0.15 | 1 | 1 | 0.46 | 0.93 | 1 | 0.31 | 0.59 | 1 | 0.91 | 0.99 | 1 | 0.46 | 0.87 | 1 | 0.00082 | 6.8e-05 | 0 | 0.00024 | 0 | 0.00062 | 0.00011 | 0 | 0.00028 | -1.1 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 5.1e-06 | 0 | 1.7e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 1.8.4.13 | L-methionine (S)-S-oxide reductase. | 0.24 | 0.55 | 1 | 0.66 | 1 | 1 | 0.39 | 0.91 | 1 | 0.31 | 0.58 | 1 | 0.35 | 0.9 | 1 | 0.26 | 0.76 | 1 | 0.00056 | 0.00016 | 0 | 0.00031 | 0 | 0.00075 | 0.00041 | 0 | 0.0011 | 0.4 | 10 / 36 (28%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 1.5e-05 | 0 | 2.9e-05 | 4.4e-05 | 0 | 8.2e-05 | 1.6 | 3 / 11 (27%) | 3 / 11 (27%) | 0.273 | 0.273 |
| 6.3.2.12 | Dihydrofolate synthase. | 0.68 | 0.85 | 1 | 0.75 | 1 | 1 | 0.83 | 1 | 1 | 0.33 | 0.6 | 1 | 0.4 | 0.9 | 1 | 0.34 | 0.8 | 1 | 0.0029 | 0.0029 | 0.0029 | 0.0027 | 0.0021 | 0.0015 | 0.0029 | 0.0036 | 0.0017 | 0.1 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0029 | 0.003 | 0.0012 | 0.003 | 0.0031 | 0.0014 | 0.049 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.8.1.12 | Molybdopterin synthase. | 0.15 | 0.5 | 1 | 0.45 | 1 | 1 | 0.55 | 0.95 | 1 | 0.19 | 0.5 | 1 | 0.12 | 0.87 | 1 | 0.19 | 0.72 | 1 | 0.00029 | 2.4e-05 | 0 | 6.7e-05 | 0 | 0.00018 | 5.6e-05 | 0 | 1e-04 | -0.26 | 3 / 36 (8.3%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 2.7.10.1 | Receptor protein-tyrosine kinase. | 0.32 | 0.6 | 1 | 0.71 | 1 | 1 | 0.24 | 0.89 | 1 | 0.26 | 0.55 | 1 | 0.87 | 0.99 | 1 | 0.57 | 0.91 | 1 | 0.0011 | 0.001 | 0.00063 | 0.0011 | 0.00069 | 0.0012 | 0.00099 | 0.00059 | 0.0012 | -0.15 | 33 / 36 (92%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00066 | 0.00049 | 0.00061 | 0.0014 | 0.00075 | 0.0017 | 1.1 | 9 / 11 (82%) | 10 / 11 (91%) | 0.818 | 0.909 |
| 2.1.2.- | 0.17 | 0.51 | 1 | 0.83 | 1 | 1 | 0.92 | 1 | 1 | 0.12 | 0.47 | 1 | 0.79 | 0.97 | 1 | 0.82 | 0.96 | 1 | 0.0028 | 0.0028 | 0.0021 | 0.0016 | 0.0016 | 0.00087 | 0.0018 | 0.0016 | 0.0015 | 0.17 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0035 | 0.0023 | 0.0045 | 0.0035 | 0.0046 | 0.0023 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 4.3.1.17 | L-serine ammonia-lyase. | 0.62 | 0.82 | 1 | 0.92 | 1 | 1 | 0.97 | 1 | 1 | 0.93 | 0.98 | 1 | 0.47 | 0.92 | 1 | 0.51 | 0.89 | 1 | 0.018 | 0.018 | 0.017 | 0.018 | 0.02 | 0.0043 | 0.019 | 0.022 | 0.0073 | 0.078 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.017 | 0.016 | 0.0063 | 0.017 | 0.016 | 0.004 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.6.1.13 | Ornithine aminotransferase. | 0.14 | 0.5 | 1 | 0.06 | 1 | 1 | 0.065 | 0.72 | 1 | 0.045 | 0.43 | 1 | 0.085 | 0.86 | 1 | 0.22 | 0.74 | 1 | 0.00064 | 0.00043 | 0.00014 | 0.00057 | 0.00056 | 0.00044 | 0.00027 | 0.00024 | 0.00025 | -1.1 | 24 / 36 (67%) | 7 / 7 (100%) | 5 / 7 (71%) | 1 | 0.714 | 0.00033 | 9.5e-05 | 0.00072 | 0.00054 | 0 | 0.0011 | 0.71 | 7 / 11 (64%) | 5 / 11 (45%) | 0.636 | 0.455 |
| 2.3.3.5 | 2-methylcitrate synthase. | 0.58 | 0.79 | 1 | 0.65 | 1 | 1 | 0.84 | 1 | 1 | 0.6 | 0.81 | 1 | 0.77 | 0.97 | 1 | 0.71 | 0.94 | 1 | 0.0028 | 0.0028 | 0.0024 | 0.0024 | 0.0024 | 0.00082 | 0.0021 | 0.0017 | 0.0013 | -0.19 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.003 | 0.0036 | 0.0014 | 0.0031 | 0.0023 | 0.0024 | 0.047 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.135 | GDP-6-deoxy-D-talose 4-dehydrogenase. | 0.22 | 0.54 | 1 | 0.39 | 1 | 1 | 0.5 | 0.95 | 1 | 0.23 | 0.53 | 1 | 0.47 | 0.92 | 1 | 0.47 | 0.87 | 1 | 0.00018 | 1.5e-05 | 0 | 1.9e-05 | 0 | 4.2e-05 | 5.7e-05 | 0 | 0.00015 | 1.6 | 3 / 36 (8.3%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 2.1.1.72 | Site-specific DNA-methyltransferase (adenine-specific). | 0.23 | 0.54 | 1 | 0.52 | 1 | 1 | 0.83 | 1 | 1 | 0.12 | 0.47 | 1 | 0.92 | 0.99 | 1 | 0.43 | 0.85 | 1 | 0.038 | 0.038 | 0.036 | 0.036 | 0.034 | 0.0064 | 0.034 | 0.036 | 0.011 | -0.082 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.042 | 0.036 | 0.014 | 0.038 | 0.037 | 0.0068 | -0.14 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.1.15 | Glycerol-3-phosphate 1-O-acyltransferase. | 0.025 | 0.49 | 1 | 0.33 | 1 | 1 | 0.38 | 0.9 | 1 | 0.12 | 0.47 | 1 | 0.46 | 0.92 | 1 | 0.46 | 0.86 | 1 | 0.00076 | 7e-04 | 0.00049 | 0.0013 | 0.00056 | 0.0012 | 0.00096 | 0.00077 | 0.00081 | -0.44 | 33 / 36 (92%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00044 | 0.00039 | 0.00035 | 0.00043 | 0.00043 | 2e-04 | -0.033 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 2.8.1.7 | Cysteine desulfurase. | 0.99 | 1 | 1 | 0.33 | 1 | 1 | 0.23 | 0.88 | 1 | 0.65 | 0.85 | 1 | 0.98 | 0.99 | 1 | 0.78 | 0.95 | 1 | 0.049 | 0.049 | 0.052 | 0.049 | 0.049 | 0.0079 | 0.046 | 0.052 | 0.014 | -0.091 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.05 | 0.053 | 0.013 | 0.051 | 0.052 | 0.0093 | 0.029 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.7.99.7 | Transferred entry: 1.7.2.5. | 0.34 | 0.62 | 1 | 0.2 | 1 | 1 | 0.46 | 0.93 | 1 | 0.46 | 0.71 | 1 | 0.15 | 0.87 | 1 | 0.37 | 0.82 | 1 | 9.3e-05 | 1e-05 | 0 | 1.2e-05 | 0 | 3.1e-05 | 3.7e-05 | 0 | 6.3e-05 | 1.6 | 4 / 36 (11%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 0 | 0 | 0 | 3e-06 | 0 | 1e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.1.1.51 | Transferred entry: 2.1.1.187 and 2.1.1.188. | 0.33 | 0.61 | 1 | 0.75 | 1 | 1 | 0.52 | 0.95 | 1 | 0.55 | 0.78 | 1 | 0.86 | 0.98 | 1 | 0.8 | 0.96 | 1 | 0.00052 | 0.00045 | 0.00039 | 0.00052 | 0.00055 | 0.00031 | 0.00049 | 0.00036 | 0.00042 | -0.086 | 31 / 36 (86%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00034 | 0.00011 | 0.00035 | 0.00049 | 3e-04 | 0.00049 | 0.53 | 10 / 11 (91%) | 9 / 11 (82%) | 0.909 | 0.818 |
| 1.2.1.3 | Aldehyde dehydrogenase (NAD(+)). | 0.024 | 0.49 | 1 | 0.039 | 0.97 | 1 | 0.26 | 0.89 | 1 | 0.046 | 0.43 | 1 | 0.22 | 0.88 | 1 | 0.88 | 0.97 | 1 | 0.00032 | 0.00024 | 0.00012 | 0.00056 | 0.00023 | 0.00073 | 0.00024 | 0.00019 | 0.00024 | -1.2 | 27 / 36 (75%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00016 | 9.7e-05 | 0.00016 | 0.00013 | 2.9e-05 | 2e-04 | -0.3 | 8 / 11 (73%) | 6 / 11 (55%) | 0.727 | 0.545 |
| 4.2.1.12 | Phosphogluconate dehydratase. | 0.069 | 0.49 | 1 | 0.09 | 1 | 1 | 0.18 | 0.85 | 1 | 0.031 | 0.43 | 1 | 0.17 | 0.87 | 1 | 0.33 | 0.8 | 1 | 0.00059 | 4.9e-05 | 0 | 0.00014 | 0 | 0.00031 | 0.00012 | 0 | 0.00031 | -0.22 | 3 / 36 (8.3%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 5.3.2.3 | TDP-4-oxo-6-deoxy-alpha-D-glucose-3.4-oxoisomerase (dTDP-3-dehydro-6- | 0.1 | 0.49 | 1 | 0.16 | 1 | 1 | 0.64 | 0.97 | 1 | 0.22 | 0.52 | 1 | 0.54 | 0.93 | 1 | 0.72 | 0.94 | 1 | 0.00027 | 4.5e-05 | 0 | 0.00016 | 0 | 0.00031 | 4.6e-05 | 0 | 1e-04 | -1.8 | 6 / 36 (17%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 1.8e-05 | 0 | 3.9e-05 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 3.3.1.1 | Adenosylhomocysteinase. | 0.92 | 0.96 | 1 | 0.7 | 1 | 1 | 0.88 | 1 | 1 | 0.73 | 0.89 | 1 | 0.52 | 0.93 | 1 | 0.91 | 0.98 | 1 | 0.0026 | 0.0026 | 0.0025 | 0.0023 | 0.0026 | 0.001 | 0.0026 | 0.0028 | 0.0013 | 0.18 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0025 | 0.0021 | 0.0015 | 0.0028 | 0.0026 | 0.0016 | 0.16 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.2.12 | S-formylglutathione hydrolase. | 0.64 | 0.83 | 1 | 0.33 | 1 | 1 | 0.04 | 0.71 | 1 | 0.95 | 0.99 | 1 | 0.74 | 0.96 | 1 | 0.45 | 0.86 | 1 | 0.0019 | 0.0018 | 0.0015 | 0.0022 | 0.0026 | 0.0018 | 0.003 | 0.0031 | 0.0025 | 0.45 | 35 / 36 (97%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.0016 | 0.0017 | 0.00071 | 0.001 | 8e-04 | 0.00069 | -0.68 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.6.1.1 | Inorganic diphosphatase. | 0.22 | 0.54 | 1 | 0.93 | 1 | 1 | 0.65 | 0.97 | 1 | 0.19 | 0.5 | 1 | 0.48 | 0.92 | 1 | 0.68 | 0.93 | 1 | 0.025 | 0.025 | 0.026 | 0.022 | 0.02 | 0.0074 | 0.022 | 0.019 | 0.01 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.026 | 0.026 | 0.0048 | 0.027 | 0.026 | 0.0049 | 0.054 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.6.3.19 | Transferred entry: 7.5.2.1. | 0.16 | 0.5 | 1 | 0.96 | 1 | 1 | 0.83 | 1 | 1 | 0.083 | 0.44 | 1 | 0.99 | 1 | 1 | 0.59 | 0.91 | 1 | 0.0057 | 0.0057 | 0.0046 | 0.0036 | 0.0033 | 0.0028 | 0.0038 | 0.0042 | 0.0032 | 0.078 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0072 | 0.0055 | 0.0089 | 0.0069 | 0.005 | 0.0041 | -0.061 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.8.26 | Adenosylcobinamide-GDP ribazoletransferase. | 0.61 | 0.81 | 1 | 0.16 | 1 | 1 | 0.17 | 0.85 | 1 | 0.93 | 0.98 | 1 | 0.42 | 0.91 | 1 | 0.44 | 0.85 | 1 | 0.0063 | 0.0063 | 0.006 | 0.0066 | 0.0067 | 0.00081 | 0.0057 | 0.0049 | 0.0015 | -0.21 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0063 | 0.0065 | 0.0018 | 0.0065 | 0.0057 | 0.0019 | 0.045 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.- | 0.44 | 0.69 | 1 | 0.9 | 1 | 1 | 0.16 | 0.85 | 1 | 0.18 | 0.5 | 1 | 0.4 | 0.9 | 1 | 0.44 | 0.85 | 1 | 0.085 | 0.085 | 0.083 | 0.072 | 0.076 | 0.017 | 0.074 | 0.074 | 0.028 | 0.04 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.083 | 0.084 | 0.035 | 0.1 | 0.089 | 0.036 | 0.27 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 6.3.2.4 | D-alanine--D-alanine ligase. | 0.092 | 0.49 | 1 | 0.9 | 1 | 1 | 0.14 | 0.83 | 1 | 0.13 | 0.47 | 1 | 0.38 | 0.9 | 1 | 0.043 | 0.64 | 1 | 0.02 | 0.02 | 0.02 | 0.019 | 0.017 | 0.0051 | 0.019 | 0.018 | 0.0058 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.023 | 0.022 | 0.0047 | 0.02 | 0.021 | 0.002 | -0.2 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.1.243 | 6(G)-fructosyltransferase. | 0.15 | 0.5 | 1 | 0.53 | 1 | 1 | 0.51 | 0.95 | 1 | 0.16 | 0.48 | 1 | 0.63 | 0.95 | 1 | 0.29 | 0.78 | 1 | 0.0011 | 0.00092 | 0.00062 | 4e-04 | 0.00055 | 0.00031 | 0.00066 | 0.00024 | 0.00072 | 0.72 | 29 / 36 (81%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.0012 | 0.00064 | 0.0015 | 0.0012 | 0.00075 | 0.0013 | 0 | 10 / 11 (91%) | 9 / 11 (82%) | 0.909 | 0.818 |
| 2.1.1.201 | 2-methoxy-6-polyprenyl-1.4-benzoquinol methylase. | 0.46 | 0.71 | 1 | 0.75 | 1 | 1 | 0.32 | 0.89 | 1 | 0.44 | 0.69 | 1 | 0.57 | 0.93 | 1 | 0.38 | 0.83 | 1 | 0.0016 | 0.0016 | 0.0014 | 0.0014 | 0.0014 | 0.00079 | 0.0015 | 0.0013 | 0.0014 | 0.1 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0018 | 0.0019 | 0.00085 | 0.0014 | 0.0014 | 0.00085 | -0.36 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.21 | Aldose reductase. | 0.69 | 0.85 | 1 | 0.41 | 1 | 1 | 0.77 | 0.99 | 1 | 0.33 | 0.6 | 1 | 0.84 | 0.98 | 1 | 0.32 | 0.79 | 1 | 0.0011 | 0.0011 | 0.0011 | 0.0013 | 0.0011 | 0.0011 | 0.00098 | 0.00093 | 0.00061 | -0.41 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0013 | 0.0013 | 0.00051 | 0.00096 | 9e-04 | 0.00039 | -0.44 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.22.4 | Crossover junction endodeoxyribonuclease. | 0.013 | 0.49 | 1 | 0.75 | 1 | 1 | 0.6 | 0.96 | 1 | 0.027 | 0.43 | 1 | 0.44 | 0.91 | 1 | 0.1 | 0.66 | 1 | 0.01 | 0.01 | 0.01 | 0.0091 | 0.0094 | 0.0011 | 0.009 | 0.0091 | 0.0019 | -0.016 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.011 | 0.0021 | 0.011 | 0.01 | 0.0019 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.8.3.5 | 3-oxoacid CoA-transferase. | 0.067 | 0.49 | 1 | 0.054 | 1 | 1 | 0.052 | 0.71 | 1 | 0.056 | 0.43 | 1 | 0.15 | 0.87 | 1 | 0.19 | 0.72 | 1 | 0.00071 | 0.00067 | 0.00052 | 4e-04 | 0.00026 | 0.00053 | 0.00079 | 0.00063 | 0.00065 | 0.98 | 34 / 36 (94%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00082 | 6e-04 | 7e-04 | 0.00063 | 0.00045 | 4e-04 | -0.38 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.2 | Alcohol dehydrogenase (NADP(+)). | 0.43 | 0.69 | 1 | 0.34 | 1 | 1 | 0.15 | 0.84 | 1 | 0.91 | 0.97 | 1 | 0.76 | 0.97 | 1 | 0.46 | 0.86 | 1 | 0.0063 | 0.0063 | 0.0057 | 0.0067 | 0.0064 | 0.0023 | 0.006 | 0.005 | 0.0025 | -0.16 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0059 | 0.0057 | 0.0014 | 0.0066 | 0.0057 | 0.0024 | 0.16 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.3.1 | Isocitrate lyase. | 0.09 | 0.49 | 1 | 0.35 | 1 | 1 | 0.22 | 0.88 | 1 | 0.054 | 0.43 | 1 | 0.38 | 0.9 | 1 | 0.25 | 0.76 | 1 | 0.00059 | 6.6e-05 | 0 | 0.00016 | 0 | 0.00035 | 0.00016 | 0 | 0.00044 | 0 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 1e-05 | 0 | 3.4e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 3.5.4.9 | Methenyltetrahydrofolate cyclohydrolase. | 0.71 | 0.86 | 1 | 0.14 | 1 | 1 | 0.24 | 0.89 | 1 | 0.31 | 0.58 | 1 | 0.87 | 0.99 | 1 | 0.54 | 0.9 | 1 | 0.017 | 0.017 | 0.017 | 0.017 | 0.019 | 0.0034 | 0.016 | 0.015 | 0.0034 | -0.087 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.018 | 0.018 | 0.0024 | 0.017 | 0.017 | 0.0017 | -0.082 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.1.241 | Kdo(2)-lipid IV(A) lauroyltransferase. | 0.099 | 0.49 | 1 | 0.26 | 1 | 1 | 0.38 | 0.9 | 1 | 0.075 | 0.43 | 1 | 0.27 | 0.88 | 1 | 0.43 | 0.85 | 1 | 0.00053 | 7.4e-05 | 0 | 0.00019 | 0 | 4e-04 | 0.00018 | 0 | 0.00049 | -0.078 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 3e-06 | 0 | 9.9e-06 | 3e-06 | 0 | 1e-05 | 0 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.3.1.191 | UDP-3-O-(3-hydroxymyristoyl)glucosamine N-acyltransferase. | 0.18 | 0.52 | 1 | 0.39 | 1 | 1 | 0.18 | 0.85 | 1 | 0.12 | 0.47 | 1 | 0.49 | 0.92 | 1 | 0.44 | 0.85 | 1 | 0.00055 | 0.00023 | 0 | 3e-04 | 0.00012 | 4e-04 | 0.00017 | 0.00012 | 2e-04 | -0.82 | 15 / 36 (42%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 9.6e-05 | 0 | 0.00017 | 0.00036 | 0 | 0.00081 | 1.9 | 3 / 11 (27%) | 3 / 11 (27%) | 0.273 | 0.273 |
| 2.1.1.74 | Methylenetetrahydrofolate--tRNA-(uracil(54)-C(5))-methyltransferase | 0.058 | 0.49 | 1 | 0.69 | 1 | 1 | 0.72 | 0.98 | 1 | 0.061 | 0.43 | 1 | 0.62 | 0.95 | 1 | 0.58 | 0.91 | 1 | 0.006 | 0.006 | 0.0061 | 0.0051 | 0.0053 | 0.0022 | 0.0049 | 0.0044 | 0.002 | -0.058 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0066 | 0.0066 | 0.0017 | 0.0067 | 0.0066 | 0.0015 | 0.022 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.8.5.3 | Respiratory dimethylsulfoxide reductase. | 0.11 | 0.5 | 1 | 0.15 | 1 | 1 | 0.75 | 0.99 | 1 | 0.074 | 0.43 | 1 | 0.24 | 0.88 | 1 | 0.82 | 0.96 | 1 | 0.00068 | 0.00038 | 5.1e-05 | 0.00089 | 0.00014 | 0.0019 | 0.00076 | 5.4e-05 | 0.0018 | -0.23 | 20 / 36 (56%) | 6 / 7 (86%) | 5 / 7 (71%) | 0.857 | 0.714 | 0.00014 | 0 | 0.00023 | 5.6e-05 | 0 | 8.8e-05 | -1.3 | 5 / 11 (45%) | 4 / 11 (36%) | 0.455 | 0.364 |
| 2.4.1.281 | 4-O-beta-D-mannosyl-D-glucose phosphorylase. | 0.37 | 0.64 | 1 | 0.68 | 1 | 1 | 0.12 | 0.82 | 1 | 0.47 | 0.72 | 1 | 0.93 | 0.99 | 1 | 0.23 | 0.75 | 1 | 0.004 | 0.004 | 0.0038 | 0.0043 | 0.0031 | 0.003 | 0.0038 | 0.0039 | 0.0018 | -0.18 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0034 | 0.0038 | 0.0017 | 0.0044 | 0.0042 | 0.0018 | 0.37 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.-.-.- | 0.87 | 0.94 | 1 | 0.47 | 1 | 1 | 0.7 | 0.98 | 1 | 0.87 | 0.95 | 1 | 0.9 | 0.99 | 1 | 0.37 | 0.82 | 1 | 0.0099 | 0.0099 | 0.0085 | 0.011 | 0.011 | 0.0048 | 0.0098 | 0.01 | 0.0047 | -0.17 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.0082 | 0.0067 | 0.0085 | 0.0083 | 0.0024 | -0.37 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 1.8.2.3 | Sulfide-cytochrome-c reductase (flavocytochrome c). | 0.62 | 0.82 | 1 | 0.69 | 1 | 1 | 0.44 | 0.93 | 1 | 0.43 | 0.69 | 1 | 0.84 | 0.98 | 1 | 0.65 | 0.92 | 1 | 8.7e-05 | 1.2e-05 | 0 | 2.3e-06 | 0 | 6.1e-06 | 0 | 0 | 0 | -Inf | 5 / 36 (14%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 1e-05 | 0 | 2.5e-05 | 2.8e-05 | 0 | 7.2e-05 | 1.5 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 3.2.1.135 | Neopullulanase. | 0.53 | 0.76 | 1 | 0.78 | 1 | 1 | 0.51 | 0.95 | 1 | 0.44 | 0.69 | 1 | 0.85 | 0.98 | 1 | 0.78 | 0.95 | 1 | 0.028 | 0.028 | 0.027 | 0.025 | 0.025 | 0.0052 | 0.025 | 0.03 | 0.011 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.029 | 0.031 | 0.012 | 0.03 | 0.026 | 0.0096 | 0.049 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.8.4.12 | Peptide-methionine (R)-S-oxide reductase. | 0.4 | 0.67 | 1 | 0.82 | 1 | 1 | 0.23 | 0.88 | 1 | 0.1 | 0.46 | 1 | 0.51 | 0.93 | 1 | 0.063 | 0.66 | 1 | 0.0018 | 0.0018 | 0.0017 | 0.0017 | 0.0017 | 0.0013 | 0.0017 | 0.001 | 0.0015 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.002 | 0.0019 | 0.00096 | 0.0016 | 0.0016 | 0.00091 | -0.32 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.3.2.12 | Oxepin-CoA hydrolase. | 0.83 | 0.92 | 1 | 0.48 | 1 | 1 | 0.91 | 1 | 1 | 0.87 | 0.96 | 1 | 0.51 | 0.93 | 1 | 0.85 | 0.96 | 1 | 0.00022 | 1.9e-05 | 0 | 1.7e-05 | 0 | 4.5e-05 | 0 | 0 | 0 | -Inf | 3 / 36 (8.3%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 4.4e-05 | 0 | 0.00015 | 6.5e-06 | 0 | 2.1e-05 | -2.8 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 1.3.1.31 | 2-enoate reductase. | 0.29 | 0.57 | 1 | 0.63 | 1 | 1 | 0.59 | 0.96 | 1 | 0.42 | 0.68 | 1 | 0.52 | 0.93 | 1 | 0.44 | 0.85 | 1 | 0.00012 | 3.7e-05 | 0 | 6.5e-05 | 0 | 0.00013 | 7.7e-05 | 0 | 0.00012 | 0.24 | 11 / 36 (31%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 1.5e-05 | 0 | 2.7e-05 | 1.6e-05 | 0 | 4.3e-05 | 0.093 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 1.1.1.215 | Gluconate 2-dehydrogenase. | 0.18 | 0.52 | 1 | 0.77 | 1 | 1 | 0.71 | 0.98 | 1 | 0.12 | 0.47 | 1 | 0.97 | 0.99 | 1 | 0.56 | 0.9 | 1 | 0.00026 | 2.9e-05 | 0 | 5.3e-05 | 0 | 0.00013 | 8.7e-05 | 0 | 0.00023 | 0.72 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.8.3.1 | Propionate CoA-transferase. | 0.31 | 0.59 | 1 | 0.47 | 1 | 1 | 0.81 | 1 | 1 | 0.45 | 0.7 | 1 | 0.68 | 0.95 | 1 | 0.91 | 0.98 | 1 | 0.0015 | 0.0015 | 0.0013 | 0.0017 | 0.0012 | 0.0012 | 0.0021 | 0.0021 | 0.0014 | 0.3 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0012 | 0.0012 | 0.00084 | 0.0014 | 0.0013 | 0.00093 | 0.22 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.11.18 | Methionyl aminopeptidase. | 0.046 | 0.49 | 1 | 1 | 1 | 1 | 0.92 | 1 | 1 | 0.14 | 0.47 | 1 | 0.11 | 0.87 | 1 | 0.11 | 0.66 | 1 | 0.023 | 0.023 | 0.023 | 0.021 | 0.021 | 0.0032 | 0.021 | 0.022 | 0.0046 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.024 | 0.025 | 0.0033 | 0.024 | 0.025 | 0.0027 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.2.1.79 | Succinate-semialdehyde dehydrogenase (NADP(+)). | 0.72 | 0.87 | 1 | 0.95 | 1 | 1 | 0.94 | 1 | 1 | 0.89 | 0.97 | 1 | 0.82 | 0.98 | 1 | 0.69 | 0.94 | 1 | 0.00039 | 0.00022 | 8.2e-05 | 0.00025 | 0.00014 | 0.00042 | 0.00025 | 0.00013 | 0.00041 | 0 | 20 / 36 (56%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.00016 | 6.5e-05 | 2e-04 | 0.00024 | 2.9e-05 | 0.00049 | 0.58 | 6 / 11 (55%) | 6 / 11 (55%) | 0.545 | 0.545 |
| 1.1.1.131 | Mannuronate reductase. | 0.87 | 0.94 | 1 | 0.18 | 1 | 1 | 0.058 | 0.71 | 1 | 0.91 | 0.97 | 1 | 0.22 | 0.88 | 1 | 0.096 | 0.66 | 1 | 0.002 | 0.002 | 0.0016 | 0.0019 | 0.0019 | 0.00085 | 0.0012 | 0.0013 | 0.00054 | -0.66 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0019 | 0.0016 | 0.0013 | 0.0026 | 0.0026 | 0.0017 | 0.45 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.125 | 2-deoxy-D-gluconate 3-dehydrogenase. | 0.36 | 0.63 | 1 | 0.07 | 1 | 1 | 0.051 | 0.71 | 1 | 0.24 | 0.54 | 1 | 0.07 | 0.86 | 1 | 0.053 | 0.65 | 1 | 0.0011 | 0.001 | 0.00069 | 0.0012 | 0.0013 | 0.00078 | 0.00065 | 0.00024 | 0.00071 | -0.88 | 34 / 36 (94%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 9e-04 | 0.00056 | 0.00096 | 0.0013 | 6e-04 | 0.0019 | 0.53 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 6.3.2.17 | Tetrahydrofolate synthase. | 0.17 | 0.5 | 1 | 0.83 | 1 | 1 | 0.91 | 1 | 1 | 0.15 | 0.47 | 1 | 0.51 | 0.93 | 1 | 0.34 | 0.8 | 1 | 0.022 | 0.022 | 0.023 | 0.02 | 0.02 | 0.0066 | 0.02 | 0.018 | 0.0076 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.024 | 0.024 | 0.0066 | 0.023 | 0.023 | 0.0038 | -0.061 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.7.67 | CDP-2.3-bis-(O-geranylgeranyl)-sn-glycerol synthase. | 0.4 | 0.66 | 1 | 0.25 | 1 | 1 | 0.78 | 1 | 1 | 0.68 | 0.87 | 1 | 0.55 | 0.93 | 1 | 0.78 | 0.95 | 1 | 0.00012 | 1e-05 | 0 | 3.2e-05 | 0 | 8.6e-05 | 4.4e-06 | 0 | 1.2e-05 | -2.9 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 9.5e-06 | 0 | 3.2e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 5.1.1.1 | Alanine racemase. | 0.0062 | 0.49 | 1 | 0.29 | 1 | 1 | 0.18 | 0.85 | 1 | 0.023 | 0.42 | 1 | 0.053 | 0.81 | 1 | 0.026 | 0.59 | 1 | 0.03 | 0.03 | 0.028 | 0.025 | 0.025 | 0.0021 | 0.027 | 0.024 | 0.0062 | 0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.033 | 0.032 | 0.0074 | 0.031 | 0.031 | 0.0069 | -0.09 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.1.7 | Sucrose phosphorylase. | 0.31 | 0.59 | 1 | 0.82 | 1 | 1 | 0.68 | 0.98 | 1 | 0.39 | 0.66 | 1 | 0.79 | 0.97 | 1 | 0.43 | 0.85 | 1 | 0.0061 | 0.0061 | 0.0052 | 0.0045 | 0.0054 | 0.0026 | 0.0044 | 0.0035 | 0.0035 | -0.032 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0073 | 0.0049 | 0.0072 | 0.0069 | 0.0074 | 0.0034 | -0.081 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.4.2.6 | Beta-phosphoglucomutase. | 0.85 | 0.94 | 1 | 0.27 | 1 | 1 | 0.66 | 0.97 | 1 | 0.8 | 0.92 | 1 | 0.24 | 0.88 | 1 | 0.5 | 0.88 | 1 | 0.003 | 0.003 | 0.0029 | 0.0029 | 0.003 | 0.0013 | 0.0039 | 0.0025 | 0.0031 | 0.43 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0027 | 0.0029 | 0.00086 | 0.003 | 0.0028 | 0.0014 | 0.15 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.7.1.1 | Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P-lyase. | 0.6 | 0.81 | 1 | 0.78 | 1 | 1 | 0.89 | 1 | 1 | 0.56 | 0.79 | 1 | 0.7 | 0.95 | 1 | 0.96 | 0.99 | 1 | 0.00019 | 7.5e-05 | 0 | 8.4e-05 | 1.6e-05 | 1e-04 | 0.00011 | 0 | 0.00023 | 0.39 | 14 / 36 (39%) | 4 / 7 (57%) | 3 / 7 (43%) | 0.571 | 0.429 | 7.2e-05 | 0 | 0.00014 | 5.1e-05 | 0 | 1e-04 | -0.5 | 4 / 11 (36%) | 3 / 11 (27%) | 0.364 | 0.273 |
| 1.2.1.59 | Glyceraldehyde-3-phosphate dehydrogenase (NAD(P)(+)) (phosphorylating). | 0.17 | 0.51 | 1 | 0.37 | 1 | 1 | 0.17 | 0.85 | 1 | 0.3 | 0.58 | 1 | 0.75 | 0.97 | 1 | 0.35 | 0.81 | 1 | 0.00023 | 1.9e-05 | 0 | 5.3e-05 | 0 | 0.00014 | 1.3e-05 | 0 | 3.5e-05 | -2 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 2.1e-05 | 0 | 6.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.1.3.15 | (S)-2-hydroxy-acid oxidase. | 0.19 | 0.53 | 1 | 0.34 | 1 | 1 | 0.31 | 0.89 | 1 | 0.2 | 0.51 | 1 | 0.38 | 0.9 | 1 | 0.27 | 0.77 | 1 | 0.012 | 0.012 | 0.012 | 0.0098 | 0.0089 | 0.0032 | 0.012 | 0.013 | 0.0074 | 0.29 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.013 | 0.011 | 0.004 | 0.012 | 0.012 | 0.0036 | -0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.1.-.- | 0.8 | 0.91 | 1 | 0.36 | 1 | 1 | 0.29 | 0.89 | 1 | 0.65 | 0.85 | 1 | 0.2 | 0.87 | 1 | 0.09 | 0.66 | 1 | 0.00024 | 0.00012 | 1.4e-05 | 0.00019 | 0 | 0.00044 | 0.00019 | 0.00011 | 0.00032 | 0 | 18 / 36 (50%) | 3 / 7 (43%) | 4 / 7 (57%) | 0.429 | 0.571 | 6.5e-05 | 6.2e-05 | 7.8e-05 | 8.2e-05 | 0 | 0.00016 | 0.34 | 7 / 11 (64%) | 4 / 11 (36%) | 0.636 | 0.364 | |
| 3.4.23.51 | HycI peptidase. | 0.085 | 0.49 | 1 | 0.36 | 1 | 1 | 0.029 | 0.71 | 1 | 0.074 | 0.43 | 1 | 0.22 | 0.88 | 1 | 0.014 | 0.55 | 1 | 0.00031 | 0.00019 | 0.00011 | 0.00012 | 0 | 0.00022 | 0.00019 | 0 | 0.00027 | 0.66 | 22 / 36 (61%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 0.00029 | 0.00028 | 0.00026 | 0.00013 | 0.00011 | 0.00013 | -1.2 | 8 / 11 (73%) | 8 / 11 (73%) | 0.727 | 0.727 |
| 1.5.1.34 | 6.7-dihydropteridine reductase. | 0.45 | 0.7 | 1 | 0.11 | 1 | 1 | 0.046 | 0.71 | 1 | 0.71 | 0.88 | 1 | 0.22 | 0.88 | 1 | 0.13 | 0.68 | 1 | 0.00032 | 0.00022 | 9.7e-05 | 0.00032 | 8.1e-05 | 0.00055 | 0.00012 | 2.7e-05 | 0.00018 | -1.4 | 24 / 36 (67%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 0.00014 | 8.7e-05 | 0.00015 | 0.00028 | 0.00015 | 0.00043 | 1 | 7 / 11 (64%) | 8 / 11 (73%) | 0.636 | 0.727 |
| 7.2.4.2 | Oxaloacetate decarboxylase (Na(+) extruding). | 0.49 | 0.73 | 1 | 0.24 | 1 | 1 | 0.54 | 0.95 | 1 | 0.92 | 0.97 | 1 | 0.62 | 0.95 | 1 | 0.93 | 0.98 | 1 | 0.0058 | 0.0058 | 0.0057 | 0.0064 | 0.0051 | 0.0023 | 0.0058 | 0.0063 | 0.0023 | -0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0058 | 0.0059 | 0.0018 | 0.0055 | 0.0054 | 0.0014 | -0.077 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.3.6 | Citrate (pro-3S)-lyase. | 0.26 | 0.56 | 1 | 0.48 | 1 | 1 | 0.66 | 0.97 | 1 | 0.47 | 0.72 | 1 | 0.81 | 0.98 | 1 | 0.9 | 0.98 | 1 | 0.0039 | 0.0039 | 0.0036 | 0.0045 | 0.0043 | 0.00066 | 0.004 | 0.0035 | 0.0013 | -0.17 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0038 | 0.0033 | 0.0022 | 0.0037 | 0.0036 | 0.002 | -0.038 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.2.1.19 | Long-chain-fatty-acid--protein ligase. | 0.27 | 0.56 | 1 | 0.36 | 1 | 1 | 0.27 | 0.89 | 1 | 0.16 | 0.48 | 1 | 0.26 | 0.88 | 1 | 0.11 | 0.66 | 1 | 0.0015 | 0.0012 | 0.00085 | 0.00085 | 0.00048 | 0.001 | 0.0013 | 0.0011 | 0.0014 | 0.61 | 30 / 36 (83%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.0014 | 0.0012 | 0.0016 | 0.0012 | 0.00087 | 0.0011 | -0.22 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 4.2.1.30 | Glycerol dehydratase. | 0.52 | 0.75 | 1 | 0.17 | 1 | 1 | 0.44 | 0.93 | 1 | 0.25 | 0.54 | 1 | 0.46 | 0.92 | 1 | 0.2 | 0.73 | 1 | 0.0035 | 0.0035 | 0.0034 | 0.0039 | 0.0035 | 0.0028 | 0.003 | 0.0015 | 0.0026 | -0.38 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0041 | 0.0043 | 0.00082 | 0.0028 | 0.0028 | 0.0016 | -0.55 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.7.99.4 | Nitrate reductase. | 0.11 | 0.5 | 1 | 0.55 | 1 | 1 | 0.72 | 0.98 | 1 | 0.089 | 0.45 | 1 | 0.94 | 0.99 | 1 | 0.82 | 0.96 | 1 | 0.001 | 0.00092 | 0.00023 | 0.002 | 0.00042 | 0.0041 | 0.0017 | 0.00032 | 0.0037 | -0.23 | 32 / 36 (89%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00031 | 0.00021 | 0.00029 | 0.00035 | 0.00015 | 0.00058 | 0.18 | 9 / 11 (82%) | 9 / 11 (82%) | 0.818 | 0.818 |
| 2.7.4.14 | UMP/CMP kinase. | 0.067 | 0.49 | 1 | 0.3 | 1 | 1 | 0.53 | 0.95 | 1 | 0.066 | 0.43 | 1 | 0.28 | 0.88 | 1 | 0.48 | 0.87 | 1 | 0.0041 | 0.0041 | 0.0039 | 0.0028 | 0.0029 | 0.0014 | 0.0039 | 0.0023 | 0.0031 | 0.48 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0045 | 0.0048 | 0.002 | 0.0046 | 0.0043 | 0.0017 | 0.032 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.4.28 | S-adenosylhomocysteine deaminase. | 0.74 | 0.88 | 1 | 0.64 | 1 | 1 | 0.97 | 1 | 1 | 0.39 | 0.65 | 1 | 0.35 | 0.9 | 1 | 0.55 | 0.9 | 1 | 0.0051 | 0.0051 | 0.0046 | 0.0049 | 0.0037 | 0.0028 | 0.0054 | 0.0031 | 0.0037 | 0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.005 | 0.0046 | 0.0013 | 0.0053 | 0.0046 | 0.0019 | 0.084 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.2.6 | Beta-lactamase. | 0.064 | 0.49 | 1 | 0.34 | 1 | 1 | 0.24 | 0.88 | 1 | 0.19 | 0.5 | 1 | 0.68 | 0.95 | 1 | 0.44 | 0.85 | 1 | 0.013 | 0.013 | 0.013 | 0.016 | 0.015 | 0.003 | 0.014 | 0.016 | 0.0038 | -0.19 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.012 | 0.013 | 0.0048 | 0.013 | 0.013 | 0.004 | 0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.86 | 6-phospho-beta-glucosidase. | 0.46 | 0.71 | 1 | 0.097 | 1 | 1 | 0.17 | 0.85 | 1 | 0.87 | 0.96 | 1 | 0.25 | 0.88 | 1 | 0.48 | 0.87 | 1 | 0.025 | 0.025 | 0.023 | 0.029 | 0.026 | 0.015 | 0.022 | 0.017 | 0.012 | -0.4 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.024 | 0.024 | 0.0095 | 0.025 | 0.022 | 0.012 | 0.059 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.5.1.54 | 3-deoxy-7-phosphoheptulonate synthase. | 0.17 | 0.51 | 1 | 0.52 | 1 | 1 | 0.89 | 1 | 1 | 0.13 | 0.47 | 1 | 0.94 | 0.99 | 1 | 0.59 | 0.91 | 1 | 0.027 | 0.027 | 0.027 | 0.025 | 0.026 | 0.005 | 0.024 | 0.023 | 0.0076 | -0.059 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.03 | 0.028 | 0.011 | 0.028 | 0.027 | 0.0049 | -0.1 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.284 | S-(hydroxymethyl)glutathione dehydrogenase. | 0.9 | 0.96 | 1 | 0.83 | 1 | 1 | 0.57 | 0.96 | 1 | 0.57 | 0.79 | 1 | 0.91 | 0.99 | 1 | 0.34 | 0.8 | 1 | 0.0013 | 0.0011 | 0.00083 | 0.0011 | 0.00082 | 0.0012 | 0.0011 | 0.00072 | 0.0012 | 0 | 30 / 36 (83%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.0011 | 0.00084 | 0.0014 | 0.00097 | 0.00085 | 0.00065 | -0.18 | 9 / 11 (82%) | 11 / 11 (100%) | 0.818 | 1 |
| 4.1.1.70 | Transferred entry: 7.2.4.5. | 0.12 | 0.5 | 1 | 0.21 | 1 | 1 | 0.089 | 0.79 | 1 | 0.19 | 0.5 | 1 | 0.37 | 0.9 | 1 | 0.19 | 0.72 | 1 | 0.0066 | 0.0066 | 0.006 | 0.0077 | 0.0082 | 0.002 | 0.006 | 0.0052 | 0.0025 | -0.36 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0057 | 0.0052 | 0.0029 | 0.0073 | 0.0051 | 0.0049 | 0.36 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.2.59 | Sulfide-dependent adenosine diphosphate thiazole synthase. | 0.22 | 0.54 | 1 | 0.9 | 1 | 1 | 0.43 | 0.93 | 1 | 0.16 | 0.48 | 1 | 0.97 | 0.99 | 1 | 0.22 | 0.74 | 1 | 3e-04 | 0.00011 | 0 | 4.7e-05 | 0 | 8.1e-05 | 3.7e-05 | 0 | 6.3e-05 | -0.35 | 13 / 36 (36%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0.00015 | 9.7e-05 | 0.00018 | 0.00015 | 0 | 0.00045 | 0 | 6 / 11 (55%) | 3 / 11 (27%) | 0.545 | 0.273 |
| 2.7.7.23 | UDP-N-acetylglucosamine diphosphorylase. | 0.042 | 0.49 | 1 | 1 | 1 | 1 | 0.87 | 1 | 1 | 0.046 | 0.43 | 1 | 0.69 | 0.95 | 1 | 0.58 | 0.91 | 1 | 0.0073 | 0.0073 | 0.0067 | 0.0053 | 0.0054 | 0.0017 | 0.0054 | 0.0047 | 0.0022 | 0.027 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0089 | 0.0081 | 0.0053 | 0.0083 | 0.0082 | 0.0035 | -0.1 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.26.11 | Ribonuclease Z. | 0.33 | 0.6 | 1 | 0.39 | 1 | 1 | 0.68 | 0.98 | 1 | 0.27 | 0.56 | 1 | 0.09 | 0.86 | 1 | 0.2 | 0.73 | 1 | 0.011 | 0.011 | 0.011 | 0.01 | 0.011 | 0.0026 | 0.011 | 0.012 | 0.0048 | 0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.011 | 0.0025 | 0.012 | 0.011 | 0.0017 | 0.13 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.1.157 | Sarcosine/dimethylglycine N-methyltransferase. | 0.13 | 0.5 | 1 | 0.18 | 1 | 1 | 0.055 | 0.71 | 1 | 0.17 | 0.49 | 1 | 0.19 | 0.87 | 1 | 0.11 | 0.66 | 1 | 0.00033 | 0.00017 | 6.4e-05 | 0.00026 | 2e-04 | 0.00026 | 0.00018 | 0 | 0.00028 | -0.53 | 19 / 36 (53%) | 5 / 7 (71%) | 3 / 7 (43%) | 0.714 | 0.429 | 8.6e-05 | 0 | 0.00013 | 2e-04 | 3e-05 | 0.00027 | 1.2 | 5 / 11 (45%) | 6 / 11 (55%) | 0.455 | 0.545 |
| 2.1.1.258 | 5-methyltetrahydrofolate:corrinoid/iron-sulfur protein | 0.069 | 0.49 | 1 | 0.73 | 1 | 1 | 0.95 | 1 | 1 | 0.036 | 0.43 | 1 | 0.93 | 0.99 | 1 | 0.76 | 0.95 | 1 | 2e-04 | 4e-05 | 0 | 5.2e-05 | 1.6e-05 | 8.6e-05 | 0.00014 | 0 | 0.00033 | 1.4 | 7 / 36 (19%) | 4 / 7 (57%) | 2 / 7 (29%) | 0.571 | 0.286 | 0 | 0 | 0 | 6.5e-06 | 0 | 2.1e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.3.99.10 | Transferred entry: 1.3.8.4. | 0.095 | 0.49 | 1 | 0.098 | 1 | 1 | 0.33 | 0.89 | 1 | 0.065 | 0.43 | 1 | 0.23 | 0.88 | 1 | 0.63 | 0.92 | 1 | 5e-04 | 5.5e-05 | 0 | 0.00015 | 0 | 0.00035 | 0.00013 | 0 | 0.00033 | -0.21 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 1.5.5.1 | Electron-transferring-flavoprotein dehydrogenase. | 0.28 | 0.56 | 1 | 0.73 | 1 | 1 | 0.68 | 0.98 | 1 | 0.47 | 0.72 | 1 | 0.91 | 0.99 | 1 | 0.58 | 0.91 | 1 | 0.00033 | 0.00016 | 0 | 0.00034 | 4.8e-05 | 0.00079 | 0.00032 | 2.7e-05 | 0.00076 | -0.087 | 17 / 36 (47%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 6.3e-05 | 0 | 9e-05 | 2.8e-05 | 0 | 4.2e-05 | -1.2 | 5 / 11 (45%) | 4 / 11 (36%) | 0.455 | 0.364 |
| 2.3.1.18 | Galactoside O-acetyltransferase. | 0.25 | 0.56 | 1 | 0.7 | 1 | 1 | 0.92 | 1 | 1 | 0.13 | 0.47 | 1 | 0.91 | 0.99 | 1 | 0.89 | 0.98 | 1 | 0.0067 | 0.0067 | 0.0053 | 0.0053 | 0.0041 | 0.0031 | 0.0049 | 0.0054 | 0.0025 | -0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0083 | 0.0059 | 0.0089 | 0.0071 | 0.0058 | 0.0032 | -0.23 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.7.72 | CCA tRNA nucleotidyltransferase. | 0.28 | 0.57 | 1 | 0.6 | 1 | 1 | 0.34 | 0.89 | 1 | 0.2 | 0.5 | 1 | 0.24 | 0.88 | 1 | 0.13 | 0.68 | 1 | 0.018 | 0.018 | 0.018 | 0.017 | 0.014 | 0.0059 | 0.018 | 0.019 | 0.0079 | 0.082 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.02 | 0.018 | 0.0093 | 0.018 | 0.019 | 0.0032 | -0.15 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.1.29 | Glycine C-acetyltransferase. | 0.48 | 0.72 | 1 | 0.17 | 1 | 1 | 0.2 | 0.87 | 1 | 0.33 | 0.6 | 1 | 0.2 | 0.87 | 1 | 0.28 | 0.78 | 1 | 0.0024 | 0.0024 | 0.0015 | 0.0026 | 0.0023 | 0.0012 | 0.0017 | 0.0015 | 0.00096 | -0.61 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0023 | 0.0013 | 0.0027 | 0.0027 | 0.0014 | 0.0036 | 0.23 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.1.- | 0.029 | 0.49 | 1 | 0.35 | 1 | 1 | 0.93 | 1 | 1 | 0.024 | 0.42 | 1 | 0.66 | 0.95 | 1 | 0.14 | 0.7 | 1 | 0.036 | 0.036 | 0.036 | 0.033 | 0.031 | 0.0061 | 0.031 | 0.034 | 0.0056 | -0.09 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.04 | 0.039 | 0.0081 | 0.037 | 0.038 | 0.0046 | -0.11 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 3.2.2.16 | Methylthioadenosine nucleosidase. | 0.12 | 0.5 | 1 | 0.56 | 1 | 1 | 0.92 | 1 | 1 | 0.14 | 0.47 | 1 | 0.66 | 0.95 | 1 | 0.82 | 0.96 | 1 | 0.00022 | 6.6e-05 | 0 | 0.00013 | 8.1e-05 | 0.00021 | 0.00011 | 0 | 2e-04 | -0.24 | 11 / 36 (31%) | 4 / 7 (57%) | 3 / 7 (43%) | 0.571 | 0.429 | 2.8e-05 | 0 | 4.9e-05 | 3.5e-05 | 0 | 0.00012 | 0.32 | 3 / 11 (27%) | 1 / 11 (9.1%) | 0.273 | 0.0909 |
| 2.8.3.- | 0.57 | 0.79 | 1 | 0.41 | 1 | 1 | 0.043 | 0.71 | 1 | 0.91 | 0.97 | 1 | 0.63 | 0.95 | 1 | 0.11 | 0.66 | 1 | 0.0089 | 0.0089 | 0.0086 | 0.0086 | 0.0084 | 0.0017 | 0.0077 | 0.0087 | 0.0031 | -0.16 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0079 | 0.0083 | 0.0024 | 0.011 | 0.011 | 0.0041 | 0.48 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 4.2.1.28 | Propanediol dehydratase. | 0.57 | 0.79 | 1 | 0.82 | 1 | 1 | 0.31 | 0.89 | 1 | 0.62 | 0.83 | 1 | 0.87 | 0.99 | 1 | 0.22 | 0.74 | 1 | 0.0017 | 0.0016 | 0.0014 | 0.0016 | 0.0018 | 0.00079 | 0.0016 | 0.0013 | 0.0012 | 0 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.002 | 0.0016 | 0.0013 | 0.0013 | 0.0014 | 0.001 | -0.62 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 1.1.1.140 | Sorbitol-6-phosphate 2-dehydrogenase. | 0.77 | 0.89 | 1 | 0.18 | 1 | 1 | 0.54 | 0.95 | 1 | 0.91 | 0.97 | 1 | 0.97 | 0.99 | 1 | 0.28 | 0.78 | 1 | 0.0013 | 0.0013 | 0.0011 | 0.00093 | 0.00083 | 5e-04 | 0.0017 | 0.0013 | 0.0017 | 0.87 | 35 / 36 (97%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00084 | 0.00081 | 0.00047 | 0.0016 | 0.0016 | 0.00082 | 0.93 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.5.1.4 | Inositol-3-phosphate synthase. | 0.46 | 0.71 | 1 | 0.62 | 1 | 1 | 0.83 | 1 | 1 | 0.95 | 0.99 | 1 | 0.65 | 0.95 | 1 | 0.31 | 0.79 | 1 | 0.0026 | 0.0025 | 0.0013 | 0.0017 | 0.0014 | 0.0014 | 0.0013 | 0.00096 | 0.001 | -0.39 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0038 | 0.0013 | 0.0072 | 0.0026 | 0.0018 | 0.0026 | -0.55 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 6.6.1.2 | Cobaltochelatase. | 0.16 | 0.5 | 1 | 0.69 | 1 | 1 | 0.17 | 0.85 | 1 | 0.07 | 0.43 | 1 | 0.76 | 0.97 | 1 | 0.31 | 0.79 | 1 | 0.0027 | 0.0027 | 0.0014 | 0.0028 | 0.0022 | 0.0016 | 0.0028 | 0.0013 | 0.0034 | 0 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0015 | 0.00078 | 0.0019 | 0.0037 | 0.0014 | 0.0056 | 1.3 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 4.1.1.48 | Indole-3-glycerol-phosphate synthase. | 0.032 | 0.49 | 1 | 0.52 | 1 | 1 | 0.37 | 0.9 | 1 | 0.021 | 0.42 | 1 | 0.13 | 0.87 | 1 | 0.11 | 0.66 | 1 | 0.011 | 0.011 | 0.0098 | 0.0084 | 0.0078 | 0.0012 | 0.0093 | 0.0088 | 0.0024 | 0.15 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.013 | 0.01 | 0.0072 | 0.011 | 0.012 | 0.0025 | -0.24 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.50 | Alpha-N-acetylglucosaminidase. | 0.25 | 0.56 | 1 | 0.98 | 1 | 1 | 0.51 | 0.95 | 1 | 0.019 | 0.42 | 1 | 0.85 | 0.98 | 1 | 0.098 | 0.66 | 1 | 0.0011 | 0.00083 | 0.00033 | 0.00094 | 0.00033 | 0.0013 | 0.001 | 0.00074 | 0.0012 | 0.089 | 27 / 36 (75%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00079 | 0 | 0.0017 | 0.00069 | 0.00038 | 0.00092 | -0.2 | 4 / 11 (36%) | 9 / 11 (82%) | 0.364 | 0.818 |
| 3.1.3.71 | 2-phosphosulfolactate phosphatase. | 0.51 | 0.74 | 1 | 0.9 | 1 | 1 | 0.63 | 0.96 | 1 | 0.5 | 0.74 | 1 | 0.93 | 0.99 | 1 | 0.62 | 0.92 | 1 | 0.00022 | 7.2e-05 | 0 | 6.2e-05 | 0 | 0.00014 | 4.4e-05 | 0 | 8.2e-05 | -0.49 | 12 / 36 (33%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0.00013 | 0 | 0.00026 | 4.2e-05 | 0 | 7.6e-05 | -1.6 | 4 / 11 (36%) | 4 / 11 (36%) | 0.364 | 0.364 |
| 1.11.1.21 | Catalase peroxidase. | 0.21 | 0.54 | 1 | 0.88 | 1 | 1 | 0.97 | 1 | 1 | 0.23 | 0.53 | 1 | 0.85 | 0.98 | 1 | 0.93 | 0.98 | 1 | 0.00078 | 0.00011 | 0 | 0.00025 | 0 | 0.00067 | 0.00029 | 0 | 0.00077 | 0.21 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 5.1e-06 | 0 | 1.7e-05 | 2.7e-06 | 0 | 8.9e-06 | -0.92 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.1.1.164 | Demethylrebeccamycin-D-glucose O-methyltransferase. | 0.23 | 0.54 | 1 | 0.36 | 1 | 1 | 0.65 | 0.97 | 1 | 0.12 | 0.47 | 1 | 0.54 | 0.93 | 1 | 0.72 | 0.94 | 1 | 0.00074 | 0.00064 | 0.00044 | 6e-04 | 0.00065 | 0.00053 | 0.00046 | 0.00012 | 0.00062 | -0.38 | 31 / 36 (86%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.00087 | 0.00089 | 0.00061 | 0.00055 | 0.00041 | 0.00058 | -0.66 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 4.2.1.7 | Altronate dehydratase. | 0.067 | 0.49 | 1 | 0.14 | 1 | 1 | 0.057 | 0.71 | 1 | 0.13 | 0.47 | 1 | 0.22 | 0.88 | 1 | 0.17 | 0.71 | 1 | 0.0032 | 0.0032 | 0.0028 | 0.0041 | 0.0039 | 0.0019 | 0.0027 | 0.0024 | 0.0017 | -0.6 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0024 | 0.0024 | 0.0012 | 0.0037 | 0.0034 | 0.0026 | 0.62 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.23 | Histidinol dehydrogenase. | 0.14 | 0.5 | 1 | 0.79 | 1 | 1 | 0.75 | 0.99 | 1 | 0.17 | 0.49 | 1 | 0.24 | 0.88 | 1 | 0.6 | 0.91 | 1 | 0.016 | 0.016 | 0.016 | 0.014 | 0.014 | 0.0036 | 0.015 | 0.015 | 0.0026 | 0.1 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.017 | 0.018 | 0.0054 | 0.018 | 0.019 | 0.0028 | 0.082 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.4.1.22 | S-(hydroxymethyl)glutathione synthase. | 0.37 | 0.65 | 1 | 0.25 | 1 | 1 | 0.79 | 1 | 1 | 0.5 | 0.74 | 1 | 0.31 | 0.9 | 1 | 0.96 | 0.99 | 1 | 8.5e-05 | 9.4e-06 | 0 | 2.2e-05 | 0 | 4.5e-05 | 3.9e-06 | 0 | 1e-05 | -2.5 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 1.5e-05 | 0 | 4.9e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 5.3.1.16 | 1-(5-phosphoribosyl)-5- | 0.01 | 0.49 | 1 | 0.26 | 1 | 1 | 0.43 | 0.93 | 1 | 0.014 | 0.42 | 1 | 0.045 | 0.8 | 1 | 0.094 | 0.66 | 1 | 0.01 | 0.01 | 0.0099 | 0.0083 | 0.0085 | 0.001 | 0.0093 | 0.0091 | 0.0028 | 0.16 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.012 | 0.003 | 0.011 | 0.011 | 0.0014 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.3.5.12 | Ni-sirohydrochlorin a.c-diamide synthase. | 0.89 | 0.96 | 1 | 0.54 | 1 | 1 | 0.55 | 0.95 | 1 | 0.73 | 0.9 | 1 | 0.82 | 0.98 | 1 | 0.31 | 0.79 | 1 | 0.00028 | 3.1e-05 | 0 | 5.3e-05 | 0 | 0.00014 | 1.3e-05 | 0 | 3.5e-05 | -2 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 5.9e-05 | 0 | 0.00016 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 3.6.3.44 | Transferred entry: 7.6.2.2. | 0.061 | 0.49 | 1 | 0.41 | 1 | 1 | 0.35 | 0.9 | 1 | 0.0098 | 0.42 | 1 | 0.049 | 0.81 | 1 | 0.084 | 0.66 | 1 | 0.032 | 0.032 | 0.023 | 0.016 | 0.01 | 0.012 | 0.022 | 0.016 | 0.019 | 0.46 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.046 | 0.024 | 0.067 | 0.036 | 0.028 | 0.025 | -0.35 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.9.2 | Pyruvate. water dikinase. | 0.21 | 0.53 | 1 | 0.69 | 1 | 1 | 0.46 | 0.93 | 1 | 0.19 | 0.5 | 1 | 0.68 | 0.95 | 1 | 0.25 | 0.76 | 1 | 0.0011 | 0.001 | 8e-04 | 0.0013 | 0.0012 | 0.0011 | 0.0014 | 0.00064 | 0.0017 | 0.11 | 34 / 36 (94%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00075 | 8e-04 | 0.00053 | 0.00082 | 8e-04 | 0.00069 | 0.13 | 9 / 11 (82%) | 11 / 11 (100%) | 0.818 | 1 |
| 1.8.4.11 | Peptide-methionine (S)-S-oxide reductase. | 0.97 | 0.99 | 1 | 0.9 | 1 | 1 | 0.86 | 1 | 1 | 0.47 | 0.72 | 1 | 0.71 | 0.95 | 1 | 0.43 | 0.85 | 1 | 0.0029 | 0.0029 | 0.0025 | 0.003 | 0.003 | 0.0015 | 0.003 | 0.0025 | 0.0018 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0029 | 0.0024 | 0.0011 | 0.0028 | 0.0025 | 0.0012 | -0.051 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 7.6.2.7 | ABC-type taurine transporter. | 0.041 | 0.49 | 1 | 0.066 | 1 | 1 | 0.031 | 0.71 | 1 | 0.035 | 0.43 | 1 | 0.21 | 0.88 | 1 | 0.081 | 0.66 | 1 | 0.00023 | 3.2e-05 | 0 | 1e-04 | 0 | 0.00022 | 3.9e-05 | 0 | 1e-04 | -1.4 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 1.3e-05 | 0 | 3.5e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 3.2.1.91 | Cellulose 1.4-beta-cellobiosidase (non-reducing end). | 0.67 | 0.84 | 1 | 0.013 | 0.89 | 1 | 0.038 | 0.71 | 1 | 0.52 | 0.75 | 1 | 0.0027 | 0.62 | 1 | 0.0073 | 0.52 | 1 | 0.0038 | 0.0038 | 0.0025 | 0.0038 | 0.002 | 0.0032 | 0.002 | 0.0012 | 0.0025 | -0.93 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0045 | 0.0028 | 0.004 | 0.0042 | 0.0027 | 0.0027 | -0.1 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.1.118 | N-hydroxyarylamine O-acetyltransferase. | 0.26 | 0.56 | 1 | 0.56 | 1 | 1 | 0.6 | 0.96 | 1 | 0.49 | 0.74 | 1 | 0.85 | 0.98 | 1 | 0.98 | 0.99 | 1 | 0.00018 | 6.5e-05 | 0 | 0.00016 | 0 | 4e-04 | 7.4e-05 | 0 | 0.00013 | -1.1 | 13 / 36 (36%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 3.4e-05 | 0 | 7e-05 | 2.7e-05 | 0 | 4.6e-05 | -0.33 | 3 / 11 (27%) | 4 / 11 (36%) | 0.273 | 0.364 |
| 2.5.1.78 | 6.7-dimethyl-8-ribityllumazine synthase. | 0.49 | 0.73 | 1 | 0.89 | 1 | 1 | 0.78 | 1 | 1 | 0.91 | 0.97 | 1 | 0.51 | 0.93 | 1 | 0.84 | 0.96 | 1 | 0.0052 | 0.0052 | 0.0052 | 0.0053 | 0.005 | 0.0015 | 0.0055 | 0.0054 | 0.0018 | 0.053 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0049 | 0.005 | 0.0014 | 0.0052 | 0.0052 | 0.0015 | 0.086 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.1.- | 0.54 | 0.76 | 1 | 0.97 | 1 | 1 | 0.96 | 1 | 1 | 0.31 | 0.58 | 1 | 0.42 | 0.91 | 1 | 0.45 | 0.86 | 1 | 0.041 | 0.041 | 0.04 | 0.039 | 0.033 | 0.013 | 0.039 | 0.034 | 0.016 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.042 | 0.04 | 0.014 | 0.042 | 0.04 | 0.007 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 3.4.14.4 | Dipeptidyl-peptidase III. | 0.084 | 0.49 | 1 | 0.91 | 1 | 1 | 0.13 | 0.82 | 1 | 0.0024 | 0.34 | 1 | 0.46 | 0.92 | 1 | 0.019 | 0.55 | 1 | 0.0015 | 0.001 | 0.00016 | 0.00095 | 0.00058 | 0.00099 | 0.0016 | 0.00021 | 0.0031 | 0.75 | 25 / 36 (69%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00034 | 0 | 0.001 | 0.0013 | 0.00018 | 0.0026 | 1.9 | 4 / 11 (36%) | 8 / 11 (73%) | 0.364 | 0.727 |
| 2.1.3.5 | Oxamate carbamoyltransferase. | 0.063 | 0.49 | 1 | 0.073 | 1 | 1 | 0.099 | 0.82 | 1 | 0.072 | 0.43 | 1 | 0.24 | 0.88 | 1 | 0.28 | 0.78 | 1 | 0.00023 | 3.2e-05 | 0 | 1e-04 | 0 | 0.00022 | 3.9e-05 | 0 | 1e-04 | -1.4 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 9.9e-06 | 0 | 3.3e-05 | 1.9 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 1.15.1.- | 0.049 | 0.49 | 1 | 0.16 | 1 | 1 | 0.28 | 0.89 | 1 | 0.15 | 0.48 | 1 | 0.38 | 0.9 | 1 | 0.53 | 0.9 | 1 | 0.00049 | 0.00049 | 0.00033 | 8e-04 | 0.00073 | 0.00074 | 0.00056 | 6e-04 | 0.00041 | -0.51 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00038 | 0.00022 | 0.00032 | 0.00036 | 0.00033 | 0.00022 | -0.078 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 3.4.11.24 | Aminopeptidase S. | 0.41 | 0.67 | 1 | 0.75 | 1 | 1 | 1 | 1 | 1 | 0.22 | 0.52 | 1 | 0.52 | 0.93 | 1 | 0.41 | 0.84 | 1 | 0.0017 | 0.0017 | 0.0012 | 0.0013 | 0.00081 | 0.0011 | 0.0014 | 0.0011 | 0.00099 | 0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0017 | 0.0019 | 0.00096 | 0.0022 | 0.0014 | 0.0024 | 0.37 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.4.2.3 | Phosphoacetylglucosamine mutase. | 0.37 | 0.65 | 1 | 0.53 | 1 | 1 | 0.53 | 0.95 | 1 | 0.37 | 0.64 | 1 | 0.57 | 0.93 | 1 | 0.48 | 0.87 | 1 | 0.00021 | 1.7e-05 | 0 | 1.2e-05 | 0 | 3.1e-05 | 0 | 0 | 0 | -Inf | 3 / 36 (8.3%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 5e-05 | 0 | 0.00012 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 3.7.1.8 | 2.6-dioxo-6-phenylhexa-3-enoate hydrolase. | 0.76 | 0.89 | 1 | 0.3 | 1 | 1 | 0.23 | 0.88 | 1 | 0.85 | 0.95 | 1 | 0.17 | 0.87 | 1 | 0.13 | 0.68 | 1 | 0.00027 | 0.00023 | 0.00016 | 0.00028 | 0.00012 | 0.00035 | 0.00031 | 0.00032 | 0.00021 | 0.15 | 31 / 36 (86%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 2e-04 | 0.00019 | 0.00012 | 0.00018 | 0.00011 | 2e-04 | -0.15 | 9 / 11 (82%) | 9 / 11 (82%) | 0.818 | 0.818 |
| 3.2.1.1 | Alpha-amylase. | 0.058 | 0.49 | 1 | 0.9 | 1 | 1 | 0.76 | 0.99 | 1 | 0.054 | 0.43 | 1 | 0.58 | 0.93 | 1 | 0.52 | 0.89 | 1 | 0.027 | 0.027 | 0.022 | 0.02 | 0.019 | 0.0085 | 0.021 | 0.019 | 0.011 | 0.07 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.032 | 0.027 | 0.016 | 0.03 | 0.025 | 0.012 | -0.093 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.1.120 | 4-hydroxybutanoyl-CoA dehydratase. | 0.065 | 0.49 | 1 | 0.93 | 1 | 1 | 0.47 | 0.93 | 1 | 0.041 | 0.43 | 1 | 0.73 | 0.96 | 1 | 0.3 | 0.78 | 1 | 0.00028 | 0.00023 | 0.00013 | 0.00014 | 0.00011 | 2e-04 | 0.00012 | 0.00011 | 0.00013 | -0.22 | 29 / 36 (81%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.00034 | 0.00019 | 0.00037 | 0.00024 | 0.00021 | 0.00025 | -0.5 | 11 / 11 (100%) | 8 / 11 (73%) | 1 | 0.727 |
| 5.99.1.- | 0.74 | 0.88 | 1 | 0.9 | 1 | 1 | 0.92 | 1 | 1 | 0.99 | 1 | 1 | 0.45 | 0.92 | 1 | 0.58 | 0.91 | 1 | 0.0064 | 0.0064 | 0.0057 | 0.0069 | 0.0051 | 0.0053 | 0.007 | 0.0058 | 0.005 | 0.021 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0062 | 0.0063 | 0.0028 | 0.0061 | 0.0047 | 0.0027 | -0.023 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 3.9.1.- | 0.15 | 0.5 | 1 | 0.55 | 1 | 1 | 0.89 | 1 | 1 | 0.2 | 0.5 | 1 | 0.16 | 0.87 | 1 | 0.43 | 0.85 | 1 | 0.00027 | 3e-05 | 0 | 8.4e-05 | 0 | 0.00022 | 6.6e-05 | 0 | 0.00013 | -0.35 | 4 / 36 (11%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 0 | 0 | 0 | 3e-06 | 0 | 1e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 | |
| 3.4.22.10 | Streptopain. | 0.58 | 0.79 | 1 | 0.66 | 1 | 1 | 0.66 | 0.97 | 1 | 0.56 | 0.79 | 1 | 0.66 | 0.95 | 1 | 0.82 | 0.96 | 1 | 0.00025 | 0.00011 | 0 | 0.00011 | 4.8e-05 | 0.00015 | 9e-05 | 0 | 0.00015 | -0.29 | 15 / 36 (42%) | 4 / 7 (57%) | 3 / 7 (43%) | 0.571 | 0.429 | 8.3e-05 | 0 | 0.00015 | 0.00014 | 0 | 0.00037 | 0.75 | 4 / 11 (36%) | 4 / 11 (36%) | 0.364 | 0.364 |
| 5.4.3.2 | Lysine 2.3-aminomutase. | 0.17 | 0.5 | 1 | 0.76 | 1 | 1 | 0.91 | 1 | 1 | 0.021 | 0.42 | 1 | 0.95 | 0.99 | 1 | 0.76 | 0.95 | 1 | 0.0015 | 0.0013 | 0.00039 | 0.0014 | 0.0011 | 0.00087 | 0.002 | 0.00083 | 0.0032 | 0.51 | 31 / 36 (86%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00085 | 0.00019 | 0.0016 | 0.0013 | 0.00021 | 0.0027 | 0.61 | 8 / 11 (73%) | 9 / 11 (82%) | 0.727 | 0.818 |
| 3.2.1.40 | Alpha-L-rhamnosidase. | 0.51 | 0.74 | 1 | 0.72 | 1 | 1 | 0.31 | 0.89 | 1 | 0.52 | 0.76 | 1 | 0.67 | 0.95 | 1 | 0.78 | 0.95 | 1 | 0.0025 | 0.0024 | 0.00097 | 0.0023 | 0.0023 | 0.0018 | 0.0017 | 0.0011 | 0.0016 | -0.44 | 34 / 36 (94%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.0019 | 0.00049 | 0.0035 | 0.0034 | 0.00068 | 0.0063 | 0.84 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 2.8.1.1 | Thiosulfate sulfurtransferase. | 0.68 | 0.85 | 1 | 0.4 | 1 | 1 | 0.56 | 0.96 | 1 | 0.89 | 0.97 | 1 | 0.63 | 0.95 | 1 | 0.86 | 0.96 | 1 | 0.011 | 0.011 | 0.0098 | 0.011 | 0.0097 | 0.0043 | 0.01 | 0.011 | 0.0051 | -0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.011 | 0.0036 | 0.01 | 0.0097 | 0.0029 | -0.14 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.1.264 | 23S rRNA (guanine(2069)-N(7))-methyltransferase. | 0.25 | 0.55 | 1 | 0.68 | 1 | 1 | 0.39 | 0.91 | 1 | 0.27 | 0.56 | 1 | 0.59 | 0.93 | 1 | 0.45 | 0.86 | 1 | 0.0012 | 0.0012 | 9e-04 | 0.0011 | 0.00083 | 0.00082 | 0.001 | 0.00047 | 0.0012 | -0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0017 | 0.0013 | 0.0014 | 0.00095 | 0.00091 | 0.00067 | -0.84 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.196 | Protein-N(pi)-phosphohistidine--N.N'-diacetylchitobiose | 0.12 | 0.5 | 1 | 0.9 | 1 | 1 | 0.92 | 1 | 1 | 0.16 | 0.48 | 1 | 0.23 | 0.88 | 1 | 0.3 | 0.78 | 1 | 0.00029 | 2.4e-05 | 0 | 8.4e-05 | 0 | 0.00022 | 4.1e-05 | 0 | 7.8e-05 | -1 | 3 / 36 (8.3%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 4.1.2.13 | Fructose-bisphosphate aldolase. | 0.91 | 0.96 | 1 | 0.87 | 1 | 1 | 0.82 | 1 | 1 | 0.49 | 0.74 | 1 | 0.37 | 0.9 | 1 | 0.52 | 0.9 | 1 | 0.026 | 0.026 | 0.026 | 0.026 | 0.023 | 0.0059 | 0.027 | 0.031 | 0.0092 | 0.054 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.026 | 0.025 | 0.0041 | 0.027 | 0.027 | 0.0046 | 0.054 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.393 | 3-beta-hydroxycholanate 3-dehydrogenase (NADP(+)). | 0.021 | 0.49 | 1 | 0.034 | 0.97 | 1 | 0.031 | 0.71 | 1 | 0.03 | 0.43 | 1 | 0.087 | 0.86 | 1 | 0.074 | 0.66 | 1 | 0.00013 | 2.6e-05 | 0 | 7.9e-05 | 0 | 0.00013 | 2.9e-05 | 0 | 7.7e-05 | -1.4 | 7 / 36 (19%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 2.8e-06 | 0 | 9.4e-06 | 1.3e-05 | 0 | 2.9e-05 | 2.2 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 4.2.3.1 | Threonine synthase. | 0.068 | 0.49 | 1 | 0.96 | 1 | 1 | 0.96 | 1 | 1 | 0.1 | 0.46 | 1 | 0.28 | 0.88 | 1 | 0.23 | 0.75 | 1 | 0.019 | 0.019 | 0.019 | 0.017 | 0.015 | 0.0034 | 0.017 | 0.018 | 0.0038 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.02 | 0.019 | 0.005 | 0.02 | 0.021 | 0.0032 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.3.82 | D-glycero-beta-D-manno-heptose 1.7-bisphosphate 7-phosphatase. | 0.54 | 0.76 | 1 | 0.52 | 1 | 1 | 0.5 | 0.95 | 1 | 0.72 | 0.89 | 1 | 0.5 | 0.93 | 1 | 0.4 | 0.83 | 1 | 0.00064 | 0.00042 | 0.00014 | 0.00058 | 0.00012 | 0.00093 | 0.00059 | 0.00011 | 0.0013 | 0.025 | 24 / 36 (67%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 0.00034 | 0.00013 | 0.00058 | 3e-04 | 2e-04 | 0.00034 | -0.18 | 7 / 11 (64%) | 8 / 11 (73%) | 0.636 | 0.727 |
| 3.1.21.4 | Type II site-specific deoxyribonuclease. | 0.89 | 0.95 | 1 | 0.82 | 1 | 1 | 0.46 | 0.93 | 1 | 0.7 | 0.88 | 1 | 0.51 | 0.93 | 1 | 0.89 | 0.97 | 1 | 0.0054 | 0.0054 | 0.0049 | 0.0055 | 0.0039 | 0.0039 | 0.005 | 0.0041 | 0.0017 | -0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0051 | 0.0052 | 0.0019 | 0.0059 | 0.005 | 0.0029 | 0.21 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.6.1.9 | Nucleotide diphosphatase. | 0.26 | 0.56 | 1 | 0.9 | 1 | 1 | 0.68 | 0.98 | 1 | 0.045 | 0.43 | 1 | 0.3 | 0.89 | 1 | 0.72 | 0.94 | 1 | 0.012 | 0.012 | 0.012 | 0.011 | 0.011 | 0.0016 | 0.011 | 0.012 | 0.0029 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.013 | 0.011 | 0.0058 | 0.014 | 0.012 | 0.0029 | 0.11 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.2.7.1 | Pyruvate synthase. | 0.75 | 0.88 | 1 | 0.43 | 1 | 1 | 0.072 | 0.73 | 1 | 0.74 | 0.9 | 1 | 0.67 | 0.95 | 1 | 0.37 | 0.82 | 1 | 0.043 | 0.043 | 0.042 | 0.042 | 0.046 | 0.0093 | 0.04 | 0.037 | 0.011 | -0.07 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.041 | 0.039 | 0.0098 | 0.046 | 0.043 | 0.0074 | 0.17 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 7.3.2.5 | ABC-type molybdate transporter. | 0.15 | 0.5 | 1 | 0.073 | 1 | 1 | 0.39 | 0.91 | 1 | 0.18 | 0.5 | 1 | 0.18 | 0.87 | 1 | 0.64 | 0.92 | 1 | 0.00035 | 6.7e-05 | 0 | 0.00019 | 0 | 0.00044 | 9.7e-05 | 0 | 0.00026 | -0.97 | 7 / 36 (19%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 3e-05 | 0 | 6.7e-05 | 6.5e-06 | 0 | 2.1e-05 | -2.2 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 3.5.1.87 | N-carbamoyl-L-amino-acid hydrolase. | 0.35 | 0.63 | 1 | 0.11 | 1 | 1 | 0.21 | 0.87 | 1 | 0.12 | 0.47 | 1 | 0.07 | 0.86 | 1 | 0.098 | 0.66 | 1 | 0.0012 | 0.0011 | 0.001 | 0.00087 | 0.0011 | 0.00051 | 0.0013 | 0.00088 | 0.00086 | 0.58 | 35 / 36 (97%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.0011 | 0.0011 | 0.00059 | 0.0012 | 0.00098 | 0.00085 | 0.13 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.49 | Hydroxymethylpyrimidine kinase. | 0.82 | 0.92 | 1 | 0.47 | 1 | 1 | 0.9 | 1 | 1 | 0.5 | 0.74 | 1 | 0.93 | 0.99 | 1 | 0.6 | 0.91 | 1 | 0.012 | 0.012 | 0.012 | 0.012 | 0.013 | 0.0038 | 0.012 | 0.011 | 0.0054 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.013 | 0.012 | 0.0045 | 0.012 | 0.011 | 0.0021 | -0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.8.1.8 | Atrazine chlorohydrolase. | 0.9 | 0.96 | 1 | 0.7 | 1 | 1 | 0.9 | 1 | 1 | 0.85 | 0.95 | 1 | 0.73 | 0.96 | 1 | 0.73 | 0.94 | 1 | 5e-04 | 0.00035 | 0.00023 | 0.00027 | 0.00023 | 3e-04 | 0.00033 | 0.00024 | 0.00035 | 0.29 | 25 / 36 (69%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.00029 | 0.00024 | 0.00029 | 0.00047 | 0.00023 | 0.00067 | 0.7 | 8 / 11 (73%) | 7 / 11 (64%) | 0.727 | 0.636 |
| 2.1.1.170 | 16S rRNA (guanine(527)-N(7))-methyltransferase. | 0.087 | 0.49 | 1 | 0.066 | 1 | 1 | 0.013 | 0.61 | 1 | 0.096 | 0.45 | 1 | 0.25 | 0.88 | 1 | 0.054 | 0.66 | 1 | 0.00023 | 5.7e-05 | 0 | 0.00014 | 0 | 0.00031 | 7.2e-05 | 0 | 0.00018 | -0.96 | 9 / 36 (25%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 1.2e-05 | 0 | 3.9e-05 | 3.9e-05 | 0 | 7.1e-05 | 1.7 | 1 / 11 (9.1%) | 3 / 11 (27%) | 0.0909 | 0.273 |
| 3.5.1.77 | N-carbamoyl-D-amino-acid hydrolase. | 0.54 | 0.77 | 1 | 0.75 | 1 | 1 | 0.83 | 1 | 1 | 0.93 | 0.98 | 1 | 0.38 | 0.9 | 1 | 0.44 | 0.85 | 1 | 0.0054 | 0.0054 | 0.0049 | 0.0057 | 0.0054 | 0.0021 | 0.006 | 0.0068 | 0.0026 | 0.074 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0051 | 0.0055 | 0.0018 | 0.0051 | 0.0047 | 0.0021 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.7.24 | Glucose-1-phosphate thymidylyltransferase. | 0.48 | 0.72 | 1 | 0.14 | 1 | 1 | 0.15 | 0.84 | 1 | 0.86 | 0.95 | 1 | 0.64 | 0.95 | 1 | 0.79 | 0.95 | 1 | 0.016 | 0.016 | 0.016 | 0.017 | 0.016 | 0.0034 | 0.015 | 0.016 | 0.0043 | -0.18 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.015 | 0.015 | 0.0023 | 0.016 | 0.016 | 0.0037 | 0.093 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.6.99.1 | NADPH dehydrogenase. | 0.49 | 0.72 | 1 | 0.69 | 1 | 1 | 0.98 | 1 | 1 | 0.99 | 1 | 1 | 0.62 | 0.95 | 1 | 0.6 | 0.91 | 1 | 0.003 | 0.0029 | 0.002 | 0.0018 | 0.002 | 0.0011 | 0.0022 | 0.0025 | 0.0014 | 0.29 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0038 | 0.0014 | 0.0064 | 0.0032 | 0.0025 | 0.0027 | -0.25 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 2.7.7.73 | Sulfur carrier protein ThiS adenylyltransferase. | 0.99 | 1 | 1 | 0.97 | 1 | 1 | 0.84 | 1 | 1 | 0.71 | 0.89 | 1 | 0.53 | 0.93 | 1 | 0.74 | 0.94 | 1 | 0.0038 | 0.0038 | 0.0035 | 0.0038 | 0.0038 | 0.0014 | 0.0038 | 0.003 | 0.0018 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0038 | 0.0035 | 0.0016 | 0.0039 | 0.0038 | 0.0013 | 0.037 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.8.99.2 | Adenylyl-sulfate reductase. | 0.48 | 0.72 | 1 | 0.91 | 1 | 1 | 0.97 | 1 | 1 | 0.46 | 0.71 | 1 | 0.45 | 0.92 | 1 | 0.53 | 0.9 | 1 | 0.01 | 0.01 | 0.0095 | 0.0092 | 0.0097 | 0.003 | 0.0094 | 0.0094 | 0.0037 | 0.031 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.011 | 0.0046 | 0.011 | 0.0095 | 0.0045 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.2.30 | NAD(+) ADP-ribosyltransferase. | 0.24 | 0.55 | 1 | 0.87 | 1 | 1 | 0.94 | 1 | 1 | 0.26 | 0.56 | 1 | 0.77 | 0.97 | 1 | 0.86 | 0.96 | 1 | 0.00016 | 8.1e-05 | 1.3e-05 | 0.00011 | 0.00012 | 0.00012 | 0.00013 | 2.7e-05 | 0.00017 | 0.24 | 18 / 36 (50%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 6.9e-05 | 0 | 0.00013 | 4e-05 | 0 | 5.4e-05 | -0.79 | 4 / 11 (36%) | 5 / 11 (45%) | 0.364 | 0.455 |
| 2.7.1.35 | Pyridoxal kinase. | 0.034 | 0.49 | 1 | 0.89 | 1 | 1 | 0.94 | 1 | 1 | 0.047 | 0.43 | 1 | 0.57 | 0.93 | 1 | 0.46 | 0.86 | 1 | 0.012 | 0.012 | 0.012 | 0.0099 | 0.009 | 0.0026 | 0.0099 | 0.011 | 0.0032 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.013 | 0.012 | 0.004 | 0.013 | 0.013 | 0.0021 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.99.1.2 | Transferred entry: 5.6.2.1. | 0.32 | 0.6 | 1 | 0.32 | 1 | 1 | 0.16 | 0.85 | 1 | 0.98 | 0.99 | 1 | 0.74 | 0.96 | 1 | 0.93 | 0.98 | 1 | 0.052 | 0.052 | 0.052 | 0.055 | 0.053 | 0.019 | 0.051 | 0.052 | 0.012 | -0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.049 | 0.047 | 0.0099 | 0.052 | 0.054 | 0.011 | 0.086 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.4.1 | Cytosine deaminase. | 0.45 | 0.7 | 1 | 0.83 | 1 | 1 | 0.96 | 1 | 1 | 0.22 | 0.52 | 1 | 0.53 | 0.93 | 1 | 0.56 | 0.9 | 1 | 0.0018 | 0.0017 | 0.0012 | 0.0013 | 0.001 | 0.0012 | 0.0013 | 0.0016 | 0.0011 | 0 | 34 / 36 (94%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.0019 | 0.0012 | 0.0025 | 0.002 | 0.0011 | 0.0022 | 0.074 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.96 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. | 0.49 | 0.73 | 1 | 0.69 | 1 | 1 | 0.88 | 1 | 1 | 0.85 | 0.95 | 1 | 0.52 | 0.93 | 1 | 0.74 | 0.95 | 1 | 0.0052 | 0.0052 | 0.0044 | 0.0054 | 0.0046 | 0.0026 | 0.006 | 0.0042 | 0.0038 | 0.15 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0046 | 0.0041 | 0.0019 | 0.0051 | 0.0053 | 0.0021 | 0.15 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.1.82 | N-acyl-D-glutamate deacylase. | 0.68 | 0.85 | 1 | 0.094 | 1 | 1 | 0.12 | 0.82 | 1 | 0.93 | 0.98 | 1 | 0.11 | 0.87 | 1 | 0.17 | 0.71 | 1 | 3e-04 | 2e-04 | 0.00011 | 0.00027 | 0.00014 | 0.00032 | 8.8e-05 | 0 | 0.00015 | -1.6 | 24 / 36 (67%) | 5 / 7 (71%) | 3 / 7 (43%) | 0.714 | 0.429 | 0.00019 | 9.7e-05 | 0.00019 | 0.00024 | 0.00022 | 0.00025 | 0.34 | 8 / 11 (73%) | 8 / 11 (73%) | 0.727 | 0.727 |
| 5.3.2.- | 0.23 | 0.54 | 1 | 0.55 | 1 | 1 | 0.77 | 1 | 1 | 0.08 | 0.44 | 1 | 0.31 | 0.9 | 1 | 0.49 | 0.88 | 1 | 2e-04 | 0.00015 | 8.7e-05 | 0.00012 | 0 | 0.00025 | 0.00012 | 0.00011 | 0.00016 | 0 | 27 / 36 (75%) | 3 / 7 (43%) | 5 / 7 (71%) | 0.429 | 0.714 | 0.00015 | 9.7e-05 | 0.00013 | 0.00018 | 8.9e-05 | 0.00018 | 0.26 | 9 / 11 (82%) | 10 / 11 (91%) | 0.818 | 0.909 | |
| 4.1.3.36 | 1.4-dihydroxy-2-naphthoyl-CoA synthase. | 0.68 | 0.85 | 1 | 0.98 | 1 | 1 | 0.49 | 0.95 | 1 | 0.61 | 0.82 | 1 | 0.79 | 0.97 | 1 | 0.79 | 0.95 | 1 | 0.00086 | 0.00084 | 0.00055 | 0.00075 | 0.00066 | 0.00045 | 0.00075 | 0.00071 | 0.00048 | 0 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00071 | 0.00032 | 0.00072 | 0.0011 | 0.00043 | 0.0012 | 0.63 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 2.3.1.189 | Mycothiol synthase. | 0.47 | 0.72 | 1 | 0.48 | 1 | 1 | 0.96 | 1 | 1 | 0.66 | 0.86 | 1 | 0.58 | 0.93 | 1 | 0.85 | 0.96 | 1 | 0.00011 | 3.3e-05 | 0 | 5.8e-05 | 0 | 9.1e-05 | 4.1e-05 | 0 | 8.1e-05 | -0.5 | 11 / 36 (31%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 2.8e-05 | 0 | 4.4e-05 | 1.7e-05 | 0 | 3.9e-05 | -0.72 | 4 / 11 (36%) | 2 / 11 (18%) | 0.364 | 0.182 |
| 2.3.2.3 | Lysyltransferase. | 0.53 | 0.76 | 1 | 0.67 | 1 | 1 | 0.58 | 0.96 | 1 | 0.48 | 0.73 | 1 | 0.97 | 0.99 | 1 | 0.79 | 0.95 | 1 | 0.017 | 0.017 | 0.016 | 0.015 | 0.014 | 0.0043 | 0.015 | 0.014 | 0.0062 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.018 | 0.017 | 0.01 | 0.018 | 0.019 | 0.0057 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.1.12 | Dihydrolipoyllysine-residue acetyltransferase. | 0.92 | 0.96 | 1 | 0.64 | 1 | 1 | 0.89 | 1 | 1 | 0.69 | 0.87 | 1 | 0.34 | 0.9 | 1 | 0.54 | 0.9 | 1 | 0.001 | 9e-04 | 0.00076 | 0.00087 | 6e-04 | 0.00076 | 0.00095 | 0.0011 | 0.00075 | 0.13 | 32 / 36 (89%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00088 | 0.00059 | 0.00096 | 0.00092 | 8e-04 | 0.00066 | 0.064 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 2.4.2.54 | Beta-ribofuranosylphenol 5'-phosphate synthase. | 0.97 | 0.99 | 1 | 0.12 | 1 | 1 | 0.35 | 0.9 | 1 | 0.81 | 0.93 | 1 | 0.59 | 0.93 | 1 | 0.97 | 0.99 | 1 | 0.00016 | 2.7e-05 | 0 | 4.1e-05 | 0 | 0.00011 | 1.3e-05 | 0 | 3.5e-05 | -1.7 | 6 / 36 (17%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3e-05 | 0 | 7e-05 | 2.3e-05 | 0 | 5.2e-05 | -0.38 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 4.1.1.4 | Acetoacetate decarboxylase. | 0.85 | 0.93 | 1 | 0.11 | 1 | 1 | 0.3 | 0.89 | 1 | 0.98 | 0.99 | 1 | 0.16 | 0.87 | 1 | 0.4 | 0.83 | 1 | 0.00014 | 1.2e-05 | 0 | 1.7e-05 | 0 | 4.5e-05 | 0 | 0 | 0 | -Inf | 3 / 36 (8.3%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 1.8e-05 | 0 | 5.9e-05 | 1e-05 | 0 | 3.4e-05 | -0.85 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 4.3.3.7 | 4-hydroxy-tetrahydrodipicolinate synthase. | 0.73 | 0.87 | 1 | 0.76 | 1 | 1 | 0.8 | 1 | 1 | 0.39 | 0.66 | 1 | 0.61 | 0.95 | 1 | 0.57 | 0.9 | 1 | 0.022 | 0.022 | 0.022 | 0.022 | 0.022 | 0.0045 | 0.021 | 0.026 | 0.0066 | -0.067 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.022 | 0.023 | 0.0053 | 0.022 | 0.022 | 0.0021 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.7.60 | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase. | 0.13 | 0.5 | 1 | 0.23 | 1 | 1 | 0.76 | 0.99 | 1 | 0.098 | 0.46 | 1 | 0.41 | 0.9 | 1 | 0.87 | 0.97 | 1 | 0.016 | 0.016 | 0.016 | 0.015 | 0.013 | 0.0038 | 0.013 | 0.013 | 0.0048 | -0.21 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.018 | 0.017 | 0.0055 | 0.016 | 0.017 | 0.0026 | -0.17 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.55 | Allose kinase. | 0.56 | 0.78 | 1 | 0.83 | 1 | 1 | 0.59 | 0.96 | 1 | 0.22 | 0.52 | 1 | 0.95 | 0.99 | 1 | 0.66 | 0.92 | 1 | 0.00075 | 0.00068 | 0.00043 | 0.00067 | 4e-04 | 0.00085 | 0.00071 | 0.00072 | 0.00064 | 0.084 | 33 / 36 (92%) | 6 / 7 (86%) | 5 / 7 (71%) | 0.857 | 0.714 | 0.00074 | 0.00044 | 0.00081 | 0.00062 | 5e-04 | 0.00051 | -0.26 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.2.25 | Dihydroneopterin aldolase. | 0.29 | 0.58 | 1 | 0.44 | 1 | 1 | 0.22 | 0.88 | 1 | 0.17 | 0.49 | 1 | 0.15 | 0.87 | 1 | 0.08 | 0.66 | 1 | 0.0077 | 0.0077 | 0.0066 | 0.0063 | 0.0068 | 0.0022 | 0.0075 | 0.0077 | 0.0027 | 0.25 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0092 | 0.0062 | 0.0085 | 0.0073 | 0.0057 | 0.0034 | -0.33 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.17.8 | Muramoylpentapeptide carboxypeptidase. | 0.0049 | 0.49 | 1 | 0.0076 | 0.7 | 1 | 0.016 | 0.67 | 1 | 0.0013 | 0.31 | 1 | 0.032 | 0.79 | 1 | 0.048 | 0.65 | 1 | 0.00022 | 4.3e-05 | 0 | 0.00015 | 3.2e-05 | 3e-04 | 6.4e-05 | 0 | 0.00013 | -1.2 | 7 / 36 (19%) | 4 / 7 (57%) | 2 / 7 (29%) | 0.571 | 0.286 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.8e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.8.1.11 | Molybdopterin synthase sulfurtransferase. | 0.17 | 0.5 | 1 | 0.48 | 1 | 1 | 0.36 | 0.9 | 1 | 0.11 | 0.47 | 1 | 0.23 | 0.88 | 1 | 0.12 | 0.66 | 1 | 0.0016 | 0.0012 | 0.00054 | 0.00053 | 0.00035 | 0.00065 | 0.00074 | 8e-04 | 0.00071 | 0.48 | 27 / 36 (75%) | 4 / 7 (57%) | 5 / 7 (71%) | 0.571 | 0.714 | 0.0018 | 0.00044 | 0.0037 | 0.0013 | 0.001 | 0.0014 | -0.47 | 10 / 11 (91%) | 8 / 11 (73%) | 0.909 | 0.727 |
| 6.3.5.2 | GMP synthase (glutamine-hydrolyzing). | 0.21 | 0.54 | 1 | 0.73 | 1 | 1 | 0.41 | 0.92 | 1 | 0.13 | 0.47 | 1 | 0.15 | 0.87 | 1 | 0.068 | 0.66 | 1 | 0.028 | 0.028 | 0.028 | 0.026 | 0.027 | 0.004 | 0.027 | 0.028 | 0.0053 | 0.054 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.03 | 0.028 | 0.0092 | 0.028 | 0.029 | 0.0045 | -0.1 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.1.1.9 | Valine--tRNA ligase. | 0.074 | 0.49 | 1 | 0.34 | 1 | 1 | 0.91 | 1 | 1 | 0.12 | 0.47 | 1 | 0.78 | 0.97 | 1 | 0.19 | 0.72 | 1 | 0.042 | 0.042 | 0.043 | 0.039 | 0.039 | 0.0062 | 0.037 | 0.037 | 0.0088 | -0.076 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.047 | 0.048 | 0.01 | 0.043 | 0.043 | 0.0057 | -0.13 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.2.15 | Guanosine phosphorylase. | 0.1 | 0.49 | 1 | 0.14 | 1 | 1 | 0.12 | 0.82 | 1 | 0.11 | 0.47 | 1 | 0.47 | 0.92 | 1 | 0.31 | 0.79 | 1 | 0.00015 | 1.3e-05 | 0 | 5e-05 | 0 | 0.00013 | 9.7e-06 | 0 | 2.6e-05 | -2.4 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 4.3.2.3 | Ureidoglycolate lyase. | 0.72 | 0.87 | 1 | 0.85 | 1 | 1 | 0.46 | 0.93 | 1 | 0.61 | 0.82 | 1 | 0.95 | 0.99 | 1 | 0.97 | 0.99 | 1 | 0.00074 | 0.00064 | 0.00035 | 0.00074 | 0.00071 | 0.00085 | 0.00071 | 0.00061 | 0.00089 | -0.06 | 31 / 36 (86%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.00047 | 0.00024 | 0.00046 | 0.00069 | 0.00023 | 0.00084 | 0.55 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 1.4.3.5 | Pyridoxal 5'-phosphate synthase. | 0.36 | 0.64 | 1 | 0.79 | 1 | 1 | 0.066 | 0.72 | 1 | 0.33 | 0.6 | 1 | 0.79 | 0.97 | 1 | 0.12 | 0.66 | 1 | 0.00082 | 0.00068 | 0.00056 | 0.00067 | 0.00056 | 0.00048 | 0.00073 | 0.00024 | 0.00089 | 0.12 | 30 / 36 (83%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00048 | 0.00021 | 0.00058 | 0.00087 | 0.00075 | 0.00082 | 0.86 | 8 / 11 (73%) | 10 / 11 (91%) | 0.727 | 0.909 |
| 2.7.2.4 | Aspartate kinase. | 0.58 | 0.79 | 1 | 0.96 | 1 | 1 | 0.95 | 1 | 1 | 0.33 | 0.6 | 1 | 0.1 | 0.86 | 1 | 0.067 | 0.66 | 1 | 0.032 | 0.032 | 0.032 | 0.032 | 0.032 | 0.0046 | 0.032 | 0.032 | 0.0053 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.033 | 0.031 | 0.0053 | 0.033 | 0.031 | 0.0045 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.1.17 | Enoyl-CoA hydratase. | 0.19 | 0.53 | 1 | 0.64 | 1 | 1 | 0.38 | 0.9 | 1 | 0.45 | 0.7 | 1 | 0.3 | 0.9 | 1 | 0.79 | 0.95 | 1 | 0.0064 | 0.0064 | 0.0061 | 0.0065 | 0.006 | 0.0016 | 0.007 | 0.0062 | 0.0016 | 0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0055 | 0.0053 | 0.0021 | 0.0068 | 0.007 | 0.0023 | 0.31 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.8.2.22 | Aryl-sulfate sulfotransferase. | 0.65 | 0.84 | 1 | 0.66 | 1 | 1 | 0.85 | 1 | 1 | 0.91 | 0.97 | 1 | 0.73 | 0.96 | 1 | 0.55 | 0.9 | 1 | 6e-04 | 0.00043 | 0.00012 | 0.00056 | 0.00027 | 0.00065 | 0.00047 | 0.00012 | 7e-04 | -0.25 | 26 / 36 (72%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 4e-04 | 0.00013 | 0.00052 | 0.00036 | 7.5e-05 | 0.00092 | -0.15 | 9 / 11 (82%) | 7 / 11 (64%) | 0.818 | 0.636 |
| 2.5.1.15 | Dihydropteroate synthase. | 0.76 | 0.89 | 1 | 0.28 | 1 | 1 | 0.28 | 0.89 | 1 | 0.82 | 0.93 | 1 | 0.043 | 0.8 | 1 | 0.05 | 0.65 | 1 | 0.0074 | 0.0074 | 0.0068 | 0.0075 | 0.0079 | 0.002 | 0.0084 | 0.0085 | 0.0028 | 0.16 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0073 | 0.0058 | 0.0035 | 0.0069 | 0.0069 | 0.0017 | -0.081 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.130 | Tetraacyldisaccharide 4'-kinase. | 0.36 | 0.64 | 1 | 0.27 | 1 | 1 | 0.012 | 0.61 | 1 | 0.22 | 0.52 | 1 | 0.59 | 0.93 | 1 | 0.052 | 0.65 | 1 | 0.0014 | 0.0014 | 0.0011 | 0.0014 | 0.0014 | 0.00075 | 0.00095 | 0.0012 | 0.00053 | -0.56 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0011 | 0.00095 | 0.0012 | 0.0021 | 0.0011 | 0.002 | 0.93 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 2.7.4.1 | Polyphosphate kinase. | 0.19 | 0.53 | 1 | 0.86 | 1 | 1 | 0.9 | 1 | 1 | 0.17 | 0.49 | 1 | 0.17 | 0.87 | 1 | 0.3 | 0.78 | 1 | 0.04 | 0.04 | 0.041 | 0.036 | 0.033 | 0.0099 | 0.037 | 0.037 | 0.013 | 0.04 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.042 | 0.041 | 0.01 | 0.042 | 0.044 | 0.0047 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.95 | Phosphoglycerate dehydrogenase. | 0.15 | 0.5 | 1 | 0.54 | 1 | 1 | 0.38 | 0.9 | 1 | 0.1 | 0.46 | 1 | 0.25 | 0.88 | 1 | 0.2 | 0.73 | 1 | 0.023 | 0.023 | 0.021 | 0.019 | 0.02 | 0.0033 | 0.022 | 0.018 | 0.0086 | 0.21 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.025 | 0.02 | 0.013 | 0.023 | 0.024 | 0.0046 | -0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.8.3.15 | Succinyl-CoA:(R)-benzylsuccinate CoA-transferase. | 0.55 | 0.77 | 1 | 0.17 | 1 | 1 | 0.22 | 0.88 | 1 | 0.52 | 0.75 | 1 | 0.23 | 0.88 | 1 | 0.21 | 0.74 | 1 | 0.00021 | 3.6e-05 | 0 | 5.1e-05 | 0 | 1e-04 | 7.7e-06 | 0 | 2e-05 | -2.7 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 4.7e-05 | 0 | 0.00016 | 3.3e-05 | 0 | 7.5e-05 | -0.51 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 1.1.1.28 | D-lactate dehydrogenase. | 0.92 | 0.96 | 1 | 0.36 | 1 | 1 | 0.15 | 0.84 | 1 | 0.57 | 0.8 | 1 | 0.74 | 0.96 | 1 | 0.37 | 0.82 | 1 | 0.0051 | 0.0051 | 0.0052 | 0.005 | 0.0053 | 0.0018 | 0.0043 | 0.0037 | 0.0015 | -0.22 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0051 | 0.0049 | 0.0016 | 0.0058 | 0.0059 | 0.0019 | 0.19 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.5.1.38 | FMN reductase (NADPH). | 0.64 | 0.83 | 1 | 0.74 | 1 | 1 | 0.86 | 1 | 1 | 0.53 | 0.76 | 1 | 0.46 | 0.92 | 1 | 0.86 | 0.96 | 1 | 0.0028 | 0.0028 | 0.0023 | 0.0022 | 0.0021 | 0.0017 | 0.0025 | 0.0016 | 0.0019 | 0.18 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0031 | 0.0021 | 0.0042 | 0.0031 | 0.0026 | 0.0023 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.1.- | 0.37 | 0.64 | 1 | 0.47 | 1 | 1 | 0.63 | 0.96 | 1 | 0.2 | 0.51 | 1 | 0.036 | 0.8 | 1 | 0.066 | 0.66 | 1 | 0.21 | 0.21 | 0.21 | 0.2 | 0.21 | 0.029 | 0.21 | 0.23 | 0.054 | 0.07 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.21 | 0.21 | 0.047 | 0.21 | 0.21 | 0.025 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 5.1.1.3 | Glutamate racemase. | 0.19 | 0.53 | 1 | 0.78 | 1 | 1 | 0.49 | 0.94 | 1 | 0.14 | 0.47 | 1 | 0.17 | 0.87 | 1 | 0.067 | 0.66 | 1 | 0.014 | 0.014 | 0.014 | 0.013 | 0.013 | 0.002 | 0.014 | 0.013 | 0.004 | 0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.015 | 0.015 | 0.0042 | 0.014 | 0.014 | 0.002 | -0.1 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.1.91 | N-substituted formamide deformylase. | 0.16 | 0.5 | 1 | 0.78 | 1 | 1 | 0.38 | 0.91 | 1 | 0.11 | 0.47 | 1 | 0.39 | 0.9 | 1 | 0.19 | 0.72 | 1 | 0.0079 | 0.0079 | 0.0052 | 0.0057 | 0.0046 | 0.0042 | 0.0061 | 0.0052 | 0.0042 | 0.098 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.0069 | 0.012 | 0.0078 | 0.0061 | 0.0047 | -0.5 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.3.- | 0.55 | 0.77 | 1 | 1 | 1 | 1 | 0.28 | 0.89 | 1 | 0.4 | 0.67 | 1 | 0.69 | 0.95 | 1 | 0.55 | 0.9 | 1 | 0.00011 | 9.2e-06 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 3 / 36 (8.3%) | 0 / 7 (0%) | 0 / 7 (0%) | 0 | 0 | 7.3e-06 | 0 | 2.4e-05 | 2.3e-05 | 0 | 5.2e-05 | 1.7 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 | |
| 3.4.21.26 | Prolyl oligopeptidase. | 0.17 | 0.5 | 1 | 0.86 | 1 | 1 | 0.52 | 0.95 | 1 | 0.13 | 0.47 | 1 | 0.89 | 0.99 | 1 | 0.87 | 0.97 | 1 | 0.00041 | 0.00024 | 9.6e-05 | 3e-04 | 0.00034 | 0.00021 | 0.00026 | 0.00013 | 0.00028 | -0.21 | 21 / 36 (58%) | 5 / 7 (71%) | 6 / 7 (86%) | 0.714 | 0.857 | 0.00015 | 0 | 0.00029 | 0.00029 | 0 | 0.00065 | 0.95 | 5 / 11 (45%) | 5 / 11 (45%) | 0.455 | 0.455 |
| 6.3.1.5 | NAD(+) synthase. | 0.31 | 0.59 | 1 | 0.73 | 1 | 1 | 0.73 | 0.99 | 1 | 0.31 | 0.58 | 1 | 0.46 | 0.92 | 1 | 0.76 | 0.95 | 1 | 0.0032 | 0.0032 | 0.0027 | 0.0022 | 0.0022 | 0.0011 | 0.0021 | 0.002 | 0.0015 | -0.067 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0046 | 0.0029 | 0.0076 | 0.0031 | 0.0028 | 0.0023 | -0.57 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.24.55 | Pitrilysin. | 0.049 | 0.49 | 1 | 0.6 | 1 | 1 | 0.26 | 0.89 | 1 | 0.013 | 0.42 | 1 | 0.53 | 0.93 | 1 | 0.25 | 0.76 | 1 | 0.002 | 0.002 | 0.0011 | 0.0025 | 0.0024 | 0.00097 | 0.0025 | 0.0012 | 0.0029 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0011 | 0.00087 | 0.0011 | 0.0021 | 0.00076 | 0.003 | 0.93 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.99.6 | Glucosamine-6-phosphate deaminase. | 0.55 | 0.77 | 1 | 0.87 | 1 | 1 | 0.81 | 1 | 1 | 0.87 | 0.95 | 1 | 0.27 | 0.88 | 1 | 0.35 | 0.81 | 1 | 0.015 | 0.015 | 0.014 | 0.015 | 0.014 | 0.0026 | 0.015 | 0.016 | 0.0035 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.014 | 0.014 | 0.0021 | 0.015 | 0.013 | 0.0054 | 0.1 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.11.6 | Exodeoxyribonuclease VII. | 0.047 | 0.49 | 1 | 0.64 | 1 | 1 | 0.76 | 0.99 | 1 | 0.071 | 0.43 | 1 | 0.43 | 0.91 | 1 | 0.32 | 0.79 | 1 | 0.023 | 0.023 | 0.022 | 0.021 | 0.02 | 0.0026 | 0.02 | 0.02 | 0.0032 | -0.07 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.025 | 0.022 | 0.0055 | 0.024 | 0.025 | 0.004 | -0.059 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.2.2 | Palmitoyl-CoA hydrolase. | 0.15 | 0.5 | 1 | 0.84 | 1 | 1 | 0.86 | 1 | 1 | 0.26 | 0.55 | 1 | 0.75 | 0.97 | 1 | 0.71 | 0.94 | 1 | 0.00024 | 9.8e-05 | 0 | 0.00017 | 4.8e-05 | 0.00022 | 0.00018 | 0.00016 | 0.00019 | 0.082 | 15 / 36 (42%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 4.6e-05 | 0 | 7.9e-05 | 5.8e-05 | 0 | 0.00012 | 0.33 | 4 / 11 (36%) | 3 / 11 (27%) | 0.364 | 0.273 |
| 6.3.2.- | 0.63 | 0.83 | 1 | 0.43 | 1 | 1 | 0.67 | 0.98 | 1 | 0.99 | 1 | 1 | 0.16 | 0.87 | 1 | 0.46 | 0.86 | 1 | 0.022 | 0.022 | 0.018 | 0.024 | 0.016 | 0.02 | 0.028 | 0.019 | 0.025 | 0.22 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.02 | 0.014 | 0.015 | 0.02 | 0.02 | 0.0061 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 3.4.17.- | 0.91 | 0.96 | 1 | 0.97 | 1 | 1 | 0.6 | 0.96 | 1 | 0.79 | 0.92 | 1 | 0.65 | 0.95 | 1 | 0.88 | 0.97 | 1 | 0.003 | 0.003 | 0.0031 | 0.0028 | 0.0029 | 0.001 | 0.0029 | 0.0034 | 0.001 | 0.051 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.003 | 0.0036 | 0.0014 | 0.0033 | 0.0034 | 0.0015 | 0.14 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 4.2.1.81 | D(-)-tartrate dehydratase. | 0.38 | 0.65 | 1 | 0.91 | 1 | 1 | 0.93 | 1 | 1 | 0.64 | 0.84 | 1 | 0.76 | 0.97 | 1 | 0.85 | 0.96 | 1 | 0.00032 | 5.4e-05 | 0 | 0.00012 | 0 | 0.00031 | 0.00013 | 0 | 0.00033 | 0.12 | 6 / 36 (17%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 8.1e-06 | 0 | 1.9e-05 | 1.3e-05 | 0 | 3.5e-05 | 0.68 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 4.1.1.33 | Diphosphomevalonate decarboxylase. | 0.59 | 0.8 | 1 | 0.4 | 1 | 1 | 0.31 | 0.89 | 1 | 0.44 | 0.69 | 1 | 0.22 | 0.88 | 1 | 0.21 | 0.74 | 1 | 0.00038 | 0.00027 | 0.00018 | 0.00026 | 4.8e-05 | 0.00044 | 0.00032 | 0.00019 | 0.00037 | 0.3 | 26 / 36 (72%) | 4 / 7 (57%) | 6 / 7 (86%) | 0.571 | 0.857 | 3e-04 | 0.00026 | 0.00033 | 0.00022 | 0.00014 | 0.00026 | -0.45 | 8 / 11 (73%) | 8 / 11 (73%) | 0.727 | 0.727 |
| 6.1.1.- | 0.27 | 0.56 | 1 | 0.1 | 1 | 1 | 0.026 | 0.71 | 1 | 0.46 | 0.71 | 1 | 0.16 | 0.87 | 1 | 0.054 | 0.65 | 1 | 0.013 | 0.013 | 0.013 | 0.015 | 0.013 | 0.004 | 0.011 | 0.012 | 0.0046 | -0.45 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.012 | 0.0099 | 0.0058 | 0.015 | 0.014 | 0.0058 | 0.32 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 3.1.6.12 | N-acetylgalactosamine-4-sulfatase. | 0.51 | 0.74 | 1 | 0.47 | 1 | 1 | 0.9 | 1 | 1 | 0.55 | 0.78 | 1 | 0.48 | 0.92 | 1 | 0.82 | 0.96 | 1 | 0.00035 | 0.00011 | 0 | 0.00012 | 0 | 0.00027 | 0.00018 | 0 | 0.00032 | 0.58 | 11 / 36 (31%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 5.4e-05 | 0 | 0.00012 | 0.00011 | 0 | 0.00027 | 1 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 1.1.1.355 | 2'-dehydrokanamycin reductase. | 0.69 | 0.86 | 1 | 0.082 | 1 | 1 | 0.047 | 0.71 | 1 | 0.55 | 0.78 | 1 | 0.081 | 0.86 | 1 | 0.038 | 0.63 | 1 | 1e-04 | 1.4e-05 | 0 | 4.1e-06 | 0 | 1.1e-05 | 4.9e-05 | 0 | 9.6e-05 | 3.6 | 5 / 36 (14%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 1.1e-05 | 0 | 3e-05 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 1.5.1.28 | Opine dehydrogenase. | 0.43 | 0.69 | 1 | 0.39 | 1 | 1 | 0.51 | 0.95 | 1 | 0.25 | 0.55 | 1 | 0.42 | 0.91 | 1 | 0.53 | 0.9 | 1 | 0.00043 | 0.00032 | 0.00013 | 0.00028 | 0.00011 | 0.00033 | 0.00013 | 0 | 0.00021 | -1.1 | 27 / 36 (75%) | 4 / 7 (57%) | 3 / 7 (43%) | 0.571 | 0.429 | 0.00041 | 0.00016 | 0.00052 | 0.00039 | 0.00015 | 0.00055 | -0.072 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 3.6.4.- | 0.018 | 0.49 | 1 | 0.98 | 1 | 1 | 0.56 | 0.96 | 1 | 0.082 | 0.44 | 1 | 0.098 | 0.86 | 1 | 0.028 | 0.61 | 1 | 0.1 | 0.1 | 0.1 | 0.094 | 0.093 | 0.011 | 0.094 | 0.094 | 0.018 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.11 | 0.11 | 0.016 | 0.11 | 0.11 | 0.013 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 2.6.1.39 | 2-aminoadipate transaminase. | 0.013 | 0.49 | 1 | 0.45 | 1 | 1 | 0.85 | 1 | 1 | 0.0031 | 0.36 | 1 | 0.1 | 0.86 | 1 | 0.58 | 0.91 | 1 | 0.0051 | 0.0051 | 0.004 | 0.0023 | 0.0025 | 0.0013 | 0.003 | 0.0034 | 0.0012 | 0.38 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0068 | 0.0059 | 0.0072 | 0.0066 | 0.0067 | 0.0029 | -0.043 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.1.304 | UDP-Gal:alpha-D-GlcNAc-diphosphoundecaprenol beta-1.4- | 0.99 | 1 | 1 | 0.95 | 1 | 1 | 0.87 | 1 | 1 | 0.96 | 0.99 | 1 | 0.88 | 0.99 | 1 | 0.5 | 0.89 | 1 | 0.00026 | 7.1e-05 | 0 | 4.2e-05 | 0 | 5.7e-05 | 5.9e-05 | 0 | 0.00012 | 0.49 | 10 / 36 (28%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 4.9e-05 | 0 | 7.6e-05 | 0.00012 | 0 | 4e-04 | 1.3 | 4 / 11 (36%) | 1 / 11 (9.1%) | 0.364 | 0.0909 |
| 3.5.99.- | 0.084 | 0.49 | 1 | 0.26 | 1 | 1 | 0.11 | 0.82 | 1 | 0.088 | 0.45 | 1 | 0.44 | 0.91 | 1 | 0.22 | 0.74 | 1 | 8.3e-05 | 9.2e-06 | 0 | 2.1e-05 | 0 | 4.4e-05 | 9.7e-06 | 0 | 2.6e-05 | -1.1 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 1e-05 | 0 | 3.4e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 | |
| 2.1.1.254 | Erythromycin 3''-O-methyltransferase. | 0.47 | 0.71 | 1 | 0.91 | 1 | 1 | 0.36 | 0.9 | 1 | 0.71 | 0.89 | 1 | 0.52 | 0.93 | 1 | 0.17 | 0.71 | 1 | 7e-05 | 5.8e-06 | 0 | 1.2e-05 | 0 | 3.2e-05 | 1.3e-05 | 0 | 3.5e-05 | 0.12 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3e-06 | 0 | 9.9e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 1.5.98.2 | 5.10-methylenetetrahydromethanopterin reductase. | 0.82 | 0.92 | 1 | 0.35 | 1 | 1 | 0.92 | 1 | 1 | 0.71 | 0.89 | 1 | 0.74 | 0.96 | 1 | 0.25 | 0.76 | 1 | 0.00024 | 4e-05 | 0 | 4.1e-05 | 0 | 0.00011 | 2.6e-05 | 0 | 7e-05 | -0.66 | 6 / 36 (17%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 5.1e-05 | 0 | 0.00011 | 3.6e-05 | 0 | 8.9e-05 | -0.5 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 6.3.5.4 | Asparagine synthase (glutamine-hydrolyzing). | 0.8 | 0.91 | 1 | 0.47 | 1 | 1 | 0.61 | 0.96 | 1 | 0.83 | 0.94 | 1 | 0.8 | 0.97 | 1 | 0.76 | 0.95 | 1 | 0.022 | 0.022 | 0.021 | 0.022 | 0.021 | 0.0058 | 0.021 | 0.02 | 0.0044 | -0.067 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.022 | 0.023 | 0.0092 | 0.021 | 0.022 | 0.005 | -0.067 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.303 | Diacetyl reductase ((R)-acetoin forming). | 0.4 | 0.66 | 1 | 0.19 | 1 | 1 | 0.19 | 0.87 | 1 | 0.46 | 0.71 | 1 | 0.1 | 0.86 | 1 | 0.16 | 0.71 | 1 | 0.00025 | 0.00015 | 8.7e-05 | 9.1e-05 | 0 | 0.00014 | 0.00018 | 0.00019 | 0.00015 | 0.98 | 21 / 36 (58%) | 3 / 7 (43%) | 6 / 7 (86%) | 0.429 | 0.857 | 0.00019 | 2.8e-05 | 0.00028 | 0.00012 | 5.9e-05 | 0.00013 | -0.66 | 6 / 11 (55%) | 6 / 11 (55%) | 0.545 | 0.545 |
| 3.4.21.116 | SpoIVB peptidase. | 0.76 | 0.89 | 1 | 0.44 | 1 | 1 | 0.36 | 0.9 | 1 | 0.62 | 0.83 | 1 | 0.7 | 0.95 | 1 | 0.55 | 0.9 | 1 | 0.0097 | 0.0097 | 0.0098 | 0.0093 | 0.0085 | 0.0025 | 0.0086 | 0.0099 | 0.0033 | -0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.01 | 0.01 | 0.0039 | 0.01 | 0.0097 | 0.0031 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.1.42 | Galactarate dehydratase. | 0.06 | 0.49 | 1 | 0.42 | 1 | 1 | 0.38 | 0.9 | 1 | 0.2 | 0.5 | 1 | 0.71 | 0.95 | 1 | 0.92 | 0.98 | 1 | 0.0056 | 0.0056 | 0.0048 | 0.0069 | 0.0076 | 0.0022 | 0.0061 | 0.0066 | 0.0026 | -0.18 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0046 | 0.0045 | 0.0016 | 0.0053 | 0.005 | 0.003 | 0.2 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.21 | Beta-glucosidase. | 0.96 | 0.99 | 1 | 0.81 | 1 | 1 | 0.92 | 1 | 1 | 0.7 | 0.88 | 1 | 0.27 | 0.88 | 1 | 0.58 | 0.91 | 1 | 0.12 | 0.12 | 0.12 | 0.12 | 0.11 | 0.049 | 0.12 | 0.13 | 0.044 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.12 | 0.11 | 0.063 | 0.12 | 0.12 | 0.028 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.3.3.6 | Methylitaconate Delta-isomerase. | 0.3 | 0.58 | 1 | 0.87 | 1 | 1 | 0.78 | 1 | 1 | 0.67 | 0.86 | 1 | 0.27 | 0.88 | 1 | 0.26 | 0.76 | 1 | 0.00023 | 4.5e-05 | 0 | 0.00013 | 0 | 0.00036 | 6.1e-05 | 0 | 1e-04 | -1.1 | 7 / 36 (19%) | 1 / 7 (14%) | 3 / 7 (43%) | 0.143 | 0.429 | 1.1e-05 | 0 | 2.9e-05 | 1e-05 | 0 | 3.4e-05 | -0.14 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 6.2.1.47 | Medium-chain-fatty-acid--(acyl-carrier-protein) ligase. | 0.17 | 0.5 | 1 | 0.27 | 1 | 1 | 0.09 | 0.79 | 1 | 0.18 | 0.5 | 1 | 0.59 | 0.93 | 1 | 0.24 | 0.76 | 1 | 0.00048 | 8e-05 | 0 | 0.00021 | 0 | 0.00053 | 0.00017 | 0 | 0.00046 | -0.3 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 3e-06 | 0 | 9.9e-06 | 1.6e-05 | 0 | 3.7e-05 | 2.4 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 1.1.1.133 | dTDP-4-dehydrorhamnose reductase. | 0.13 | 0.5 | 1 | 0.22 | 1 | 1 | 0.047 | 0.71 | 1 | 0.63 | 0.84 | 1 | 0.71 | 0.95 | 1 | 0.2 | 0.73 | 1 | 0.018 | 0.018 | 0.019 | 0.019 | 0.021 | 0.0047 | 0.017 | 0.017 | 0.0039 | -0.16 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.017 | 0.018 | 0.0042 | 0.019 | 0.019 | 0.003 | 0.16 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.201 | Protein-N(pi)-phosphohistidine--trehalose phosphotransferase. | 0.25 | 0.56 | 1 | 0.56 | 1 | 1 | 0.95 | 1 | 1 | 0.43 | 0.69 | 1 | 0.67 | 0.95 | 1 | 0.84 | 0.96 | 1 | 0.00046 | 2e-04 | 0 | 4e-04 | 8e-05 | 0.00056 | 0.00031 | 0 | 0.00052 | -0.37 | 16 / 36 (44%) | 4 / 7 (57%) | 3 / 7 (43%) | 0.571 | 0.429 | 0.00015 | 0 | 0.00023 | 7.1e-05 | 0 | 0.00017 | -1.1 | 5 / 11 (45%) | 4 / 11 (36%) | 0.455 | 0.364 |
| 3.5.5.1 | Nitrilase. | 0.32 | 0.6 | 1 | 0.26 | 1 | 1 | 0.37 | 0.9 | 1 | 0.32 | 0.59 | 1 | 0.37 | 0.9 | 1 | 0.35 | 0.81 | 1 | 0.00045 | 0.00032 | 0.00023 | 0.00014 | 0.00014 | 0.00014 | 0.00037 | 0.00024 | 0.00044 | 1.4 | 26 / 36 (72%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.00037 | 0.00019 | 0.00064 | 0.00036 | 0.00028 | 0.00034 | -0.04 | 9 / 11 (82%) | 7 / 11 (64%) | 0.818 | 0.636 |
| 2.8.3.19 | CoA:oxalate CoA-transferase. | 0.087 | 0.49 | 1 | 0.089 | 1 | 1 | 0.12 | 0.82 | 1 | 0.13 | 0.47 | 1 | 0.32 | 0.9 | 1 | 0.32 | 0.8 | 1 | 3e-04 | 5.1e-05 | 0 | 0.00016 | 0 | 0.00035 | 7.7e-05 | 0 | 2e-04 | -1.1 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 6.6e-06 | 0 | 2.2e-05 | 9.5e-06 | 0 | 2.3e-05 | 0.53 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 1.3.8.13 | Crotonobetainyl-CoA reductase. | 0.081 | 0.49 | 1 | 0.029 | 0.97 | 1 | 0.22 | 0.88 | 1 | 0.13 | 0.47 | 1 | 0.16 | 0.87 | 1 | 0.76 | 0.95 | 1 | 0.00048 | 6.7e-05 | 0 | 0.00024 | 0 | 0.00053 | 9.7e-05 | 0 | 0.00026 | -1.3 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 8.5e-06 | 0 | 2.1e-05 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 4.1.3.- | 0.21 | 0.54 | 1 | 0.5 | 1 | 1 | 0.31 | 0.89 | 1 | 0.11 | 0.47 | 1 | 0.7 | 0.95 | 1 | 0.45 | 0.86 | 1 | 0.0047 | 0.0047 | 0.0047 | 0.004 | 0.0045 | 0.0011 | 0.0036 | 0.0032 | 0.0018 | -0.15 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.005 | 0.0049 | 0.0023 | 0.0054 | 0.0049 | 0.0014 | 0.11 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 1.3.3.3 | Coproporphyrinogen oxidase. | 0.013 | 0.49 | 1 | 0.0085 | 0.7 | 1 | 0.0054 | 0.61 | 1 | 0.0015 | 0.31 | 1 | 0.038 | 0.8 | 1 | 0.02 | 0.55 | 1 | 0.00038 | 9.5e-05 | 0 | 0.00028 | 2.8e-05 | 0.00061 | 0.00018 | 0 | 0.00046 | -0.64 | 9 / 36 (25%) | 5 / 7 (71%) | 2 / 7 (29%) | 0.714 | 0.286 | 0 | 0 | 0 | 1.8e-05 | 0 | 4e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 1.2.2.2 | Deleted entry. | 0.065 | 0.49 | 1 | 0.1 | 1 | 1 | 0.012 | 0.61 | 1 | 0.019 | 0.42 | 1 | 0.067 | 0.86 | 1 | 0.015 | 0.55 | 1 | 0.00039 | 0.00019 | 0 | 0.00025 | 0.00014 | 0.00025 | 0.00021 | 0 | 0.00038 | -0.25 | 17 / 36 (47%) | 6 / 7 (86%) | 3 / 7 (43%) | 0.857 | 0.429 | 7.5e-05 | 0 | 0.00022 | 0.00024 | 0 | 0.00054 | 1.7 | 3 / 11 (27%) | 5 / 11 (45%) | 0.273 | 0.455 |
| 2.3.1.35 | Glutamate N-acetyltransferase. | 0.12 | 0.5 | 1 | 0.39 | 1 | 1 | 0.32 | 0.89 | 1 | 0.15 | 0.47 | 1 | 0.12 | 0.87 | 1 | 0.093 | 0.66 | 1 | 0.019 | 0.019 | 0.019 | 0.017 | 0.017 | 0.0027 | 0.019 | 0.02 | 0.0054 | 0.16 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.02 | 0.02 | 0.0048 | 0.019 | 0.019 | 0.0036 | -0.074 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.3.4.20 | 7-cyano-7-deazaguanine synthase. | 0.16 | 0.5 | 1 | 0.15 | 1 | 1 | 0.62 | 0.96 | 1 | 0.49 | 0.74 | 1 | 0.51 | 0.93 | 1 | 0.67 | 0.93 | 1 | 0.0034 | 0.0034 | 0.0034 | 0.0043 | 0.0048 | 0.0016 | 0.0034 | 0.0036 | 0.0011 | -0.34 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0033 | 0.003 | 0.001 | 0.0029 | 0.0032 | 0.0015 | -0.19 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.4.53 | 3'.5'-cyclic-AMP phosphodiesterase. | 0.14 | 0.5 | 1 | 0.019 | 0.92 | 1 | 0.14 | 0.83 | 1 | 0.38 | 0.65 | 1 | 0.072 | 0.86 | 1 | 0.48 | 0.87 | 1 | 0.00043 | 0.00018 | 0 | 0.00061 | 6.4e-05 | 0.0015 | 0.00018 | 0 | 0.00046 | -1.8 | 15 / 36 (42%) | 4 / 7 (57%) | 2 / 7 (29%) | 0.571 | 0.286 | 5.3e-05 | 0 | 6.8e-05 | 3e-05 | 0 | 5.2e-05 | -0.82 | 5 / 11 (45%) | 4 / 11 (36%) | 0.455 | 0.364 |
| 1.2.1.88 | L-glutamate gamma-semialdehyde dehydrogenase. | 0.82 | 0.92 | 1 | 0.47 | 1 | 1 | 0.25 | 0.89 | 1 | 0.85 | 0.95 | 1 | 0.57 | 0.93 | 1 | 0.54 | 0.9 | 1 | 0.0012 | 0.0012 | 0.00069 | 0.0011 | 0.00069 | 0.00092 | 8e-04 | 7e-04 | 0.00046 | -0.46 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0011 | 0.00053 | 0.0011 | 0.0017 | 0.00068 | 0.0023 | 0.63 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.3.2.2 | Adenylosuccinate lyase. | 0.16 | 0.5 | 1 | 0.4 | 1 | 1 | 0.45 | 0.93 | 1 | 0.19 | 0.5 | 1 | 0.65 | 0.95 | 1 | 0.56 | 0.9 | 1 | 0.025 | 0.025 | 0.025 | 0.023 | 0.022 | 0.0059 | 0.022 | 0.021 | 0.0056 | -0.064 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.026 | 0.026 | 0.0045 | 0.027 | 0.028 | 0.0042 | 0.054 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.13.-.- | 0.049 | 0.49 | 1 | 0.042 | 0.97 | 1 | 0.027 | 0.71 | 1 | 0.034 | 0.43 | 1 | 0.18 | 0.87 | 1 | 0.086 | 0.66 | 1 | 0.00026 | 3.6e-05 | 0 | 0.00012 | 0 | 0.00026 | 5.8e-05 | 0 | 0.00015 | -1 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.2e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 | |
| 3.1.1.29 | Aminoacyl-tRNA hydrolase. | 0.0042 | 0.49 | 1 | 0.81 | 1 | 1 | 0.12 | 0.82 | 1 | 0.034 | 0.43 | 1 | 0.16 | 0.87 | 1 | 0.012 | 0.55 | 1 | 0.012 | 0.012 | 0.011 | 0.01 | 0.01 | 0.0012 | 0.011 | 0.011 | 0.0027 | 0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.013 | 0.013 | 0.0016 | 0.012 | 0.011 | 0.0013 | -0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.2.43 | Lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. | 0.12 | 0.5 | 1 | 0.18 | 1 | 1 | 0.11 | 0.82 | 1 | 0.096 | 0.45 | 1 | 0.092 | 0.86 | 1 | 0.17 | 0.71 | 1 | 0.0014 | 0.0012 | 0.00046 | 0.0015 | 0.0012 | 0.0011 | 0.0012 | 0.00032 | 0.0018 | -0.32 | 30 / 36 (83%) | 7 / 7 (100%) | 5 / 7 (71%) | 1 | 0.714 | 0.00063 | 0.00044 | 0.00074 | 0.0014 | 0.00034 | 0.0025 | 1.2 | 9 / 11 (82%) | 9 / 11 (82%) | 0.818 | 0.818 |
| 3.11.1.3 | Phosphonopyruvate hydrolase. | 0.058 | 0.49 | 1 | 0.44 | 1 | 1 | 0.11 | 0.82 | 1 | 0.032 | 0.43 | 1 | 0.4 | 0.9 | 1 | 0.12 | 0.66 | 1 | 0.00039 | 0.00026 | 0.00011 | 0.00035 | 0.00028 | 0.00032 | 0.00028 | 0.00012 | 4e-04 | -0.32 | 24 / 36 (67%) | 6 / 7 (86%) | 5 / 7 (71%) | 0.857 | 0.714 | 8e-05 | 0 | 0.00012 | 0.00037 | 9.9e-05 | 0.00094 | 2.2 | 5 / 11 (45%) | 8 / 11 (73%) | 0.455 | 0.727 |
| 2.7.7.89 | [Glutamine synthetase]-adenylyl-L-tyrosine phosphorylase. | 0.16 | 0.5 | 1 | 0.81 | 1 | 1 | 0.89 | 1 | 1 | 0.19 | 0.5 | 1 | 0.71 | 0.95 | 1 | 0.86 | 0.96 | 1 | 0.0044 | 0.0038 | 0.0017 | 0.0014 | 0.00062 | 0.0015 | 0.0017 | 0.0016 | 0.002 | 0.28 | 31 / 36 (86%) | 6 / 7 (86%) | 5 / 7 (71%) | 0.857 | 0.714 | 0.006 | 0.0023 | 0.012 | 0.0045 | 0.0029 | 0.0045 | -0.42 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 2.4.1.329 | Sucrose 6(F)-phosphate phosphorylase. | 0.48 | 0.72 | 1 | 0.17 | 1 | 1 | 0.084 | 0.77 | 1 | 0.34 | 0.61 | 1 | 0.42 | 0.91 | 1 | 0.17 | 0.71 | 1 | 0.0012 | 0.0012 | 0.0011 | 0.0013 | 0.0011 | 0.00085 | 0.00091 | 0.00084 | 0.00078 | -0.51 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0012 | 0.00095 | 0.0015 | 0.0013 | 0.0013 | 0.00084 | 0.12 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 2.3.1.269 | Apolipoprotein N-acyltransferase. | 0.53 | 0.76 | 1 | 0.23 | 1 | 1 | 0.85 | 1 | 1 | 0.8 | 0.93 | 1 | 0.89 | 0.99 | 1 | 0.66 | 0.92 | 1 | 0.00075 | 0.00052 | 0.00015 | 0.00087 | 0.00011 | 0.0013 | 0.00046 | 0.00032 | 0.00066 | -0.92 | 25 / 36 (69%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.00053 | 0.00014 | 0.00086 | 0.00032 | 0.00015 | 0.00054 | -0.73 | 8 / 11 (73%) | 7 / 11 (64%) | 0.727 | 0.636 |
| 1.1.1.29 | Glycerate dehydrogenase. | 0.83 | 0.92 | 1 | 0.85 | 1 | 1 | 0.96 | 1 | 1 | 0.66 | 0.85 | 1 | 0.7 | 0.95 | 1 | 0.7 | 0.94 | 1 | 0.0076 | 0.0076 | 0.0071 | 0.0073 | 0.0071 | 0.0021 | 0.0073 | 0.0056 | 0.0029 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.008 | 0.0067 | 0.0051 | 0.0076 | 0.0074 | 0.0024 | -0.074 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.1.3.23 | UDP-2.3-diacetamido-2.3-dideoxyglucuronic acid 2-epimerase. | 0.55 | 0.77 | 1 | 0.3 | 1 | 1 | 0.044 | 0.71 | 1 | 0.27 | 0.56 | 1 | 0.15 | 0.87 | 1 | 0.026 | 0.59 | 1 | 0.00059 | 0.00041 | 0.00012 | 3e-04 | 0.00034 | 0.00024 | 0.00024 | 0.00012 | 0.00035 | -0.32 | 25 / 36 (69%) | 6 / 7 (86%) | 4 / 7 (57%) | 0.857 | 0.571 | 0.00031 | 2.8e-05 | 0.00061 | 0.00068 | 0.00011 | 0.0014 | 1.1 | 6 / 11 (55%) | 9 / 11 (82%) | 0.545 | 0.818 |
| 2.3.1.157 | Glucosamine-1-phosphate N-acetyltransferase. | 0.18 | 0.52 | 1 | 1 | 1 | 1 | 0.87 | 1 | 1 | 0.11 | 0.47 | 1 | 0.65 | 0.95 | 1 | 0.56 | 0.9 | 1 | 0.0053 | 0.0053 | 0.0046 | 0.0042 | 0.0041 | 0.0015 | 0.0041 | 0.0041 | 0.0015 | -0.035 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0063 | 0.0048 | 0.0052 | 0.0058 | 0.006 | 0.0031 | -0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.5.1.12 | Transferred entry: 1.2.1.88. | 0.21 | 0.53 | 1 | 0.041 | 0.97 | 1 | 0.073 | 0.73 | 1 | 0.43 | 0.69 | 1 | 0.18 | 0.87 | 1 | 0.17 | 0.71 | 1 | 0.00048 | 0.00019 | 0 | 0.00049 | 0 | 0.0011 | 0.00023 | 0 | 0.00059 | -1.1 | 14 / 36 (39%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 8.4e-05 | 0 | 0.00019 | 6.9e-05 | 0 | 8.9e-05 | -0.28 | 4 / 11 (36%) | 5 / 11 (45%) | 0.364 | 0.455 |
| 1.16.3.2 | Bacterial non-heme ferritin. | 0.67 | 0.84 | 1 | 0.12 | 1 | 1 | 0.11 | 0.82 | 1 | 0.66 | 0.85 | 1 | 0.16 | 0.87 | 1 | 0.24 | 0.76 | 1 | 0.0022 | 0.0022 | 0.0019 | 0.0023 | 0.0018 | 0.001 | 0.0014 | 0.0011 | 0.00077 | -0.72 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0022 | 0.0015 | 0.0017 | 0.0028 | 0.002 | 0.0024 | 0.35 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.292 | 1.5-anhydro-D-fructose reductase (1.5-anhydro-D-mannitol-forming). | 0.052 | 0.49 | 1 | 0.59 | 1 | 1 | 0.94 | 1 | 1 | 0.061 | 0.43 | 1 | 1 | 1 | 1 | 0.59 | 0.91 | 1 | 0.013 | 0.013 | 0.013 | 0.011 | 0.0095 | 0.003 | 0.011 | 0.011 | 0.0042 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.015 | 0.015 | 0.007 | 0.014 | 0.014 | 0.0024 | -0.1 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.1.21 | Starch synthase. | 0.13 | 0.5 | 1 | 0.84 | 1 | 1 | 0.99 | 1 | 1 | 0.15 | 0.47 | 1 | 0.58 | 0.93 | 1 | 0.43 | 0.85 | 1 | 0.032 | 0.032 | 0.034 | 0.029 | 0.028 | 0.0065 | 0.029 | 0.032 | 0.01 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.034 | 0.034 | 0.0078 | 0.034 | 0.035 | 0.0053 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.3.1.12 | Ornithine cyclodeaminase. | 0.22 | 0.54 | 1 | 0.011 | 0.81 | 1 | 0.0029 | 0.55 | 1 | 0.31 | 0.58 | 1 | 0.047 | 0.81 | 1 | 0.0088 | 0.55 | 1 | 0.001 | 0.00093 | 0.00071 | 0.0013 | 0.0012 | 0.0013 | 0.00078 | 9e-04 | 0.00061 | -0.74 | 32 / 36 (89%) | 7 / 7 (100%) | 5 / 7 (71%) | 1 | 0.714 | 0.00078 | 0.00049 | 0.00092 | 0.00092 | 0.00075 | 0.00085 | 0.24 | 9 / 11 (82%) | 11 / 11 (100%) | 0.818 | 1 |
| 4.2.2.7 | Heparin lyase. | 0.082 | 0.49 | 1 | 0.9 | 1 | 1 | 0.31 | 0.89 | 1 | 0.18 | 0.5 | 1 | 0.63 | 0.95 | 1 | 0.54 | 0.9 | 1 | 0.00078 | 0.00044 | 9.3e-05 | 0.00084 | 0.00016 | 0.0014 | 0.00063 | 0.00024 | 0.00091 | -0.42 | 20 / 36 (56%) | 4 / 7 (57%) | 5 / 7 (71%) | 0.571 | 0.714 | 0.00011 | 0 | 0.00023 | 0.00038 | 0.00011 | 0.00079 | 1.8 | 4 / 11 (36%) | 7 / 11 (64%) | 0.364 | 0.636 |
| 1.6.5.8 | Transferred entry: 7.2.1.1. | 0.34 | 0.62 | 1 | 0.3 | 1 | 1 | 0.41 | 0.92 | 1 | 0.58 | 0.8 | 1 | 0.67 | 0.95 | 1 | 0.77 | 0.95 | 1 | 2e-04 | 2.3e-05 | 0 | 6.7e-05 | 0 | 0.00018 | 9.7e-06 | 0 | 2.6e-05 | -2.8 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 1.2e-05 | 0 | 3.9e-05 | 1.3e-05 | 0 | 4.3e-05 | 0.12 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.5.1.2 | Thiamine pyridinylase. | 0.4 | 0.66 | 1 | 0.27 | 1 | 1 | 0.33 | 0.89 | 1 | 0.47 | 0.72 | 1 | 0.41 | 0.9 | 1 | 0.39 | 0.83 | 1 | 0.00013 | 2.9e-05 | 0 | 5.3e-05 | 0 | 9.5e-05 | 2.9e-05 | 0 | 7.7e-05 | -0.87 | 8 / 36 (22%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.5e-05 | 0 | 7.5e-05 | 1.8e-05 | 0 | 3e-05 | -0.47 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 1.4.3.16 | L-aspartate oxidase. | 0.37 | 0.64 | 1 | 0.6 | 1 | 1 | 0.3 | 0.89 | 1 | 0.28 | 0.57 | 1 | 0.27 | 0.88 | 1 | 0.088 | 0.66 | 1 | 0.024 | 0.024 | 0.024 | 0.022 | 0.021 | 0.0067 | 0.024 | 0.026 | 0.01 | 0.13 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.025 | 0.024 | 0.0053 | 0.023 | 0.023 | 0.0034 | -0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.59 | N-acetylglucosamine kinase. | 0.79 | 0.9 | 1 | 0.47 | 1 | 1 | 0.77 | 0.99 | 1 | 0.84 | 0.94 | 1 | 0.46 | 0.92 | 1 | 0.9 | 0.98 | 1 | 0.0016 | 0.0016 | 0.0016 | 0.0015 | 0.0012 | 0.001 | 0.0019 | 0.0019 | 0.0013 | 0.34 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0014 | 0.001 | 0.00069 | 0.0017 | 0.0017 | 0.00068 | 0.28 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.2.17 | N-acetylmuramoyl-L-alanyl-D-glutamyl-L-lysyl-(N(6)-glycyl)-D-alanyl-D- | 0.19 | 0.53 | 1 | 0.87 | 1 | 1 | 0.18 | 0.86 | 1 | 0.41 | 0.68 | 1 | 0.67 | 0.95 | 1 | 0.38 | 0.83 | 1 | 0.00038 | 0.00027 | 0.00012 | 0.00014 | 0.00012 | 0.00012 | 0.00026 | 6.8e-05 | 4e-04 | 0.89 | 26 / 36 (72%) | 6 / 7 (86%) | 4 / 7 (57%) | 0.857 | 0.571 | 0.00048 | 0.00021 | 0.00067 | 0.00017 | 0.00011 | 0.00023 | -1.5 | 9 / 11 (82%) | 7 / 11 (64%) | 0.818 | 0.636 |
| 1.2.1.85 | 2-hydroxymuconate-6-semialdehyde dehydrogenase. | 0.62 | 0.82 | 1 | 0.096 | 1 | 1 | 0.18 | 0.85 | 1 | 0.31 | 0.58 | 1 | 0.045 | 0.8 | 1 | 0.087 | 0.66 | 1 | 6.6e-05 | 5.5e-06 | 0 | 0 | 0 | 0 | 1.9e-05 | 0 | 5.1e-05 | Inf | 3 / 36 (8.3%) | 0 / 7 (0%) | 1 / 7 (14%) | 0 | 0.143 | 3e-06 | 0 | 9.9e-06 | 2.7e-06 | 0 | 8.9e-06 | -0.15 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 1.14.13.48 | Transferred entry: 1.14.14.51. | 0.12 | 0.5 | 1 | 0.62 | 1 | 1 | 0.87 | 1 | 1 | 0.18 | 0.5 | 1 | 0.58 | 0.93 | 1 | 0.85 | 0.96 | 1 | 4e-04 | 0.00026 | 8.4e-05 | 5e-04 | 0.00026 | 0.00088 | 0.00047 | 0.00015 | 0.00078 | -0.089 | 23 / 36 (64%) | 6 / 7 (86%) | 5 / 7 (71%) | 0.857 | 0.714 | 0.00013 | 8.7e-05 | 0.00019 | 8.4e-05 | 0 | 0.00011 | -0.63 | 7 / 11 (64%) | 5 / 11 (45%) | 0.636 | 0.455 |
| 1.1.1.69 | Gluconate 5-dehydrogenase. | 0.21 | 0.54 | 1 | 0.53 | 1 | 1 | 0.13 | 0.82 | 1 | 0.57 | 0.8 | 1 | 0.99 | 1 | 1 | 0.4 | 0.83 | 1 | 0.0049 | 0.0049 | 0.0043 | 0.0054 | 0.0052 | 0.0025 | 0.0046 | 0.0038 | 0.0019 | -0.23 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0041 | 0.0042 | 0.0014 | 0.0055 | 0.0057 | 0.0022 | 0.42 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.1.3.32 | L-rhamnose mutarotase. | 0.52 | 0.75 | 1 | 0.77 | 1 | 1 | 0.57 | 0.96 | 1 | 0.69 | 0.87 | 1 | 0.96 | 0.99 | 1 | 0.88 | 0.97 | 1 | 0.0017 | 0.0017 | 0.0017 | 0.0017 | 0.0017 | 0.00069 | 0.0016 | 0.0018 | 0.00063 | -0.087 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0015 | 0.0015 | 0.00075 | 0.0018 | 0.0014 | 0.0014 | 0.26 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.2.5 | Allantoinase. | 0.13 | 0.5 | 1 | 0.42 | 1 | 1 | 0.6 | 0.96 | 1 | 0.081 | 0.44 | 1 | 0.097 | 0.86 | 1 | 0.48 | 0.87 | 1 | 0.0012 | 0.0011 | 0.00067 | 0.00044 | 0.00024 | 0.00047 | 0.00057 | 0.00043 | 0.00041 | 0.37 | 34 / 36 (94%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.0017 | 0.00084 | 0.0034 | 0.0013 | 0.00068 | 0.0013 | -0.39 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 1.3.8.6 | Glutaryl-CoA dehydrogenase (ETF). | 0.23 | 0.55 | 1 | 0.68 | 1 | 1 | 0.77 | 1 | 1 | 0.45 | 0.7 | 1 | 0.12 | 0.87 | 1 | 0.25 | 0.76 | 1 | 0.00079 | 0.00013 | 0 | 0.00037 | 0 | 0.00098 | 0.00027 | 0 | 0.00063 | -0.45 | 6 / 36 (17%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 1.5e-05 | 0 | 4.9e-05 | 8.4e-06 | 0 | 2e-05 | -0.84 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 2.1.1.13 | Methionine synthase. | 0.7 | 0.86 | 1 | 0.46 | 1 | 1 | 0.078 | 0.74 | 1 | 0.71 | 0.89 | 1 | 0.77 | 0.97 | 1 | 0.32 | 0.8 | 1 | 0.031 | 0.031 | 0.032 | 0.031 | 0.029 | 0.0053 | 0.029 | 0.03 | 0.006 | -0.096 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.03 | 0.029 | 0.0063 | 0.034 | 0.034 | 0.0044 | 0.18 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.6.1.26 | CDP-diacylglycerol diphosphatase. | 0.018 | 0.49 | 1 | 0.0034 | 0.64 | 1 | 0.0034 | 0.55 | 1 | 0.0046 | 0.36 | 1 | 0.018 | 0.76 | 1 | 0.0097 | 0.55 | 1 | 0.00054 | 1e-04 | 0 | 0.00038 | 1.6e-05 | 0.00088 | 0.00014 | 0 | 0.00036 | -1.4 | 7 / 36 (19%) | 4 / 7 (57%) | 1 / 7 (14%) | 0.571 | 0.143 | 0 | 0 | 0 | 1.7e-05 | 0 | 3.9e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 5.3.1.26 | Galactose-6-phosphate isomerase. | 0.04 | 0.49 | 1 | 0.85 | 1 | 1 | 0.2 | 0.87 | 1 | 0.041 | 0.43 | 1 | 0.62 | 0.95 | 1 | 0.22 | 0.74 | 1 | 0.00087 | 0.00085 | 0.00069 | 0.00056 | 0.00056 | 0.00047 | 0.00066 | 0.00013 | 0.00075 | 0.24 | 35 / 36 (97%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.0013 | 0.0011 | 0.001 | 0.00073 | 0.00057 | 0.00049 | -0.83 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.5.1.74 | 1.4-dihydroxy-2-naphthoate polyprenyltransferase. | 0.42 | 0.68 | 1 | 0.53 | 1 | 1 | 0.63 | 0.96 | 1 | 0.57 | 0.79 | 1 | 0.54 | 0.93 | 1 | 0.36 | 0.82 | 1 | 0.0029 | 0.0028 | 0.0017 | 0.0018 | 0.0012 | 0.0012 | 0.0023 | 0.0027 | 0.0016 | 0.35 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0035 | 0.0015 | 0.0052 | 0.0032 | 0.0036 | 0.0026 | -0.13 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 4.1.1.98 | 4-hydroxy-3-polyprenylbenzoate decarboxylase. | 0.17 | 0.51 | 1 | 0.42 | 1 | 1 | 0.32 | 0.89 | 1 | 0.35 | 0.62 | 1 | 0.65 | 0.95 | 1 | 0.93 | 0.98 | 1 | 0.00054 | 9e-05 | 0 | 3e-04 | 0 | 8e-04 | 0.00013 | 0 | 0.00033 | -1.2 | 6 / 36 (17%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 5.1e-06 | 0 | 1.7e-05 | 1.6e-05 | 0 | 3.7e-05 | 1.6 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 6.3.4.22 | tRNA(Ile)(2)-agmatinylcytidine synthase. | 0.41 | 0.67 | 1 | 0.77 | 1 | 1 | 0.55 | 0.95 | 1 | 0.55 | 0.78 | 1 | 0.55 | 0.93 | 1 | 0.79 | 0.96 | 1 | 9.7e-05 | 8.1e-06 | 0 | 8.1e-06 | 0 | 2.1e-05 | 1.3e-05 | 0 | 3.5e-05 | 0.68 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 1.3e-05 | 0 | 4.3e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.8.99.- | 0.029 | 0.49 | 1 | 0.064 | 1 | 1 | 0.42 | 0.93 | 1 | 0.047 | 0.43 | 1 | 0.1 | 0.86 | 1 | 0.57 | 0.9 | 1 | 0.00071 | 0.00057 | 0.00023 | 0.0013 | 0.00051 | 0.0021 | 0.00088 | 0.00036 | 0.0016 | -0.56 | 29 / 36 (81%) | 7 / 7 (100%) | 5 / 7 (71%) | 1 | 0.714 | 3e-04 | 9.5e-05 | 0.00037 | 0.00016 | 0.00011 | 0.00021 | -0.91 | 9 / 11 (82%) | 8 / 11 (73%) | 0.818 | 0.727 | |
| 2.2.1.10 | 2-amino-3.7-dideoxy-D-threo-hept-6-ulosonate synthase. | 0.61 | 0.81 | 1 | 0.67 | 1 | 1 | 0.73 | 0.99 | 1 | 0.68 | 0.87 | 1 | 0.68 | 0.95 | 1 | 0.87 | 0.97 | 1 | 0.00018 | 5.1e-05 | 0 | 5.9e-05 | 0 | 1e-04 | 5.5e-05 | 0 | 0.00013 | -0.1 | 10 / 36 (28%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 4e-05 | 0 | 9.5e-05 | 5.4e-05 | 0 | 0.00014 | 0.43 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 3.6.3.41 | Transferred entry: 7.6.2.5. | 0.89 | 0.96 | 1 | 0.7 | 1 | 1 | 0.48 | 0.93 | 1 | 0.84 | 0.94 | 1 | 0.39 | 0.9 | 1 | 0.2 | 0.73 | 1 | 0.0086 | 0.0086 | 0.006 | 0.0069 | 0.007 | 0.0037 | 0.0072 | 0.0061 | 0.0071 | 0.061 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0096 | 0.0048 | 0.014 | 0.0098 | 0.0058 | 0.0097 | 0.03 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.3.2.6 | 2-hydroxymuconate tautomerase. | 0.02 | 0.49 | 1 | 0.4 | 1 | 1 | 0.023 | 0.71 | 1 | 0.02 | 0.42 | 1 | 0.31 | 0.9 | 1 | 0.019 | 0.55 | 1 | 0.00013 | 1.9e-05 | 0 | 0 | 0 | 0 | 1.8e-05 | 0 | 4.8e-05 | Inf | 5 / 36 (14%) | 0 / 7 (0%) | 1 / 7 (14%) | 0 | 0.143 | 4.9e-05 | 0 | 7.6e-05 | 0 | 0 | 0 | -Inf | 4 / 11 (36%) | 0 / 11 (0%) | 0.364 | 0 |
| 2.7.1.156 | Adenosylcobinamide kinase. | 0.83 | 0.92 | 1 | 0.73 | 1 | 1 | 0.95 | 1 | 1 | 0.65 | 0.85 | 1 | 0.6 | 0.94 | 1 | 0.84 | 0.96 | 1 | 0.0064 | 0.0064 | 0.0061 | 0.006 | 0.006 | 0.0011 | 0.0067 | 0.007 | 0.0037 | 0.16 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0064 | 0.0064 | 0.0025 | 0.0065 | 0.006 | 0.0017 | 0.022 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.2.2.1 | Formate dehydrogenase (cytochrome). | 0.05 | 0.49 | 1 | 0.12 | 1 | 1 | 0.091 | 0.79 | 1 | 0.024 | 0.42 | 1 | 0.13 | 0.87 | 1 | 0.11 | 0.66 | 1 | 0.00034 | 7.5e-05 | 0 | 0.00018 | 1.6e-05 | 0.00035 | 0.00015 | 0 | 0.00041 | -0.26 | 8 / 36 (22%) | 4 / 7 (57%) | 1 / 7 (14%) | 0.571 | 0.143 | 5.1e-06 | 0 | 1.7e-05 | 2.6e-05 | 0 | 6.9e-05 | 2.3 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 5.1.3.11 | Cellobiose epimerase. | 0.45 | 0.7 | 1 | 0.72 | 1 | 1 | 0.046 | 0.71 | 1 | 0.51 | 0.74 | 1 | 0.92 | 0.99 | 1 | 0.096 | 0.66 | 1 | 0.0043 | 0.0043 | 0.0042 | 0.0041 | 0.0034 | 0.0016 | 0.004 | 0.0026 | 0.0024 | -0.036 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0037 | 0.0035 | 0.0021 | 0.0053 | 0.0055 | 0.0019 | 0.52 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.14.14.9 | 4-hydroxyphenylacetate 3-monooxygenase. | 0.31 | 0.59 | 1 | 0.52 | 1 | 1 | 0.8 | 1 | 1 | 0.44 | 0.69 | 1 | 0.37 | 0.9 | 1 | 0.98 | 1 | 1 | 9.1e-05 | 1.3e-05 | 0 | 1.7e-05 | 0 | 4.5e-05 | 2.5e-05 | 0 | 4.4e-05 | 0.56 | 5 / 36 (14%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 0 | 0 | 0 | 1.5e-05 | 0 | 3.3e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 3.1.3.41 | 4-nitrophenylphosphatase. | 0.16 | 0.5 | 1 | 0.58 | 1 | 1 | 0.23 | 0.88 | 1 | 0.12 | 0.47 | 1 | 0.41 | 0.9 | 1 | 0.078 | 0.66 | 1 | 0.0018 | 0.0018 | 0.00083 | 0.00087 | 0.00036 | 0.00093 | 0.0011 | 0.00075 | 0.0012 | 0.34 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0028 | 0.001 | 0.0052 | 0.0018 | 0.0015 | 0.0019 | -0.64 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 2.6.1.5 | Tyrosine transaminase. | 0.43 | 0.69 | 1 | 0.84 | 1 | 1 | 0.86 | 1 | 1 | 0.58 | 0.8 | 1 | 0.98 | 0.99 | 1 | 0.91 | 0.98 | 1 | 0.00032 | 0.00012 | 0 | 2e-04 | 0 | 0.00036 | 0.00017 | 0 | 0.00035 | -0.23 | 13 / 36 (36%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 8.4e-05 | 0 | 0.00017 | 6.3e-05 | 0 | 9.8e-05 | -0.42 | 3 / 11 (27%) | 4 / 11 (36%) | 0.273 | 0.364 |
| 3.1.3.- | 0.39 | 0.66 | 1 | 0.8 | 1 | 1 | 0.99 | 1 | 1 | 0.25 | 0.54 | 1 | 0.61 | 0.95 | 1 | 0.45 | 0.86 | 1 | 0.083 | 0.083 | 0.085 | 0.078 | 0.073 | 0.022 | 0.078 | 0.078 | 0.032 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.088 | 0.084 | 0.026 | 0.085 | 0.088 | 0.014 | -0.05 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 2.1.1.193 | 16S rRNA (uracil(1498)-N(3))-methyltransferase. | 0.081 | 0.49 | 1 | 0.83 | 1 | 1 | 0.59 | 0.96 | 1 | 0.074 | 0.43 | 1 | 0.18 | 0.87 | 1 | 0.097 | 0.66 | 1 | 0.011 | 0.011 | 0.011 | 0.01 | 0.0099 | 0.0018 | 0.011 | 0.011 | 0.0025 | 0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.012 | 0.011 | 0.0033 | 0.012 | 0.012 | 0.0019 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.3.3.13 | Albonoursin synthase. | 0.83 | 0.92 | 1 | 0.77 | 1 | 1 | 0.23 | 0.88 | 1 | 0.87 | 0.95 | 1 | 0.7 | 0.95 | 1 | 0.32 | 0.8 | 1 | 0.00032 | 0.00014 | 0 | 9.6e-05 | 0 | 0.00016 | 0.00011 | 0 | 0.00024 | 0.2 | 15 / 36 (42%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 8.1e-05 | 0 | 0.00015 | 0.00023 | 2.9e-05 | 0.00044 | 1.5 | 4 / 11 (36%) | 6 / 11 (55%) | 0.364 | 0.545 |
| 1.1.1.395 | 3-alpha-hydroxybile acid CoA 3-dehydrogenase. | 0.23 | 0.54 | 1 | 0.61 | 1 | 1 | 0.17 | 0.85 | 1 | 0.14 | 0.47 | 1 | 0.38 | 0.9 | 1 | 0.068 | 0.66 | 1 | 0.0022 | 0.0022 | 0.0021 | 0.0019 | 0.0022 | 0.0012 | 0.0022 | 0.002 | 0.0014 | 0.21 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0026 | 0.0025 | 0.0011 | 0.002 | 0.002 | 0.0013 | -0.38 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.2.1.11 | Aspartate-semialdehyde dehydrogenase. | 0.092 | 0.49 | 1 | 0.32 | 1 | 1 | 0.36 | 0.9 | 1 | 0.095 | 0.45 | 1 | 0.79 | 0.97 | 1 | 0.83 | 0.96 | 1 | 0.017 | 0.017 | 0.017 | 0.015 | 0.014 | 0.0024 | 0.014 | 0.012 | 0.0042 | -0.1 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.019 | 0.018 | 0.0064 | 0.019 | 0.019 | 0.0032 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.24.78 | GPR endopeptidase. | 0.85 | 0.94 | 1 | 0.32 | 1 | 1 | 0.95 | 1 | 1 | 0.84 | 0.94 | 1 | 0.86 | 0.99 | 1 | 0.39 | 0.83 | 1 | 0.009 | 0.009 | 0.0094 | 0.0094 | 0.0095 | 0.0014 | 0.0088 | 0.0094 | 0.0026 | -0.095 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0094 | 0.0099 | 0.003 | 0.0086 | 0.0093 | 0.0026 | -0.13 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.1.95 | L-glutamyl-[BtrI acyl-carrier protein] decarboxylase. | 0.38 | 0.65 | 1 | 0.58 | 1 | 1 | 0.43 | 0.93 | 1 | 0.32 | 0.59 | 1 | 0.61 | 0.95 | 1 | 0.33 | 0.8 | 1 | 0.00086 | 0.00058 | 0.00041 | 0.00042 | 0.00011 | 0.00056 | 0.00054 | 0.00032 | 0.00068 | 0.36 | 24 / 36 (67%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.00065 | 0.00068 | 0.00066 | 0.00063 | 5e-04 | 0.00077 | -0.045 | 9 / 11 (82%) | 7 / 11 (64%) | 0.818 | 0.636 |
| 1.1.1.159 | 7-alpha-hydroxysteroid dehydrogenase. | 0.53 | 0.76 | 1 | 0.74 | 1 | 1 | 0.3 | 0.89 | 1 | 0.37 | 0.63 | 1 | 0.44 | 0.91 | 1 | 0.12 | 0.66 | 1 | 0.002 | 0.002 | 0.0017 | 0.0019 | 0.0022 | 0.0011 | 0.002 | 0.0019 | 0.0012 | 0.074 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0022 | 0.0017 | 0.0012 | 0.0018 | 0.0016 | 0.0013 | -0.29 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.99.17 | Transferred entry: 1.1.5.2. | 0.16 | 0.5 | 1 | 0.14 | 1 | 1 | 0.24 | 0.88 | 1 | 0.25 | 0.54 | 1 | 0.97 | 0.99 | 1 | 0.94 | 0.99 | 1 | 0.0011 | 0.00012 | 0 | 0.00042 | 0 | 0.0011 | 0.00021 | 0 | 0.00056 | -1 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 2.7e-06 | 0 | 8.9e-06 | 0.054 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 4.1.2.48 | Low-specificity L-threonine aldolase. | 0.29 | 0.57 | 1 | 0.48 | 1 | 1 | 0.52 | 0.95 | 1 | 0.15 | 0.47 | 1 | 0.92 | 0.99 | 1 | 0.84 | 0.96 | 1 | 0.012 | 0.012 | 0.011 | 0.011 | 0.011 | 0.0045 | 0.0098 | 0.011 | 0.0041 | -0.17 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.013 | 0.011 | 0.0069 | 0.013 | 0.011 | 0.0033 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.16.1.3 | Aquacobalamin reductase. | 0.21 | 0.53 | 1 | 0.99 | 1 | 1 | 0.41 | 0.92 | 1 | 0.22 | 0.52 | 1 | 0.66 | 0.95 | 1 | 0.69 | 0.93 | 1 | 0.00034 | 2.9e-05 | 0 | 6.7e-05 | 0 | 0.00018 | 6.8e-05 | 0 | 0.00018 | 0.021 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 8.1e-06 | 0 | 2.7e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.7.7.77 | Molybdenum cofactor guanylyltransferase. | 0.4 | 0.66 | 1 | 0.31 | 1 | 1 | 0.44 | 0.93 | 1 | 0.76 | 0.91 | 1 | 0.49 | 0.92 | 1 | 0.16 | 0.71 | 1 | 0.002 | 0.002 | 0.002 | 0.0025 | 0.0024 | 0.00094 | 0.0022 | 0.0018 | 0.0013 | -0.18 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0021 | 0.0021 | 0.00078 | 0.0015 | 0.0013 | 0.00087 | -0.49 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.4.9 | dTMP kinase. | 0.066 | 0.49 | 1 | 0.35 | 1 | 1 | 0.92 | 1 | 1 | 0.018 | 0.42 | 1 | 0.86 | 0.99 | 1 | 0.49 | 0.88 | 1 | 0.006 | 0.006 | 0.0055 | 0.0052 | 0.0054 | 0.0016 | 0.0045 | 0.0047 | 0.00082 | -0.21 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0072 | 0.0063 | 0.0037 | 0.0063 | 0.0066 | 0.0018 | -0.19 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.127 | 2-dehydro-3-deoxy-D-gluconate 5-dehydrogenase. | 0.053 | 0.49 | 1 | 0.00063 | 0.34 | 1 | 0.0073 | 0.61 | 1 | 0.18 | 0.49 | 1 | 0.0021 | 0.62 | 1 | 0.013 | 0.55 | 1 | 0.00063 | 0.00061 | 0.00048 | 0.0011 | 0.00089 | 0.0011 | 0.00048 | 0.00035 | 0.00057 | -1.2 | 35 / 36 (97%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00054 | 0.00039 | 0.00038 | 0.00047 | 0.00043 | 0.00018 | -0.2 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.1.3.14 | UDP-N-acetylglucosamine 2-epimerase (non-hydrolyzing). | 0.73 | 0.87 | 1 | 0.34 | 1 | 1 | 0.71 | 0.98 | 1 | 0.38 | 0.65 | 1 | 0.17 | 0.87 | 1 | 0.44 | 0.86 | 1 | 0.0073 | 0.0073 | 0.0071 | 0.0066 | 0.0072 | 0.0016 | 0.0078 | 0.0077 | 0.0033 | 0.24 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0072 | 0.0061 | 0.0034 | 0.0076 | 0.0074 | 0.0023 | 0.078 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.1.47 | 8-amino-7-oxononanoate synthase. | 0.19 | 0.53 | 1 | 0.51 | 1 | 1 | 0.24 | 0.88 | 1 | 0.11 | 0.47 | 1 | 0.49 | 0.92 | 1 | 0.44 | 0.86 | 1 | 0.0021 | 0.002 | 0.00091 | 0.0023 | 0.002 | 0.0016 | 0.0021 | 0.00085 | 0.0027 | -0.13 | 34 / 36 (94%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0013 | 0.00053 | 0.0017 | 0.0024 | 0.0011 | 0.0041 | 0.88 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 3.6.3.27 | Transferred entry: 7.3.2.1. | 0.22 | 0.54 | 1 | 0.84 | 1 | 1 | 0.68 | 0.98 | 1 | 0.12 | 0.47 | 1 | 0.82 | 0.98 | 1 | 0.81 | 0.96 | 1 | 0.0027 | 0.0027 | 0.0018 | 0.0019 | 0.0015 | 0.0023 | 0.0019 | 0.0011 | 0.0022 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0036 | 0.0027 | 0.0045 | 0.0027 | 0.0026 | 0.0019 | -0.42 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.18.1.- | 0.7 | 0.86 | 1 | 0.019 | 0.92 | 1 | 0.022 | 0.71 | 1 | 0.81 | 0.93 | 1 | 0.0021 | 0.62 | 1 | 0.014 | 0.55 | 1 | 0.00065 | 0.00045 | 0.00013 | 0.00071 | 8.1e-05 | 0.0015 | 0.00051 | 0 | 0.0012 | -0.48 | 25 / 36 (69%) | 6 / 7 (86%) | 2 / 7 (29%) | 0.857 | 0.286 | 0.00029 | 0.00024 | 0.00024 | 0.00041 | 0.00023 | 0.00045 | 0.5 | 9 / 11 (82%) | 8 / 11 (73%) | 0.818 | 0.727 | |
| 1.3.99.5 | 3-oxo-5-alpha-steroid 4-dehydrogenase (acceptor). | 0.014 | 0.49 | 1 | 0.43 | 1 | 1 | 0.047 | 0.71 | 1 | 0.0013 | 0.31 | 1 | 0.041 | 0.8 | 1 | 0.0044 | 0.48 | 1 | 0.00046 | 0.00022 | 0 | 0.00026 | 0.00022 | 0.00021 | 4e-04 | 0 | 0.00074 | 0.62 | 17 / 36 (47%) | 7 / 7 (100%) | 3 / 7 (43%) | 1 | 0.429 | 2.1e-05 | 0 | 4.8e-05 | 0.00027 | 0 | 0.00052 | 3.7 | 2 / 11 (18%) | 5 / 11 (45%) | 0.182 | 0.455 |
| 2.4.2.4 | Thymidine phosphorylase. | 0.6 | 0.8 | 1 | 0.92 | 1 | 1 | 0.2 | 0.87 | 1 | 0.76 | 0.91 | 1 | 0.98 | 0.99 | 1 | 0.46 | 0.87 | 1 | 0.0038 | 0.0038 | 0.0036 | 0.0035 | 0.0033 | 0.00096 | 0.0036 | 0.0036 | 0.0019 | 0.041 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0031 | 0.0034 | 0.0012 | 0.0048 | 0.0044 | 0.0023 | 0.63 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.4.14 | [Acyl-carrier-protein] phosphodiesterase. | 0.058 | 0.49 | 1 | 0.3 | 1 | 1 | 0.42 | 0.92 | 1 | 0.014 | 0.42 | 1 | 0.12 | 0.87 | 1 | 0.25 | 0.76 | 1 | 0.00036 | 4e-05 | 0 | 8.9e-05 | 0 | 0.00018 | 0.00012 | 0 | 0.00031 | 0.43 | 4 / 36 (11%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 1.11.1.- | 0.19 | 0.53 | 1 | 0.58 | 1 | 1 | 0.75 | 0.99 | 1 | 0.27 | 0.56 | 1 | 0.84 | 0.98 | 1 | 0.58 | 0.91 | 1 | 0.00057 | 0.00055 | 0.00029 | 0.001 | 0.00033 | 0.0018 | 0.00079 | 0.00025 | 0.0012 | -0.34 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00039 | 0.00032 | 0.00029 | 0.00025 | 0.00022 | 0.00015 | -0.64 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 | |
| 3.2.2.n1 | Cytokinin riboside 5'-monophosphate phosphoribohydrolase. | 0.3 | 0.58 | 1 | 0.9 | 1 | 1 | 0.41 | 0.92 | 1 | 0.16 | 0.48 | 1 | 0.58 | 0.93 | 1 | 0.8 | 0.96 | 1 | 0.007 | 0.007 | 0.0062 | 0.006 | 0.0057 | 0.00088 | 0.006 | 0.0053 | 0.0018 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0073 | 0.0062 | 0.0035 | 0.0079 | 0.0078 | 0.002 | 0.11 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.-.- | 0.06 | 0.49 | 1 | 0.9 | 1 | 1 | 0.29 | 0.89 | 1 | 0.073 | 0.43 | 1 | 0.94 | 0.99 | 1 | 0.32 | 0.8 | 1 | 0.0093 | 0.0093 | 0.0096 | 0.0079 | 0.0064 | 0.0027 | 0.0081 | 0.0087 | 0.0041 | 0.036 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.012 | 0.01 | 0.0056 | 0.0088 | 0.0097 | 0.0019 | -0.45 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 1.3.1.26 | Transferred entry: 1.17.1.8. | 0.41 | 0.67 | 1 | 0.88 | 1 | 1 | 0.53 | 0.95 | 1 | 0.35 | 0.62 | 1 | 1 | 1 | 1 | 0.55 | 0.9 | 1 | 0.0011 | 0.00099 | 0.00078 | 0.00065 | 0.00059 | 0.00051 | 0.00078 | 0.00038 | 0.00083 | 0.26 | 33 / 36 (92%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.001 | 0.00072 | 0.0011 | 0.0013 | 0.0011 | 0.0011 | 0.38 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 1.3.1.12 | Prephenate dehydrogenase. | 0.22 | 0.54 | 1 | 0.32 | 1 | 1 | 0.24 | 0.89 | 1 | 0.14 | 0.47 | 1 | 0.62 | 0.95 | 1 | 0.44 | 0.85 | 1 | 0.0085 | 0.0085 | 0.0083 | 0.0073 | 0.008 | 0.0018 | 0.0064 | 0.0064 | 0.0021 | -0.19 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0094 | 0.0085 | 0.0051 | 0.0096 | 0.0092 | 0.0022 | 0.03 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.1.9 | Acetyl-CoA C-acetyltransferase. | 0.52 | 0.75 | 1 | 0.64 | 1 | 1 | 0.57 | 0.96 | 1 | 0.94 | 0.98 | 1 | 0.87 | 0.99 | 1 | 0.94 | 0.99 | 1 | 0.0066 | 0.0066 | 0.0062 | 0.007 | 0.0062 | 0.0028 | 0.0066 | 0.006 | 0.0023 | -0.085 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0063 | 0.0065 | 0.0018 | 0.0065 | 0.0059 | 0.0023 | 0.045 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.5.1.24 | N(5)-(carboxyethyl)ornithine synthase. | 0.046 | 0.49 | 1 | 0.49 | 1 | 1 | 0.053 | 0.71 | 1 | 0.044 | 0.43 | 1 | 0.34 | 0.9 | 1 | 0.034 | 0.63 | 1 | 9.6e-05 | 1.1e-05 | 0 | 0 | 0 | 0 | 7.7e-06 | 0 | 2e-05 | Inf | 4 / 36 (11%) | 0 / 7 (0%) | 1 / 7 (14%) | 0 | 0.143 | 3e-05 | 0 | 5.5e-05 | 0 | 0 | 0 | -Inf | 3 / 11 (27%) | 0 / 11 (0%) | 0.273 | 0 |
| 5.1.3.6 | UDP-glucuronate 4-epimerase. | 0.31 | 0.59 | 1 | 0.11 | 1 | 1 | 0.33 | 0.89 | 1 | 0.22 | 0.52 | 1 | 0.037 | 0.8 | 1 | 0.12 | 0.66 | 1 | 0.00014 | 2e-05 | 0 | 0 | 0 | 0 | 1.5e-05 | 0 | 4e-05 | Inf | 5 / 36 (14%) | 0 / 7 (0%) | 1 / 7 (14%) | 0 | 0.143 | 2.4e-05 | 0 | 6e-05 | 3.1e-05 | 0 | 7.4e-05 | 0.37 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 6.1.1.14 | Glycine--tRNA ligase. | 0.0093 | 0.49 | 1 | 0.29 | 1 | 1 | 0.29 | 0.89 | 1 | 0.084 | 0.44 | 1 | 0.56 | 0.93 | 1 | 0.052 | 0.65 | 1 | 0.026 | 0.026 | 0.025 | 0.024 | 0.024 | 0.0029 | 0.023 | 0.023 | 0.0034 | -0.061 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.028 | 0.027 | 0.0037 | 0.025 | 0.025 | 0.0028 | -0.16 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.2.23 | 4-hydroxybenzoyl-CoA thioesterase. | 0.73 | 0.87 | 1 | 0.91 | 1 | 1 | 0.69 | 0.98 | 1 | 0.83 | 0.94 | 1 | 0.82 | 0.98 | 1 | 0.96 | 0.99 | 1 | 0.00039 | 0.00022 | 6.1e-05 | 2e-04 | 6.4e-05 | 0.00028 | 0.00021 | 0.00011 | 0.00028 | 0.07 | 20 / 36 (56%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.00014 | 2.8e-05 | 0.00024 | 0.00031 | 5.9e-05 | 0.00056 | 1.1 | 6 / 11 (55%) | 6 / 11 (55%) | 0.545 | 0.545 |
| 2.4.99.16 | Starch synthase (maltosyl-transferring). | 0.088 | 0.49 | 1 | 0.84 | 1 | 1 | 0.7 | 0.98 | 1 | 0.068 | 0.43 | 1 | 0.62 | 0.95 | 1 | 0.39 | 0.83 | 1 | 0.0053 | 0.005 | 0.0032 | 0.0024 | 0.0023 | 0.002 | 0.0027 | 0.0032 | 0.0023 | 0.17 | 34 / 36 (94%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.0074 | 0.0038 | 0.012 | 0.0057 | 0.0065 | 0.0043 | -0.38 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.1.n4 | 1-acyl-sn-glycerol-3-phosphate acyltransferase. | 0.62 | 0.82 | 1 | 0.64 | 1 | 1 | 0.98 | 1 | 1 | 0.77 | 0.92 | 1 | 0.92 | 0.99 | 1 | 0.71 | 0.94 | 1 | 2e-04 | 3.9e-05 | 0 | 7.2e-05 | 0 | 0.00018 | 3.3e-05 | 0 | 7.6e-05 | -1.1 | 7 / 36 (19%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 4.2e-05 | 0 | 0.00012 | 2.1e-05 | 0 | 6.8e-05 | -1 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 1.8.7.1 | Assimilatory sulfite reductase (ferredoxin). | 0.65 | 0.84 | 1 | 0.45 | 1 | 1 | 0.081 | 0.76 | 1 | 0.82 | 0.93 | 1 | 0.55 | 0.93 | 1 | 0.13 | 0.67 | 1 | 0.0014 | 0.0014 | 0.0012 | 0.0014 | 0.0011 | 0.00099 | 0.0013 | 0.00074 | 0.0012 | -0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0012 | 0.0011 | 0.00083 | 0.0016 | 0.0014 | 0.00081 | 0.42 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.1.109 | Dolichyl-phosphate-mannose--protein mannosyltransferase. | 0.032 | 0.49 | 1 | 0.045 | 0.97 | 1 | 0.014 | 0.65 | 1 | 0.042 | 0.43 | 1 | 0.081 | 0.86 | 1 | 0.068 | 0.66 | 1 | 0.00098 | 0.00085 | 0.00039 | 0.0013 | 0.0015 | 0.00099 | 0.00072 | 6e-04 | 0.00086 | -0.85 | 31 / 36 (86%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00041 | 0.00026 | 0.00045 | 0.001 | 0.00023 | 0.0016 | 1.3 | 9 / 11 (82%) | 9 / 11 (82%) | 0.818 | 0.818 |
| 3.4.17.14 | Zinc D-Ala-D-Ala carboxypeptidase. | 0.74 | 0.88 | 1 | 0.13 | 1 | 1 | 0.037 | 0.71 | 1 | 0.82 | 0.93 | 1 | 0.25 | 0.88 | 1 | 0.17 | 0.71 | 1 | 0.0033 | 0.0033 | 0.0027 | 0.0035 | 0.0025 | 0.0026 | 0.0021 | 0.0013 | 0.0013 | -0.74 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0029 | 0.0025 | 0.0014 | 0.0045 | 0.0034 | 0.0037 | 0.63 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.3.1.9 | Trypanothione synthase. | 0.0088 | 0.49 | 1 | 0.0073 | 0.7 | 1 | 0.03 | 0.71 | 1 | 0.0019 | 0.31 | 1 | 0.0081 | 0.71 | 1 | 0.038 | 0.63 | 1 | 0.00026 | 5e-05 | 0 | 0.00015 | 2.8e-05 | 0.00022 | 9.7e-05 | 0 | 0.00026 | -0.63 | 7 / 36 (19%) | 4 / 7 (57%) | 1 / 7 (14%) | 0.571 | 0.143 | 3e-06 | 0 | 9.9e-06 | 2.7e-06 | 0 | 8.9e-06 | -0.15 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 1.1.1.193 | 5-amino-6-(5-phosphoribosylamino)uracil reductase. | 0.088 | 0.49 | 1 | 0.84 | 1 | 1 | 0.8 | 1 | 1 | 0.061 | 0.43 | 1 | 0.56 | 0.93 | 1 | 0.63 | 0.92 | 1 | 0.011 | 0.011 | 0.011 | 0.0094 | 0.0093 | 0.0023 | 0.0092 | 0.0092 | 0.0028 | -0.031 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.012 | 0.011 | 0.0039 | 0.012 | 0.013 | 0.0023 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.-.-.- | 0.11 | 0.5 | 1 | 0.45 | 1 | 1 | 0.049 | 0.71 | 1 | 0.49 | 0.74 | 1 | 0.57 | 0.93 | 1 | 0.25 | 0.76 | 1 | 0.028 | 0.028 | 0.028 | 0.03 | 0.029 | 0.0072 | 0.028 | 0.028 | 0.0052 | -0.1 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.025 | 0.026 | 0.0087 | 0.028 | 0.028 | 0.0048 | 0.16 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 1.1.1.411 | L-threonate 2-dehydrogenase. | 0.15 | 0.5 | 1 | 0.12 | 1 | 1 | 0.35 | 0.9 | 1 | 0.2 | 0.5 | 1 | 0.3 | 0.9 | 1 | 0.59 | 0.91 | 1 | 0.00032 | 4.4e-05 | 0 | 0.00013 | 0 | 0.00031 | 4.8e-05 | 0 | 0.00013 | -1.4 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.2e-05 | 0 | 7.3e-05 | 5.4e-06 | 0 | 1.8e-05 | -2 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.7.3.- | 0.5 | 0.74 | 1 | 0.91 | 1 | 1 | 0.84 | 1 | 1 | 0.91 | 0.97 | 1 | 0.53 | 0.93 | 1 | 0.62 | 0.92 | 1 | 0.028 | 0.028 | 0.027 | 0.03 | 0.028 | 0.014 | 0.03 | 0.028 | 0.013 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.026 | 0.026 | 0.0095 | 0.026 | 0.026 | 0.0077 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 6.3.3.2 | 5-formyltetrahydrofolate cyclo-ligase. | 0.014 | 0.49 | 1 | 0.57 | 1 | 1 | 0.48 | 0.94 | 1 | 0.031 | 0.43 | 1 | 0.1 | 0.86 | 1 | 0.1 | 0.66 | 1 | 0.0082 | 0.0082 | 0.0079 | 0.0067 | 0.0064 | 0.0015 | 0.0071 | 0.0076 | 0.0013 | 0.084 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0092 | 0.0085 | 0.0032 | 0.0088 | 0.0085 | 0.0015 | -0.064 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.12.1.3 | Hydrogen dehydrogenase (NADP(+)). | 0.34 | 0.62 | 1 | 0.21 | 1 | 1 | 0.32 | 0.89 | 1 | 0.41 | 0.68 | 1 | 0.34 | 0.9 | 1 | 0.84 | 0.96 | 1 | 0.0012 | 0.00087 | 5e-04 | 0.0011 | 0.00072 | 0.00083 | 0.00074 | 0.00073 | 7e-04 | -0.57 | 27 / 36 (75%) | 6 / 7 (86%) | 5 / 7 (71%) | 0.857 | 0.714 | 7e-04 | 0.00035 | 0.00099 | 0.001 | 8.8e-05 | 0.0018 | 0.51 | 9 / 11 (82%) | 7 / 11 (64%) | 0.818 | 0.636 |
| 2.7.9.3 | Selenide. water dikinase. | 0.9 | 0.96 | 1 | 0.42 | 1 | 1 | 0.47 | 0.93 | 1 | 0.6 | 0.81 | 1 | 0.57 | 0.93 | 1 | 0.78 | 0.95 | 1 | 0.0087 | 0.0087 | 0.0085 | 0.0087 | 0.007 | 0.0034 | 0.0084 | 0.01 | 0.0044 | -0.051 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0088 | 0.0084 | 0.0024 | 0.0089 | 0.0087 | 0.0024 | 0.016 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.2.1.5 | Linoleate isomerase. | 0.22 | 0.54 | 1 | 0.32 | 1 | 1 | 0.47 | 0.93 | 1 | 0.14 | 0.47 | 1 | 0.28 | 0.88 | 1 | 0.14 | 0.7 | 1 | 0.0011 | 0.00095 | 0.00065 | 5e-04 | 0.00042 | 5e-04 | 0.00075 | 0.00072 | 0.00077 | 0.58 | 32 / 36 (89%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.0011 | 0.00056 | 0.0015 | 0.0012 | 0.0011 | 0.0011 | 0.13 | 11 / 11 (100%) | 9 / 11 (82%) | 1 | 0.818 |
| 2.6.1.2 | Alanine transaminase. | 0.023 | 0.49 | 1 | 0.57 | 1 | 1 | 0.68 | 0.98 | 1 | 0.013 | 0.42 | 1 | 0.13 | 0.87 | 1 | 0.39 | 0.83 | 1 | 0.0077 | 0.0077 | 0.007 | 0.0045 | 0.005 | 0.0018 | 0.0052 | 0.0043 | 0.0023 | 0.21 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.01 | 0.008 | 0.0099 | 0.009 | 0.0077 | 0.0033 | -0.15 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.3.1.23 | Tyrosine ammonia-lyase. | 0.2 | 0.53 | 1 | 0.34 | 1 | 1 | 0.99 | 1 | 1 | 0.24 | 0.54 | 1 | 0.3 | 0.89 | 1 | 0.91 | 0.98 | 1 | 0.00012 | 1.7e-05 | 0 | 2.1e-05 | 0 | 4.3e-05 | 4.7e-05 | 0 | 1e-04 | 1.2 | 5 / 36 (14%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0 | 0 | 0 | 1.3e-05 | 0 | 4.3e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.2.1.77 | 3.4-dehydroadipyl-CoA semialdehyde dehydrogenase (NADP(+)). | 0.83 | 0.92 | 1 | 0.48 | 1 | 1 | 0.91 | 1 | 1 | 0.87 | 0.96 | 1 | 0.51 | 0.93 | 1 | 0.85 | 0.96 | 1 | 0.00022 | 1.9e-05 | 0 | 1.7e-05 | 0 | 4.5e-05 | 0 | 0 | 0 | -Inf | 3 / 36 (8.3%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 4.4e-05 | 0 | 0.00015 | 6.5e-06 | 0 | 2.1e-05 | -2.8 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 5.4.2.1 | Transferred entry: 5.4.2.11 and 5.4.2.12. | 0.15 | 0.5 | 1 | 0.18 | 1 | 1 | 0.32 | 0.89 | 1 | 0.08 | 0.44 | 1 | 0.04 | 0.8 | 1 | 0.081 | 0.66 | 1 | 0.0063 | 0.0063 | 0.006 | 0.005 | 0.005 | 0.0018 | 0.0061 | 0.0062 | 0.0029 | 0.29 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0068 | 0.0072 | 0.003 | 0.0068 | 0.0075 | 0.0024 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.6.1.17 | Cyclic pyranopterin monophosphate synthase. | 0.13 | 0.5 | 1 | 0.9 | 1 | 1 | 0.22 | 0.88 | 1 | 0.16 | 0.48 | 1 | 0.98 | 0.99 | 1 | 0.16 | 0.71 | 1 | 0.0059 | 0.0059 | 0.0057 | 0.0052 | 0.0043 | 0.002 | 0.0055 | 0.0043 | 0.0034 | 0.081 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0068 | 0.0059 | 0.0018 | 0.0057 | 0.0055 | 0.0019 | -0.25 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.4.6 | Nucleoside-diphosphate kinase. | 0.95 | 0.98 | 1 | 0.12 | 1 | 1 | 0.56 | 0.96 | 1 | 0.84 | 0.94 | 1 | 0.11 | 0.87 | 1 | 0.75 | 0.95 | 1 | 0.00079 | 0.00079 | 0.00072 | 0.00083 | 0.00081 | 3e-04 | 0.00061 | 0.00049 | 6e-04 | -0.44 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00088 | 0.00078 | 0.00048 | 0.00078 | 0.00055 | 0.00073 | -0.17 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.2.1.80 | Long-chain acyl-[acyl-carrier-protein] reductase. | 0.32 | 0.6 | 1 | 0.29 | 1 | 1 | 0.7 | 0.98 | 1 | 0.34 | 0.61 | 1 | 0.4 | 0.9 | 1 | 0.6 | 0.91 | 1 | 0.00022 | 7.4e-05 | 0 | 4.3e-05 | 0 | 7.4e-05 | 8.3e-06 | 0 | 1.4e-05 | -2.4 | 12 / 36 (33%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0.00014 | 0 | 3e-04 | 6.8e-05 | 0 | 0.00014 | -1 | 5 / 11 (45%) | 3 / 11 (27%) | 0.455 | 0.273 |
| 2.7.1.144 | Tagatose-6-phosphate kinase. | 0.49 | 0.73 | 1 | 0.36 | 1 | 1 | 0.87 | 1 | 1 | 0.38 | 0.65 | 1 | 0.71 | 0.95 | 1 | 0.48 | 0.87 | 1 | 0.015 | 0.015 | 0.015 | 0.015 | 0.015 | 0.0035 | 0.014 | 0.016 | 0.0053 | -0.1 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.016 | 0.018 | 0.0038 | 0.015 | 0.015 | 0.003 | -0.093 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.10.2.- | 0.018 | 0.49 | 1 | 0.00025 | 0.34 | 0.56 | 0.0034 | 0.55 | 1 | 0.003 | 0.36 | 1 | 0.00029 | 0.62 | 0.66 | 0.0093 | 0.55 | 1 | 0.00059 | 0.00024 | 0 | 0.00069 | 2e-04 | 0.0014 | 0.00037 | 0 | 0.00092 | -0.9 | 15 / 36 (42%) | 7 / 7 (100%) | 2 / 7 (29%) | 1 | 0.286 | 5.2e-05 | 0 | 0.00011 | 7.8e-05 | 0 | 0.00022 | 0.58 | 4 / 11 (36%) | 2 / 11 (18%) | 0.364 | 0.182 | |
| 1.4.1.4 | Glutamate dehydrogenase (NADP(+)). | 0.11 | 0.5 | 1 | 0.47 | 1 | 1 | 0.098 | 0.81 | 1 | 0.031 | 0.43 | 1 | 0.38 | 0.9 | 1 | 0.081 | 0.66 | 1 | 0.0018 | 0.0017 | 0.00064 | 0.0019 | 0.0016 | 0.0012 | 0.0018 | 0.00064 | 0.0021 | -0.078 | 33 / 36 (92%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00097 | 0.00038 | 0.0017 | 0.0022 | 5e-04 | 0.004 | 1.2 | 9 / 11 (82%) | 11 / 11 (100%) | 0.818 | 1 |
| 5.1.2.2 | Mandelate racemase. | 0.43 | 0.69 | 1 | 0.79 | 1 | 1 | 0.76 | 0.99 | 1 | 0.99 | 1 | 1 | 0.71 | 0.95 | 1 | 0.6 | 0.91 | 1 | 0.00042 | 0.00015 | 0 | 0.00034 | 0 | 0.00079 | 0.00018 | 0 | 0.00037 | -0.92 | 13 / 36 (36%) | 2 / 7 (29%) | 3 / 7 (43%) | 0.286 | 0.429 | 5.3e-05 | 0 | 7e-05 | 0.00012 | 0 | 0.00025 | 1.2 | 5 / 11 (45%) | 3 / 11 (27%) | 0.455 | 0.273 |
| 4.2.1.52 | Transferred entry: 4.3.3.7. | 0.97 | 0.99 | 1 | 0.27 | 1 | 1 | 0.0043 | 0.57 | 1 | 0.64 | 0.84 | 1 | 0.39 | 0.9 | 1 | 0.0057 | 0.48 | 1 | 0.00052 | 0.00044 | 0.00018 | 0.00057 | 0.00034 | 0.00061 | 0.00078 | 0.00096 | 0.00067 | 0.45 | 30 / 36 (83%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00047 | 0.00035 | 0.00043 | 9.8e-05 | 1e-04 | 7.5e-05 | -2.3 | 10 / 11 (91%) | 8 / 11 (73%) | 0.909 | 0.727 |
| 2.7.1.31 | Glycerate 3-kinase. | 0.28 | 0.57 | 1 | 0.29 | 1 | 1 | 0.17 | 0.85 | 1 | 0.2 | 0.5 | 1 | 0.087 | 0.86 | 1 | 0.056 | 0.66 | 1 | 0.0099 | 0.0099 | 0.0096 | 0.0089 | 0.0093 | 0.0025 | 0.01 | 0.012 | 0.004 | 0.17 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.01 | 0.0037 | 0.0095 | 0.0095 | 0.0014 | -0.21 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.5.1.61 | Hydroxymethylbilane synthase. | 0.89 | 0.95 | 1 | 0.72 | 1 | 1 | 0.82 | 1 | 1 | 0.79 | 0.92 | 1 | 0.32 | 0.9 | 1 | 0.43 | 0.85 | 1 | 0.0081 | 0.0081 | 0.0077 | 0.008 | 0.0071 | 0.0017 | 0.0086 | 0.009 | 0.0031 | 0.1 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.008 | 0.0069 | 0.0027 | 0.008 | 0.008 | 0.0017 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.1.107 | Uroporphyrinogen-III C-methyltransferase. | 0.74 | 0.88 | 1 | 0.11 | 1 | 1 | 0.19 | 0.87 | 1 | 0.88 | 0.96 | 1 | 0.15 | 0.87 | 1 | 0.25 | 0.76 | 1 | 0.0074 | 0.0074 | 0.0071 | 0.0078 | 0.008 | 0.0016 | 0.0064 | 0.0067 | 0.0033 | -0.29 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0077 | 0.008 | 0.0038 | 0.0076 | 0.0078 | 0.0032 | -0.019 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.1.1.16 | Cysteine--tRNA ligase. | 0.051 | 0.49 | 1 | 0.29 | 1 | 1 | 0.69 | 0.98 | 1 | 0.11 | 0.47 | 1 | 0.8 | 0.98 | 1 | 0.4 | 0.83 | 1 | 0.025 | 0.025 | 0.026 | 0.024 | 0.023 | 0.0042 | 0.022 | 0.023 | 0.005 | -0.13 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.028 | 0.027 | 0.0052 | 0.027 | 0.027 | 0.0027 | -0.052 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.1 | Alcohol dehydrogenase. | 0.3 | 0.58 | 1 | 0.54 | 1 | 1 | 0.35 | 0.9 | 1 | 0.21 | 0.51 | 1 | 0.24 | 0.88 | 1 | 0.14 | 0.69 | 1 | 0.031 | 0.031 | 0.032 | 0.029 | 0.029 | 0.0065 | 0.032 | 0.04 | 0.013 | 0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.033 | 0.037 | 0.0091 | 0.031 | 0.029 | 0.0052 | -0.09 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.3.5.9 | Hydrogenobyrinic acid a.c-diamide synthase (glutamine-hydrolyzing). | 0.48 | 0.72 | 1 | 0.95 | 1 | 1 | 0.77 | 0.99 | 1 | 0.29 | 0.58 | 1 | 0.57 | 0.93 | 1 | 0.46 | 0.86 | 1 | 0.00098 | 0.00098 | 0.00069 | 0.00088 | 0.00067 | 0.00083 | 0.00084 | 0.00076 | 0.00063 | -0.067 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0011 | 7e-04 | 0.00095 | 0.001 | 5e-04 | 8e-04 | -0.14 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.1.150 | Short-chain-enoyl-CoA hydratase. | 0.074 | 0.49 | 1 | 0.1 | 1 | 1 | 0.32 | 0.89 | 1 | 0.066 | 0.43 | 1 | 0.17 | 0.87 | 1 | 0.53 | 0.9 | 1 | 0.00044 | 0.00012 | 0 | 0.00032 | 3.2e-05 | 0.00065 | 0.00024 | 0 | 0.00061 | -0.42 | 10 / 36 (28%) | 4 / 7 (57%) | 2 / 7 (29%) | 0.571 | 0.286 | 2.3e-05 | 0 | 4.9e-05 | 2.1e-05 | 0 | 6.8e-05 | -0.13 | 3 / 11 (27%) | 1 / 11 (9.1%) | 0.273 | 0.0909 |
| 2.1.1.10 | Homocysteine S-methyltransferase. | 0.97 | 0.99 | 1 | 0.56 | 1 | 1 | 0.35 | 0.9 | 1 | 0.65 | 0.85 | 1 | 0.99 | 1 | 1 | 0.78 | 0.95 | 1 | 0.004 | 0.004 | 0.0036 | 0.0039 | 0.0036 | 0.0013 | 0.0036 | 0.0031 | 0.0013 | -0.12 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.004 | 0.0036 | 0.0016 | 0.0044 | 0.004 | 0.002 | 0.14 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.1.5 | Dextransucrase. | 0.75 | 0.88 | 1 | 0.89 | 1 | 1 | 0.81 | 1 | 1 | 0.88 | 0.96 | 1 | 0.62 | 0.95 | 1 | 0.69 | 0.94 | 1 | 0.0071 | 0.0071 | 0.0065 | 0.0078 | 0.0067 | 0.007 | 0.0072 | 0.0081 | 0.0047 | -0.12 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0065 | 0.0069 | 0.0038 | 0.0072 | 0.0048 | 0.0066 | 0.15 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.2.10 | Pectin lyase. | 0.77 | 0.89 | 1 | 0.43 | 1 | 1 | 0.41 | 0.92 | 1 | 0.73 | 0.9 | 1 | 0.31 | 0.9 | 1 | 0.14 | 0.69 | 1 | 0.00033 | 4.6e-05 | 0 | 2e-05 | 0 | 5.4e-05 | 3.5e-05 | 0 | 9.3e-05 | 0.81 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 5.1e-05 | 0 | 0.00016 | 6.5e-05 | 0 | 0.00021 | 0.35 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 4.3.2.10 | Imidazole glycerol-phosphate synthase. | 0.015 | 0.49 | 1 | 0.24 | 1 | 1 | 0.31 | 0.89 | 1 | 0.038 | 0.43 | 1 | 0.029 | 0.76 | 1 | 0.041 | 0.63 | 1 | 0.021 | 0.021 | 0.021 | 0.018 | 0.018 | 0.003 | 0.02 | 0.021 | 0.0051 | 0.15 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.023 | 0.022 | 0.0047 | 0.022 | 0.023 | 0.0026 | -0.064 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.1.8 | Mannonate dehydratase. | 0.098 | 0.49 | 1 | 0.54 | 1 | 1 | 0.56 | 0.95 | 1 | 0.2 | 0.5 | 1 | 0.43 | 0.91 | 1 | 0.7 | 0.94 | 1 | 0.0064 | 0.0064 | 0.0066 | 0.0068 | 0.0067 | 0.0021 | 0.0077 | 0.0067 | 0.0027 | 0.18 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0052 | 0.0051 | 0.0025 | 0.0066 | 0.0066 | 0.0024 | 0.34 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.22.34 | Legumain. | 0.39 | 0.65 | 1 | 0.96 | 1 | 1 | 0.35 | 0.9 | 1 | 0.2 | 0.51 | 1 | 0.82 | 0.98 | 1 | 0.35 | 0.8 | 1 | 0.00094 | 0.00016 | 0 | 8.4e-05 | 0 | 0.00016 | 9.1e-05 | 0 | 0.00016 | 0.12 | 6 / 36 (17%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0 | 0 | 0 | 4e-04 | 0 | 0.0013 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 3.5.3.6 | Arginine deiminase. | 0.00095 | 0.49 | 1 | 0.8 | 1 | 1 | 0.0091 | 0.61 | 1 | 0.01 | 0.42 | 1 | 0.58 | 0.93 | 1 | 0.14 | 0.69 | 1 | 0.0021 | 0.002 | 0.0018 | 0.0012 | 0.001 | 0.00072 | 0.0015 | 0.0013 | 0.0011 | 0.32 | 35 / 36 (97%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.0034 | 0.0033 | 0.0014 | 0.0016 | 0.00091 | 0.0014 | -1.1 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.3.101 | Validoxylamine A 7'-phosphate phosphatase. | 0.28 | 0.56 | 1 | 0.27 | 1 | 1 | 0.8 | 1 | 1 | 0.44 | 0.7 | 1 | 0.84 | 0.98 | 1 | 0.51 | 0.89 | 1 | 0.00018 | 1.5e-05 | 0 | 5e-05 | 0 | 0.00013 | 1.9e-05 | 0 | 5.1e-05 | -1.4 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 6e-06 | 0 | 2e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.6.1.19 | 4-aminobutyrate--2-oxoglutarate transaminase. | 0.55 | 0.77 | 1 | 0.33 | 1 | 1 | 0.68 | 0.98 | 1 | 0.96 | 0.99 | 1 | 0.98 | 0.99 | 1 | 0.4 | 0.83 | 1 | 0.0011 | 9e-04 | 0.00041 | 0.0012 | 0.00094 | 0.0014 | 8e-04 | 5e-04 | 0.00081 | -0.58 | 30 / 36 (83%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00082 | 0.00029 | 0.001 | 0.00083 | 0.00033 | 0.0011 | 0.017 | 10 / 11 (91%) | 8 / 11 (73%) | 0.909 | 0.727 |
| 1.3.3.4 | Protoporphyrinogen oxidase. | 0.035 | 0.49 | 1 | 0.27 | 1 | 1 | 0.17 | 0.85 | 1 | 0.047 | 0.43 | 1 | 0.33 | 0.9 | 1 | 0.27 | 0.77 | 1 | 3e-04 | 0.00016 | 5.9e-05 | 0.00032 | 0.00016 | 0.00042 | 0.00027 | 6.2e-05 | 6e-04 | -0.25 | 19 / 36 (53%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 3.7e-05 | 0 | 4.6e-05 | 1e-04 | 0 | 0.00015 | 1.4 | 5 / 11 (45%) | 5 / 11 (45%) | 0.455 | 0.455 |
| 1.21.4.1 | D-proline reductase. | 0.31 | 0.59 | 1 | 0.62 | 1 | 1 | 0.31 | 0.89 | 1 | 0.14 | 0.47 | 1 | 0.98 | 0.99 | 1 | 0.16 | 0.71 | 1 | 0.0017 | 0.0017 | 0.0013 | 0.0018 | 0.0011 | 0.0015 | 0.0016 | 0.0011 | 0.0021 | -0.17 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0022 | 0.0019 | 0.0012 | 0.0012 | 0.0011 | 7e-04 | -0.87 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.21.88 | Repressor LexA. | 0.19 | 0.53 | 1 | 0.34 | 1 | 1 | 0.85 | 1 | 1 | 0.13 | 0.47 | 1 | 0.83 | 0.98 | 1 | 0.49 | 0.87 | 1 | 0.014 | 0.014 | 0.014 | 0.013 | 0.014 | 0.0019 | 0.012 | 0.011 | 0.0032 | -0.12 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.015 | 0.015 | 0.004 | 0.014 | 0.015 | 0.0034 | -0.1 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.3.1.19 | Prokaryotic ubiquitin-like protein ligase. | 0.11 | 0.5 | 1 | 0.42 | 1 | 1 | 0.41 | 0.92 | 1 | 0.077 | 0.44 | 1 | 0.058 | 0.83 | 1 | 0.25 | 0.76 | 1 | 0.0027 | 0.0024 | 0.0015 | 0.00093 | 0.00054 | 0.0011 | 0.0014 | 0.0014 | 0.0017 | 0.59 | 32 / 36 (89%) | 5 / 7 (71%) | 6 / 7 (86%) | 0.714 | 0.857 | 0.0038 | 0.0016 | 0.0071 | 0.0026 | 0.0016 | 0.0025 | -0.55 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 4.4.1.11 | Methionine gamma-lyase. | 0.11 | 0.5 | 1 | 0.95 | 1 | 1 | 0.82 | 1 | 1 | 0.18 | 0.5 | 1 | 0.39 | 0.9 | 1 | 0.45 | 0.86 | 1 | 0.039 | 0.039 | 0.039 | 0.034 | 0.034 | 0.0062 | 0.034 | 0.036 | 0.011 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.041 | 0.047 | 0.01 | 0.042 | 0.039 | 0.0084 | 0.035 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.1.1.23 | Aspartate--tRNA(Asn) ligase. | 0.14 | 0.5 | 1 | 0.37 | 1 | 1 | 0.076 | 0.74 | 1 | 0.15 | 0.47 | 1 | 0.1 | 0.86 | 1 | 0.014 | 0.55 | 1 | 0.024 | 0.024 | 0.023 | 0.022 | 0.021 | 0.004 | 0.024 | 0.022 | 0.0064 | 0.13 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.026 | 0.027 | 0.0065 | 0.022 | 0.022 | 0.0046 | -0.24 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.-.- | 0.67 | 0.84 | 1 | 0.37 | 1 | 1 | 0.03 | 0.71 | 1 | 0.18 | 0.5 | 1 | 1 | 1 | 1 | 0.16 | 0.71 | 1 | 0.11 | 0.11 | 0.096 | 0.098 | 0.097 | 0.022 | 0.091 | 0.092 | 0.023 | -0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.11 | 0.096 | 0.041 | 0.13 | 0.12 | 0.055 | 0.24 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 2.7.7.75 | Molybdopterin adenylyltransferase. | 0.74 | 0.88 | 1 | 0.57 | 1 | 1 | 0.78 | 1 | 1 | 0.56 | 0.78 | 1 | 0.72 | 0.95 | 1 | 1 | 1 | 1 | 0.0063 | 0.0063 | 0.006 | 0.0061 | 0.0053 | 0.0021 | 0.0059 | 0.0051 | 0.0033 | -0.048 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0066 | 0.0071 | 0.0024 | 0.0063 | 0.0058 | 0.0021 | -0.067 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.6.1.40 | Guanosine-5'-triphosphate.3'-diphosphate diphosphatase. | 0.39 | 0.65 | 1 | 0.58 | 1 | 1 | 0.27 | 0.89 | 1 | 0.22 | 0.52 | 1 | 0.48 | 0.92 | 1 | 0.1 | 0.66 | 1 | 0.005 | 0.005 | 0.0036 | 0.0035 | 0.0024 | 0.0027 | 0.003 | 0.0022 | 0.0023 | -0.22 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0059 | 0.0035 | 0.0086 | 0.0062 | 0.0046 | 0.0037 | 0.072 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.3.68 | 2-deoxyglucose-6-phosphatase. | 0.13 | 0.5 | 1 | 0.65 | 1 | 1 | 0.74 | 0.99 | 1 | 0.1 | 0.46 | 1 | 0.32 | 0.9 | 1 | 0.84 | 0.96 | 1 | 0.0031 | 0.0026 | 0.0012 | 0.00075 | 0.00057 | 0.00078 | 0.0011 | 6e-04 | 0.0013 | 0.55 | 30 / 36 (83%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.0043 | 0.0015 | 0.0097 | 0.003 | 0.0011 | 0.0033 | -0.52 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 2.4.1.82 | Galactinol--sucrose galactosyltransferase. | 0.35 | 0.63 | 1 | 0.3 | 1 | 1 | 0.59 | 0.96 | 1 | 0.48 | 0.72 | 1 | 0.54 | 0.93 | 1 | 0.45 | 0.86 | 1 | 0.0013 | 0.00085 | 0.00029 | 3e-04 | 0.00014 | 0.00046 | 0.00073 | 0.00036 | 0.00096 | 1.3 | 24 / 36 (67%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 0.00097 | 0.00029 | 0.0017 | 0.0011 | 0.00057 | 0.0014 | 0.18 | 8 / 11 (73%) | 7 / 11 (64%) | 0.727 | 0.636 |
| 2.3.1.89 | Tetrahydrodipicolinate N-acetyltransferase. | 0.3 | 0.58 | 1 | 0.72 | 1 | 1 | 0.58 | 0.96 | 1 | 0.18 | 0.5 | 1 | 0.89 | 0.99 | 1 | 0.33 | 0.8 | 1 | 0.0012 | 0.0012 | 0.001 | 0.0012 | 0.001 | 0.00075 | 0.0011 | 0.00072 | 0.00094 | -0.13 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0015 | 0.0013 | 0.00062 | 0.0011 | 0.00091 | 8e-04 | -0.45 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.25 | Shikimate dehydrogenase. | 0.68 | 0.85 | 1 | 0.79 | 1 | 1 | 0.97 | 1 | 1 | 0.4 | 0.67 | 1 | 0.77 | 0.97 | 1 | 0.54 | 0.9 | 1 | 0.02 | 0.02 | 0.021 | 0.02 | 0.019 | 0.0048 | 0.02 | 0.022 | 0.0078 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.02 | 0.021 | 0.0035 | 0.02 | 0.021 | 0.0031 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.-.-.- | 0.94 | 0.97 | 1 | 0.87 | 1 | 1 | 0.83 | 1 | 1 | 0.48 | 0.72 | 1 | 0.21 | 0.88 | 1 | 0.21 | 0.74 | 1 | 0.13 | 0.13 | 0.13 | 0.13 | 0.14 | 0.036 | 0.13 | 0.14 | 0.037 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.13 | 0.12 | 0.03 | 0.13 | 0.12 | 0.013 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 4.1.1.104 | 3-dehydro-4-phosphotetronate decarboxylase. | 0.12 | 0.5 | 1 | 0.78 | 1 | 1 | 0.36 | 0.9 | 1 | 0.092 | 0.45 | 1 | 0.67 | 0.95 | 1 | 0.3 | 0.78 | 1 | 0.00017 | 2.4e-05 | 0 | 4e-05 | 0 | 7e-05 | 5.8e-05 | 0 | 0.00015 | 0.54 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 1.6e-05 | 0 | 4.5e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 2.5.1.18 | Glutathione transferase. | 0.66 | 0.84 | 1 | 0.045 | 0.97 | 1 | 0.023 | 0.71 | 1 | 0.59 | 0.81 | 1 | 0.025 | 0.76 | 1 | 0.012 | 0.55 | 1 | 0.00058 | 0.00037 | 7.3e-05 | 0.00062 | 0 | 0.0014 | 0.00081 | 0.00032 | 0.0015 | 0.39 | 23 / 36 (64%) | 3 / 7 (43%) | 6 / 7 (86%) | 0.429 | 0.857 | 0.00019 | 7.2e-05 | 0.00025 | 0.00011 | 2.9e-05 | 0.00014 | -0.79 | 8 / 11 (73%) | 6 / 11 (55%) | 0.727 | 0.545 |
| 6.3.4.4 | Adenylosuccinate synthase. | 0.012 | 0.49 | 1 | 0.76 | 1 | 1 | 0.93 | 1 | 1 | 0.06 | 0.43 | 1 | 0.4 | 0.9 | 1 | 0.27 | 0.77 | 1 | 0.026 | 0.026 | 0.026 | 0.023 | 0.023 | 0.0028 | 0.023 | 0.021 | 0.0049 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.029 | 0.028 | 0.006 | 0.028 | 0.028 | 0.0043 | -0.051 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.2.3 | Phosphoglycerate kinase. | 0.15 | 0.5 | 1 | 0.45 | 1 | 1 | 0.84 | 1 | 1 | 0.13 | 0.47 | 1 | 0.87 | 0.99 | 1 | 0.36 | 0.81 | 1 | 0.018 | 0.018 | 0.018 | 0.017 | 0.017 | 0.0026 | 0.016 | 0.017 | 0.0046 | -0.087 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.019 | 0.019 | 0.0022 | 0.018 | 0.02 | 0.003 | -0.078 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.237 | Hydroxyphenylpyruvate reductase. | 0.19 | 0.53 | 1 | 0.22 | 1 | 1 | 0.33 | 0.89 | 1 | 0.29 | 0.57 | 1 | 0.96 | 0.99 | 1 | 0.95 | 0.99 | 1 | 0.00032 | 3.6e-05 | 0 | 0.00012 | 0 | 0.00031 | 5.8e-05 | 0 | 0.00015 | -1 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 2.7e-06 | 0 | 8.9e-06 | 0.054 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 1.3.1.76 | Precorrin-2 dehydrogenase. | 0.12 | 0.5 | 1 | 0.086 | 1 | 1 | 0.24 | 0.89 | 1 | 0.3 | 0.58 | 1 | 0.36 | 0.9 | 1 | 0.53 | 0.9 | 1 | 0.0015 | 0.0015 | 0.0015 | 0.002 | 0.0019 | 0.001 | 0.0015 | 0.0014 | 0.00055 | -0.42 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0015 | 0.0016 | 0.00066 | 0.0014 | 0.0015 | 0.00058 | -0.1 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 7.2.2.14 | P-type Mg(2+) transporter. | 0.2 | 0.53 | 1 | 0.65 | 1 | 1 | 0.37 | 0.9 | 1 | 0.24 | 0.53 | 1 | 0.53 | 0.93 | 1 | 0.39 | 0.83 | 1 | 0.00074 | 0.00014 | 0 | 0.00031 | 0 | 0.00075 | 0.00035 | 0 | 0.00092 | 0.18 | 7 / 36 (19%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 8.1e-06 | 0 | 1.9e-05 | 4.9e-05 | 0 | 0.00014 | 2.6 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 2.5.1.76 | Cysteate synthase. | 0.63 | 0.83 | 1 | 0.13 | 1 | 1 | 0.43 | 0.93 | 1 | 0.84 | 0.94 | 1 | 0.22 | 0.88 | 1 | 0.93 | 0.98 | 1 | 0.00032 | 0.00012 | 0 | 0.00016 | 0 | 0.00022 | 8.9e-05 | 0 | 0.00017 | -0.85 | 13 / 36 (36%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 9.7e-05 | 0 | 0.00018 | 0.00013 | 0 | 0.00035 | 0.42 | 5 / 11 (45%) | 3 / 11 (27%) | 0.455 | 0.273 |
| 1.2.4.- | 0.85 | 0.94 | 1 | 0.63 | 1 | 1 | 0.71 | 0.98 | 1 | 0.81 | 0.93 | 1 | 0.61 | 0.95 | 1 | 0.9 | 0.98 | 1 | 0.00069 | 0.00061 | 0.00047 | 0.00057 | 0.00045 | 0.00057 | 0.00071 | 0.00071 | 0.00061 | 0.32 | 32 / 36 (89%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00062 | 0.00056 | 0.00063 | 0.00057 | 0.00029 | 0.00057 | -0.12 | 9 / 11 (82%) | 11 / 11 (100%) | 0.818 | 1 | |
| 2.3.1.n5 | Glycerol-3-phosphate acyltransferase (acyl-[acyl-carrier-protein]- | 0.22 | 0.54 | 1 | 0.36 | 1 | 1 | 0.46 | 0.93 | 1 | 0.32 | 0.59 | 1 | 0.95 | 0.99 | 1 | 0.88 | 0.97 | 1 | 2e-04 | 2.2e-05 | 0 | 6.7e-05 | 0 | 0.00018 | 3.9e-05 | 0 | 1e-04 | -0.78 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 2.7e-06 | 0 | 8.9e-06 | 0.054 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.1.1.45 | Carboxymethylenebutenolidase. | 0.24 | 0.55 | 1 | 0.53 | 1 | 1 | 0.86 | 1 | 1 | 0.35 | 0.62 | 1 | 0.53 | 0.93 | 1 | 0.72 | 0.94 | 1 | 0.00046 | 0.00011 | 0 | 0.00026 | 0 | 0.00062 | 0.00027 | 0 | 0.00072 | 0.054 | 9 / 36 (25%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 2.2e-05 | 0 | 4.1e-05 | 1.3e-05 | 0 | 4.5e-05 | -0.76 | 3 / 11 (27%) | 1 / 11 (9.1%) | 0.273 | 0.0909 |
| 1.1.1.363 | Glucose-6-phosphate dehydrogenase (NAD(P)(+)). | 0.27 | 0.56 | 1 | 1 | 1 | 1 | 0.63 | 0.96 | 1 | 0.5 | 0.74 | 1 | 0.81 | 0.98 | 1 | 0.84 | 0.96 | 1 | 7.9e-05 | 6.6e-06 | 0 | 1.2e-05 | 0 | 3.1e-05 | 1.2e-05 | 0 | 3.1e-05 | 0 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 6.8e-06 | 0 | 2.2e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 3.1.1.17 | Gluconolactonase. | 0.65 | 0.84 | 1 | 0.26 | 1 | 1 | 0.56 | 0.95 | 1 | 0.55 | 0.78 | 1 | 0.32 | 0.9 | 1 | 0.7 | 0.94 | 1 | 0.00015 | 2.8e-05 | 0 | 2.2e-05 | 0 | 5.2e-05 | 0 | 0 | 0 | -Inf | 7 / 36 (19%) | 2 / 7 (29%) | 0 / 7 (0%) | 0.286 | 0 | 4.1e-05 | 0 | 7.5e-05 | 3.7e-05 | 0 | 9.2e-05 | -0.15 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 4.4.1.28 | L-cysteine desulfidase. | 0.16 | 0.5 | 1 | 0.26 | 1 | 1 | 0.75 | 0.99 | 1 | 0.13 | 0.47 | 1 | 0.5 | 0.93 | 1 | 0.9 | 0.98 | 1 | 0.00031 | 3.5e-05 | 0 | 8.9e-05 | 0 | 0.00022 | 8.7e-05 | 0 | 0.00023 | -0.033 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 6.3.1.8 | Glutathionylspermidine synthase. | 0.035 | 0.49 | 1 | 0.031 | 0.97 | 1 | 0.01 | 0.61 | 1 | 0.0078 | 0.38 | 1 | 0.019 | 0.76 | 1 | 0.012 | 0.55 | 1 | 0.00067 | 0.00041 | 8.6e-05 | 0.00072 | 0.00024 | 0.00098 | 0.00043 | 0 | 0.00091 | -0.74 | 22 / 36 (61%) | 7 / 7 (100%) | 3 / 7 (43%) | 1 | 0.429 | 0.00015 | 0 | 0.00028 | 0.00046 | 7.5e-05 | 0.00085 | 1.6 | 5 / 11 (45%) | 7 / 11 (64%) | 0.455 | 0.636 |
| 3.5.2.2 | Dihydropyrimidinase. | 0.9 | 0.96 | 1 | 0.44 | 1 | 1 | 0.13 | 0.82 | 1 | 0.78 | 0.92 | 1 | 0.43 | 0.91 | 1 | 0.12 | 0.66 | 1 | 0.0041 | 0.0041 | 0.0036 | 0.0043 | 0.0036 | 0.002 | 0.0051 | 0.0039 | 0.0033 | 0.25 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0041 | 0.0038 | 0.0017 | 0.0033 | 0.0031 | 0.0013 | -0.31 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.7.53 | ATP adenylyltransferase. | 0.62 | 0.82 | 1 | 0.63 | 1 | 1 | 0.91 | 1 | 1 | 0.42 | 0.68 | 1 | 1 | 1 | 1 | 0.48 | 0.87 | 1 | 0.00047 | 0.00033 | 0.00029 | 0.00032 | 0.00031 | 0.00034 | 0.00026 | 0.00034 | 0.00026 | -0.3 | 25 / 36 (69%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.00038 | 0.00026 | 0.00049 | 0.00031 | 0.00028 | 0.00028 | -0.29 | 9 / 11 (82%) | 8 / 11 (73%) | 0.818 | 0.727 |
| 3.5.1.44 | Protein-glutamine glutaminase. | 0.033 | 0.49 | 1 | 0.78 | 1 | 1 | 0.53 | 0.95 | 1 | 0.04 | 0.43 | 1 | 0.93 | 0.99 | 1 | 0.51 | 0.89 | 1 | 0.0018 | 0.0018 | 0.0014 | 0.0023 | 0.0028 | 0.00088 | 0.0022 | 0.0016 | 0.0014 | -0.064 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0013 | 0.00072 | 0.0014 | 0.0015 | 0.0012 | 0.0012 | 0.21 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.1.86 | Diaminobutyrate decarboxylase. | 0.12 | 0.5 | 1 | 0.0094 | 0.72 | 1 | 0.016 | 0.67 | 1 | 0.19 | 0.5 | 1 | 0.0074 | 0.71 | 1 | 0.019 | 0.55 | 1 | 0.00025 | 9.1e-05 | 0 | 0.00022 | 5.7e-05 | 0.00043 | 7.7e-05 | 0 | 2e-04 | -1.5 | 13 / 36 (36%) | 4 / 7 (57%) | 1 / 7 (14%) | 0.571 | 0.143 | 3.6e-05 | 0 | 7e-05 | 7.1e-05 | 0 | 0.00017 | 0.98 | 4 / 11 (36%) | 4 / 11 (36%) | 0.364 | 0.364 |
| 2.6.1.- | 0.047 | 0.49 | 1 | 0.89 | 1 | 1 | 0.52 | 0.95 | 1 | 0.016 | 0.42 | 1 | 0.19 | 0.87 | 1 | 0.089 | 0.66 | 1 | 0.083 | 0.083 | 0.076 | 0.071 | 0.07 | 0.0093 | 0.072 | 0.067 | 0.013 | 0.02 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.094 | 0.075 | 0.043 | 0.086 | 0.082 | 0.016 | -0.13 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 2.7.8.35 | UDP-N-acetylglucosamine--decaprenyl-phosphate | 0.082 | 0.49 | 1 | 0.69 | 1 | 1 | 0.36 | 0.9 | 1 | 0.084 | 0.44 | 1 | 0.41 | 0.9 | 1 | 0.17 | 0.71 | 1 | 0.0031 | 0.0026 | 0.0013 | 0.0011 | 0.00077 | 0.0014 | 0.0014 | 0.00085 | 0.0018 | 0.35 | 30 / 36 (83%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.0042 | 0.0017 | 0.0071 | 0.0026 | 0.0019 | 0.0024 | -0.69 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 5.1.1.20 | L-Ala-D/L-Glu epimerase. | 0.005 | 0.49 | 1 | 0.086 | 1 | 1 | 0.0045 | 0.57 | 1 | 0.0013 | 0.31 | 1 | 0.14 | 0.87 | 1 | 0.007 | 0.52 | 1 | 0.00026 | 0.00013 | 2.9e-05 | 0.00024 | 0.00028 | 2e-04 | 0.00011 | 9.3e-05 | 0.00013 | -1.1 | 19 / 36 (53%) | 6 / 7 (86%) | 4 / 7 (57%) | 0.857 | 0.571 | 2.6e-05 | 0 | 7.9e-05 | 0.00019 | 7.1e-05 | 0.00039 | 2.9 | 2 / 11 (18%) | 7 / 11 (64%) | 0.182 | 0.636 |
| 1.1.1.205 | IMP dehydrogenase. | 0.12 | 0.5 | 1 | 0.47 | 1 | 1 | 0.94 | 1 | 1 | 0.12 | 0.47 | 1 | 0.85 | 0.98 | 1 | 0.43 | 0.85 | 1 | 0.025 | 0.025 | 0.024 | 0.023 | 0.023 | 0.0042 | 0.022 | 0.024 | 0.0054 | -0.064 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.028 | 0.027 | 0.0089 | 0.026 | 0.027 | 0.0041 | -0.11 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.1.19 | Nicotinamidase. | 0.24 | 0.55 | 1 | 0.65 | 1 | 1 | 0.33 | 0.89 | 1 | 0.2 | 0.5 | 1 | 0.48 | 0.92 | 1 | 0.33 | 0.8 | 1 | 0.0015 | 0.0014 | 0.00079 | 0.00069 | 0.00071 | 0.00059 | 0.00088 | 0.0012 | 0.00057 | 0.35 | 34 / 36 (94%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.0024 | 0.00086 | 0.0054 | 0.0011 | 0.00045 | 0.0013 | -1.1 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.6.1.37 | 2-aminoethylphosphonate--pyruvate transaminase. | 0.19 | 0.53 | 1 | 0.82 | 1 | 1 | 0.76 | 0.99 | 1 | 0.26 | 0.56 | 1 | 0.83 | 0.98 | 1 | 0.76 | 0.95 | 1 | 0.0011 | 0.0011 | 0.00081 | 0.0013 | 0.00084 | 9e-04 | 0.0014 | 0.0013 | 0.00091 | 0.11 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00072 | 0.00042 | 0.00061 | 0.0012 | 7e-04 | 0.0016 | 0.74 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 2.6.1.81 | Succinylornithine transaminase. | 0.66 | 0.84 | 1 | 0.88 | 1 | 1 | 0.96 | 1 | 1 | 0.44 | 0.69 | 1 | 0.68 | 0.95 | 1 | 0.51 | 0.89 | 1 | 0.00037 | 0.00012 | 0 | 0.00012 | 0 | 0.00031 | 8.5e-05 | 0 | 2e-04 | -0.5 | 12 / 36 (33%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0.00013 | 0 | 0.00021 | 0.00014 | 0 | 0.00027 | 0.11 | 4 / 11 (36%) | 4 / 11 (36%) | 0.364 | 0.364 |
| 1.1.1.4 | (R.R)-butanediol dehydrogenase. | 0.38 | 0.65 | 1 | 0.61 | 1 | 1 | 0.35 | 0.9 | 1 | 0.31 | 0.59 | 1 | 0.45 | 0.92 | 1 | 0.28 | 0.78 | 1 | 0.00023 | 0.00013 | 6.5e-05 | 0.00011 | 0 | 0.00022 | 0.00013 | 0.00011 | 0.00014 | 0.24 | 20 / 36 (56%) | 3 / 7 (43%) | 4 / 7 (57%) | 0.429 | 0.571 | 0.00017 | 0.00012 | 2e-04 | 9.2e-05 | 5.9e-05 | 0.00012 | -0.89 | 7 / 11 (64%) | 6 / 11 (55%) | 0.636 | 0.545 |
| 4.3.1.18 | D-serine ammonia-lyase. | 0.048 | 0.49 | 1 | 0.41 | 1 | 1 | 0.91 | 1 | 1 | 0.064 | 0.43 | 1 | 0.13 | 0.87 | 1 | 0.51 | 0.89 | 1 | 0.0011 | 0.0011 | 0.0011 | 0.00086 | 0.00079 | 0.00079 | 0.00073 | 0.00059 | 0.00064 | -0.24 | 34 / 36 (94%) | 7 / 7 (100%) | 5 / 7 (71%) | 1 | 0.714 | 0.0014 | 0.0011 | 0.00065 | 0.0011 | 0.0011 | 0.00061 | -0.35 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.7.1.17 | FMN-dependent NADH-azoreductase. | 0.51 | 0.74 | 1 | 0.56 | 1 | 1 | 0.85 | 1 | 1 | 0.39 | 0.66 | 1 | 0.49 | 0.92 | 1 | 0.97 | 0.99 | 1 | 0.00017 | 8e-05 | 0 | 1e-04 | 8.1e-05 | 0.00011 | 9.6e-05 | 0 | 0.00015 | -0.059 | 17 / 36 (47%) | 5 / 7 (71%) | 3 / 7 (43%) | 0.714 | 0.429 | 1e-04 | 0 | 0.00018 | 3.3e-05 | 0 | 6.9e-05 | -1.6 | 5 / 11 (45%) | 4 / 11 (36%) | 0.455 | 0.364 |
| 1.1.1.34 | Hydroxymethylglutaryl-CoA reductase (NADPH). | 0.76 | 0.89 | 1 | 0.8 | 1 | 1 | 0.23 | 0.88 | 1 | 0.98 | 0.99 | 1 | 0.96 | 0.99 | 1 | 0.21 | 0.74 | 1 | 0.00032 | 0.00019 | 1e-04 | 0.00019 | 0.00016 | 0.00017 | 0.00016 | 0.00014 | 0.00014 | -0.25 | 22 / 36 (61%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.00033 | 0.00011 | 0.00051 | 8.7e-05 | 0 | 0.00018 | -1.9 | 7 / 11 (64%) | 5 / 11 (45%) | 0.636 | 0.455 |
| 1.17.1.- | 0.35 | 0.63 | 1 | 0.63 | 1 | 1 | 0.94 | 1 | 1 | 0.2 | 0.5 | 1 | 0.53 | 0.93 | 1 | 0.46 | 0.86 | 1 | 0.0025 | 0.0025 | 0.0022 | 0.0024 | 0.0018 | 0.0022 | 0.0019 | 0.0019 | 0.0012 | -0.34 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0028 | 0.0023 | 0.0015 | 0.0025 | 0.0023 | 0.0011 | -0.16 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 1.1.1.82 | Malate dehydrogenase (NADP(+)). | 0.14 | 0.5 | 1 | 0.25 | 1 | 1 | 0.14 | 0.83 | 1 | 0.28 | 0.57 | 1 | 0.52 | 0.93 | 1 | 0.28 | 0.78 | 1 | 0.00015 | 1.3e-05 | 0 | 4.1e-05 | 0 | 0.00011 | 4.4e-06 | 0 | 1.2e-05 | -3.2 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 1.3e-05 | 0 | 4.3e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 6.2.1.20 | Long-chain-fatty-acid--[acyl-carrier-protein] ligase. | 0.28 | 0.56 | 1 | 0.82 | 1 | 1 | 0.68 | 0.98 | 1 | 0.44 | 0.69 | 1 | 0.84 | 0.98 | 1 | 0.92 | 0.98 | 1 | 0.00034 | 8.6e-05 | 0 | 0.00021 | 0 | 0.00053 | 0.00018 | 0 | 0.00046 | -0.22 | 9 / 36 (25%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 1.8e-05 | 0 | 4.9e-05 | 1.8e-05 | 0 | 3.7e-05 | 0 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 2.4.1.352 | Glucosylglycerate phosphorylase. | 0.22 | 0.54 | 1 | 0.52 | 1 | 1 | 0.82 | 1 | 1 | 0.27 | 0.56 | 1 | 0.4 | 0.9 | 1 | 0.78 | 0.95 | 1 | 0.00051 | 7e-05 | 0 | 0.00015 | 0 | 0.00031 | 0.00018 | 0 | 0.00049 | 0.26 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 1.7e-05 | 0 | 4e-05 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 4.2.1.45 | CDP-glucose 4.6-dehydratase. | 0.36 | 0.63 | 1 | 0.83 | 1 | 1 | 0.99 | 1 | 1 | 0.35 | 0.62 | 1 | 0.95 | 0.99 | 1 | 0.92 | 0.98 | 1 | 0.0021 | 0.0021 | 0.0018 | 0.0018 | 0.0017 | 0.0012 | 0.0017 | 0.0018 | 0.0012 | -0.082 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0025 | 0.0023 | 0.0017 | 0.0023 | 0.002 | 0.0013 | -0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.8.1.2 | (S)-2-haloacid dehalogenase. | 0.4 | 0.66 | 1 | 0.82 | 1 | 1 | 0.49 | 0.94 | 1 | 0.27 | 0.56 | 1 | 0.34 | 0.9 | 1 | 0.27 | 0.77 | 1 | 0.0044 | 0.0044 | 0.0033 | 0.0036 | 0.0019 | 0.0025 | 0.0037 | 0.0035 | 0.0023 | 0.04 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0057 | 0.0029 | 0.0077 | 0.0041 | 0.0035 | 0.0026 | -0.48 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.8.-.- | 0.89 | 0.96 | 1 | 0.64 | 1 | 1 | 0.48 | 0.93 | 1 | 0.72 | 0.89 | 1 | 0.46 | 0.92 | 1 | 0.42 | 0.85 | 1 | 0.0014 | 0.0014 | 0.0011 | 0.0012 | 0.0012 | 0.00074 | 0.0012 | 0.00072 | 0.0012 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0013 | 0.0011 | 0.00092 | 0.0015 | 0.0011 | 0.0012 | 0.21 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 4.2.1.75 | Uroporphyrinogen-III synthase. | 0.32 | 0.6 | 1 | 0.44 | 1 | 1 | 0.49 | 0.95 | 1 | 0.64 | 0.85 | 1 | 0.24 | 0.88 | 1 | 0.76 | 0.95 | 1 | 0.0021 | 0.0021 | 0.0018 | 0.0023 | 0.0026 | 0.0014 | 0.0027 | 0.0021 | 0.0022 | 0.23 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0016 | 0.0018 | 0.00064 | 0.0022 | 0.0018 | 0.0011 | 0.46 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.1.3.4 | L-ribulose-5-phosphate 4-epimerase. | 0.26 | 0.56 | 1 | 0.029 | 0.97 | 1 | 0.065 | 0.72 | 1 | 0.15 | 0.47 | 1 | 0.013 | 0.76 | 1 | 0.025 | 0.58 | 1 | 0.012 | 0.012 | 0.011 | 0.01 | 0.0086 | 0.0034 | 0.013 | 0.014 | 0.007 | 0.38 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.012 | 0.011 | 0.0029 | 0.012 | 0.012 | 0.0017 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.2.1 | Acetyl-CoA hydrolase. | 0.22 | 0.54 | 1 | 1 | 1 | 1 | 0.3 | 0.89 | 1 | 0.12 | 0.47 | 1 | 0.97 | 0.99 | 1 | 0.58 | 0.91 | 1 | 0.001 | 7e-04 | 0.00015 | 0.00066 | 0.00032 | 0.00072 | 7e-04 | 0.00041 | 0.00088 | 0.085 | 25 / 36 (69%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00028 | 8.7e-05 | 0.00059 | 0.0011 | 0.00011 | 0.002 | 2 | 6 / 11 (55%) | 7 / 11 (64%) | 0.545 | 0.636 |
| 3.2.1.6 | Endo-1.3(4)-beta-glucanase. | 0.0099 | 0.49 | 1 | 0.0044 | 0.67 | 1 | 0.0014 | 0.51 | 1 | 0.0074 | 0.38 | 1 | 0.0067 | 0.7 | 1 | 0.0029 | 0.48 | 1 | 0.00016 | 5.4e-05 | 0 | 0.00011 | 8.1e-05 | 0.00012 | 4.5e-05 | 0 | 9.4e-05 | -1.3 | 12 / 36 (33%) | 6 / 7 (86%) | 2 / 7 (29%) | 0.857 | 0.286 | 1.8e-05 | 0 | 5.9e-05 | 6.2e-05 | 0 | 0.00014 | 1.8 | 1 / 11 (9.1%) | 3 / 11 (27%) | 0.0909 | 0.273 |
| 4.6.1.12 | 2-C-methyl-D-erythritol 2.4-cyclodiphosphate synthase. | 0.016 | 0.49 | 1 | 0.37 | 1 | 1 | 0.84 | 1 | 1 | 0.036 | 0.43 | 1 | 0.98 | 0.99 | 1 | 0.38 | 0.83 | 1 | 0.0099 | 0.0099 | 0.0097 | 0.0091 | 0.0091 | 0.0014 | 0.0084 | 0.0086 | 0.002 | -0.12 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.011 | 0.002 | 0.01 | 0.0097 | 0.0015 | -0.14 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.1.248 | Cycloisomaltooligosaccharide glucanotransferase. | 0.74 | 0.88 | 1 | 0.63 | 1 | 1 | 0.3 | 0.89 | 1 | 0.9 | 0.97 | 1 | 0.69 | 0.95 | 1 | 0.21 | 0.74 | 1 | 0.0012 | 0.0012 | 0.00093 | 0.0011 | 0.00071 | 0.0011 | 0.0013 | 0.00065 | 0.0013 | 0.24 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00096 | 0.00065 | 0.00084 | 0.0014 | 0.0016 | 0.00086 | 0.54 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.1.275 | Acyl phosphate:glycerol-3-phosphate acyltransferase. | 0.34 | 0.62 | 1 | 0.56 | 1 | 1 | 0.87 | 1 | 1 | 0.29 | 0.57 | 1 | 0.92 | 0.99 | 1 | 0.5 | 0.88 | 1 | 0.009 | 0.009 | 0.009 | 0.0085 | 0.0081 | 0.0011 | 0.0083 | 0.0079 | 0.0027 | -0.034 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0095 | 0.0093 | 0.0019 | 0.0092 | 0.0088 | 0.0021 | -0.046 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.14.13.239 | Carnitine monooxygenase. | 0.014 | 0.49 | 1 | 0.025 | 0.94 | 1 | 0.021 | 0.71 | 1 | 0.041 | 0.43 | 1 | 0.056 | 0.82 | 1 | 0.039 | 0.63 | 1 | 0.00012 | 1.3e-05 | 0 | 5.5e-05 | 0 | 9.8e-05 | 0 | 0 | 0 | -Inf | 4 / 36 (11%) | 2 / 7 (29%) | 0 / 7 (0%) | 0.286 | 0 | 0 | 0 | 0 | 9.2e-06 | 0 | 2.2e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 1.1.1.26 | Glyoxylate reductase. | 0.4 | 0.66 | 1 | 0.78 | 1 | 1 | 0.28 | 0.89 | 1 | 0.18 | 0.5 | 1 | 0.91 | 0.99 | 1 | 0.63 | 0.92 | 1 | 0.00045 | 0.00032 | 2e-04 | 0.00028 | 0.00022 | 0.00024 | 2e-04 | 0.00014 | 0.00016 | -0.49 | 26 / 36 (72%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.00023 | 8e-05 | 0.00043 | 0.00051 | 0.00021 | 0.00094 | 1.1 | 6 / 11 (55%) | 7 / 11 (64%) | 0.545 | 0.636 |
| 2.7.7.1 | Nicotinamide-nucleotide adenylyltransferase. | 0.88 | 0.95 | 1 | 0.98 | 1 | 1 | 0.4 | 0.91 | 1 | 0.95 | 0.98 | 1 | 0.83 | 0.98 | 1 | 0.5 | 0.88 | 1 | 0.00042 | 0.00032 | 0.00012 | 0.00046 | 0.00012 | 0.00094 | 4e-04 | 0.00016 | 0.00067 | -0.2 | 28 / 36 (78%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00036 | 0.00021 | 0.00052 | 0.00015 | 0.00011 | 0.00017 | -1.3 | 8 / 11 (73%) | 8 / 11 (73%) | 0.727 | 0.727 |
| 5.3.1.29 | Ribose 1.5-bisphosphate isomerase. | 0.11 | 0.5 | 1 | 0.12 | 1 | 1 | 0.017 | 0.69 | 1 | 0.068 | 0.43 | 1 | 0.092 | 0.86 | 1 | 0.014 | 0.55 | 1 | 0.00033 | 0.00016 | 0 | 0.00022 | 0.00011 | 0.00027 | 0.00012 | 0 | 0.00023 | -0.87 | 17 / 36 (47%) | 5 / 7 (71%) | 3 / 7 (43%) | 0.714 | 0.429 | 6.1e-05 | 0 | 0.00015 | 0.00024 | 2.9e-05 | 0.00057 | 2 | 3 / 11 (27%) | 6 / 11 (55%) | 0.273 | 0.545 |
| 3.4.11.1 | Leucyl aminopeptidase. | 0.15 | 0.5 | 1 | 0.86 | 1 | 1 | 0.61 | 0.96 | 1 | 0.26 | 0.55 | 1 | 0.98 | 0.99 | 1 | 0.32 | 0.79 | 1 | 0.004 | 0.0039 | 0.0022 | 0.0021 | 0.0016 | 0.0017 | 0.0019 | 0.0015 | 0.0015 | -0.14 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0062 | 0.0029 | 0.01 | 0.004 | 0.0024 | 0.0042 | -0.63 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 7.3.2.2 | ABC-type phosphonate transporter. | 0.82 | 0.92 | 1 | 0.78 | 1 | 1 | 0.33 | 0.89 | 1 | 0.75 | 0.9 | 1 | 0.85 | 0.98 | 1 | 0.12 | 0.66 | 1 | 0.00064 | 0.00064 | 0.00059 | 0.00067 | 6e-04 | 0.00039 | 0.00065 | 0.00059 | 0.00045 | -0.044 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00074 | 0.00077 | 0.00041 | 0.00053 | 0.00028 | 0.00047 | -0.48 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.23 | Beta-galactosidase. | 0.46 | 0.71 | 1 | 0.95 | 1 | 1 | 0.26 | 0.89 | 1 | 0.2 | 0.5 | 1 | 0.28 | 0.89 | 1 | 0.84 | 0.96 | 1 | 0.17 | 0.17 | 0.15 | 0.15 | 0.14 | 0.042 | 0.15 | 0.15 | 0.044 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.17 | 0.15 | 0.056 | 0.19 | 0.17 | 0.063 | 0.16 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.18.1.2 | Ferredoxin--NADP(+) reductase. | 0.23 | 0.54 | 1 | 0.81 | 1 | 1 | 0.81 | 1 | 1 | 0.12 | 0.47 | 1 | 0.92 | 0.99 | 1 | 1 | 1 | 1 | 0.01 | 0.01 | 0.0096 | 0.0085 | 0.0073 | 0.0034 | 0.0083 | 0.0076 | 0.0041 | -0.034 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.012 | 0.0087 | 0.0081 | 0.011 | 0.011 | 0.004 | -0.13 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.7.1.- | 0.17 | 0.5 | 1 | 0.38 | 1 | 1 | 0.17 | 0.85 | 1 | 0.17 | 0.49 | 1 | 0.7 | 0.95 | 1 | 0.72 | 0.94 | 1 | 0.00043 | 3.6e-05 | 0 | 1e-04 | 0 | 0.00027 | 7.7e-05 | 0 | 2e-04 | -0.38 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 | |
| 3.6.1.- | 0.37 | 0.65 | 1 | 0.88 | 1 | 1 | 0.72 | 0.98 | 1 | 0.082 | 0.44 | 1 | 0.4 | 0.9 | 1 | 0.99 | 1 | 1 | 0.077 | 0.077 | 0.071 | 0.065 | 0.066 | 0.016 | 0.068 | 0.062 | 0.024 | 0.065 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.082 | 0.06 | 0.057 | 0.084 | 0.085 | 0.024 | 0.035 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 2.7.-.- | 0.2 | 0.53 | 1 | 0.99 | 1 | 1 | 0.3 | 0.89 | 1 | 0.61 | 0.82 | 1 | 0.35 | 0.9 | 1 | 0.92 | 0.98 | 1 | 0.03 | 0.03 | 0.028 | 0.031 | 0.032 | 0.0069 | 0.032 | 0.038 | 0.009 | 0.046 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.027 | 0.022 | 0.0084 | 0.03 | 0.028 | 0.0074 | 0.15 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 2.4.99.12 | Lipid IV(A) 3-deoxy-D-manno-octulosonic acid transferase. | 0.098 | 0.49 | 1 | 0.27 | 1 | 1 | 0.01 | 0.61 | 1 | 0.045 | 0.43 | 1 | 0.21 | 0.88 | 1 | 0.019 | 0.55 | 1 | 0.0019 | 0.0016 | 0.00096 | 0.0019 | 0.0013 | 0.0012 | 0.0017 | 0.00096 | 0.0018 | -0.16 | 31 / 36 (86%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00096 | 0.00042 | 0.0013 | 0.0021 | 0.00071 | 0.0024 | 1.1 | 8 / 11 (73%) | 10 / 11 (91%) | 0.727 | 0.909 |
| 3.5.3.9 | Allantoate deiminase. | 0.96 | 0.98 | 1 | 0.45 | 1 | 1 | 0.6 | 0.96 | 1 | 0.38 | 0.65 | 1 | 0.53 | 0.93 | 1 | 0.53 | 0.9 | 1 | 0.00059 | 0.00053 | 0.00044 | 0.00051 | 0.00057 | 0.00039 | 0.00083 | 0.00054 | 0.0012 | 0.7 | 32 / 36 (89%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.00041 | 0.00039 | 0.00027 | 0.00046 | 0.00037 | 0.00045 | 0.17 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.3.2.1 | Isochorismatase. | 0.054 | 0.49 | 1 | 0.56 | 1 | 1 | 0.38 | 0.9 | 1 | 0.11 | 0.47 | 1 | 0.54 | 0.93 | 1 | 0.27 | 0.77 | 1 | 0.00026 | 0.00016 | 5.2e-05 | 0.00031 | 0.00022 | 0.00036 | 0.00031 | 0.00013 | 0.00042 | 0 | 22 / 36 (61%) | 7 / 7 (100%) | 5 / 7 (71%) | 1 | 0.714 | 9.4e-05 | 7.2e-05 | 9.8e-05 | 2.7e-05 | 0 | 7e-05 | -1.8 | 8 / 11 (73%) | 2 / 11 (18%) | 0.727 | 0.182 |
| 2.7.1.76 | Deoxyadenosine kinase. | 0.18 | 0.52 | 1 | 0.81 | 1 | 1 | 0.86 | 1 | 1 | 0.093 | 0.45 | 1 | 0.95 | 0.99 | 1 | 0.53 | 0.9 | 1 | 0.00014 | 4.3e-05 | 0 | 2e-05 | 0 | 5.2e-05 | 7.7e-06 | 0 | 2e-05 | -1.4 | 11 / 36 (31%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 5.3e-05 | 3.3e-05 | 6e-05 | 7.1e-05 | 0 | 0.00017 | 0.42 | 6 / 11 (55%) | 3 / 11 (27%) | 0.545 | 0.273 |
| 1.14.11.33 | DNA oxidative demethylase. | 0.36 | 0.63 | 1 | 0.63 | 1 | 1 | 0.98 | 1 | 1 | 0.43 | 0.69 | 1 | 0.55 | 0.93 | 1 | 0.91 | 0.98 | 1 | 0.00011 | 9.5e-06 | 0 | 1.2e-05 | 0 | 3.1e-05 | 2.9e-05 | 0 | 7.7e-05 | 1.3 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 6.2.1.35 | Acetate--[acyl-carrier protein] ligase. | 0.49 | 0.73 | 1 | 0.41 | 1 | 1 | 0.79 | 1 | 1 | 0.46 | 0.71 | 1 | 0.49 | 0.92 | 1 | 0.99 | 1 | 1 | 0.00022 | 6e-05 | 0 | 3.7e-05 | 0 | 6.9e-05 | 8.8e-06 | 0 | 2.3e-05 | -2.1 | 10 / 36 (28%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 8.4e-05 | 0 | 0.00013 | 8.3e-05 | 0 | 2e-04 | -0.017 | 4 / 11 (36%) | 3 / 11 (27%) | 0.364 | 0.273 |
| 1.1.1.87 | Homoisocitrate dehydrogenase. | 0.43 | 0.69 | 1 | 0.064 | 1 | 1 | 0.028 | 0.71 | 1 | 0.34 | 0.61 | 1 | 0.043 | 0.8 | 1 | 0.014 | 0.55 | 1 | 8.3e-05 | 1.6e-05 | 0 | 4.6e-06 | 0 | 1.2e-05 | 4.2e-05 | 0 | 7.6e-05 | 3.2 | 7 / 36 (19%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 1.7e-05 | 0 | 3.3e-05 | 5.3e-06 | 0 | 1.8e-05 | -1.7 | 3 / 11 (27%) | 1 / 11 (9.1%) | 0.273 | 0.0909 |
| 1.1.5.6 | Transferred entry: 1.17.5.3. | 0.33 | 0.6 | 1 | 0.94 | 1 | 1 | 0.5 | 0.95 | 1 | 0.36 | 0.63 | 1 | 0.99 | 1 | 1 | 0.6 | 0.91 | 1 | 8.5e-05 | 1.4e-05 | 0 | 1.9e-05 | 0 | 4.4e-05 | 2.9e-05 | 0 | 7.7e-05 | 0.61 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1.3e-05 | 0 | 3.5e-05 | 2.3 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 1.1.1.9 | D-xylulose reductase. | 0.38 | 0.65 | 1 | 0.81 | 1 | 1 | 0.98 | 1 | 1 | 0.31 | 0.59 | 1 | 0.92 | 0.99 | 1 | 0.92 | 0.98 | 1 | 0.0027 | 0.0024 | 0.0011 | 0.0013 | 0.00068 | 0.0015 | 0.0016 | 0.00084 | 0.0017 | 0.3 | 31 / 36 (86%) | 6 / 7 (86%) | 5 / 7 (71%) | 0.857 | 0.714 | 0.0032 | 0.00087 | 0.0069 | 0.0027 | 0.0014 | 0.0027 | -0.25 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 6.3.3.- | 0.94 | 0.97 | 1 | 0.75 | 1 | 1 | 0.34 | 0.89 | 1 | 0.81 | 0.93 | 1 | 0.91 | 0.99 | 1 | 0.18 | 0.71 | 1 | 0.00011 | 1.2e-05 | 0 | 2.3e-05 | 0 | 6.1e-05 | 1.5e-05 | 0 | 4e-05 | -0.62 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 1.6e-05 | 0 | 3.7e-05 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 | |
| 1.1.1.287 | D-arabinitol dehydrogenase (NADP(+)). | 0.95 | 0.98 | 1 | 0.36 | 1 | 1 | 0.55 | 0.95 | 1 | 0.87 | 0.95 | 1 | 0.52 | 0.93 | 1 | 0.91 | 0.98 | 1 | 0.00068 | 0.00047 | 0.00034 | 0.00038 | 0.00033 | 0.00036 | 0.00055 | 0.00064 | 0.00044 | 0.53 | 25 / 36 (69%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.00051 | 0.00021 | 7e-04 | 0.00043 | 0.00053 | 0.00037 | -0.25 | 7 / 11 (64%) | 8 / 11 (73%) | 0.636 | 0.727 |
| 4.2.3.3 | Methylglyoxal synthase. | 0.65 | 0.84 | 1 | 0.89 | 1 | 1 | 0.46 | 0.93 | 1 | 0.98 | 0.99 | 1 | 0.4 | 0.9 | 1 | 0.88 | 0.97 | 1 | 0.0084 | 0.0084 | 0.008 | 0.0082 | 0.0078 | 0.0016 | 0.0083 | 0.0082 | 0.0013 | 0.017 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0079 | 0.0079 | 0.0029 | 0.009 | 0.0083 | 0.0025 | 0.19 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.19.- | 0.75 | 0.88 | 1 | 0.78 | 1 | 1 | 0.56 | 0.95 | 1 | 0.6 | 0.82 | 1 | 0.85 | 0.98 | 1 | 0.27 | 0.77 | 1 | 0.0098 | 0.0098 | 0.0093 | 0.0096 | 0.0093 | 0.0031 | 0.0095 | 0.01 | 0.0044 | -0.015 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.01 | 0.0053 | 0.0093 | 0.0087 | 0.0047 | -0.24 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 4.1.1.35 | UDP-glucuronate decarboxylase. | 0.79 | 0.9 | 1 | 0.21 | 1 | 1 | 0.25 | 0.89 | 1 | 0.75 | 0.9 | 1 | 0.24 | 0.88 | 1 | 0.32 | 0.79 | 1 | 0.00099 | 0.00039 | 0 | 0.00013 | 3.2e-05 | 0.00018 | 3e-05 | 0 | 5.9e-05 | -2.1 | 14 / 36 (39%) | 4 / 7 (57%) | 2 / 7 (29%) | 0.571 | 0.286 | 0.00057 | 0 | 0.0017 | 0.00059 | 0 | 0.0016 | 0.05 | 4 / 11 (36%) | 4 / 11 (36%) | 0.364 | 0.364 |
| 1.8.4.- | 0.33 | 0.6 | 1 | 0.22 | 1 | 1 | 0.45 | 0.93 | 1 | 0.61 | 0.82 | 1 | 0.098 | 0.86 | 1 | 0.23 | 0.75 | 1 | 0.00038 | 3e-04 | 0.00013 | 0.00042 | 0.00014 | 0.00062 | 0.00048 | 0.00036 | 4e-04 | 0.19 | 28 / 36 (78%) | 5 / 7 (71%) | 7 / 7 (100%) | 0.714 | 1 | 0.00018 | 9.7e-05 | 0.00025 | 0.00022 | 0.00011 | 3e-04 | 0.29 | 8 / 11 (73%) | 8 / 11 (73%) | 0.727 | 0.727 | |
| 3.4.13.20 | Beta-Ala-His dipeptidase. | 0.47 | 0.71 | 1 | 0.6 | 1 | 1 | 0.35 | 0.9 | 1 | 0.41 | 0.68 | 1 | 0.62 | 0.95 | 1 | 0.29 | 0.78 | 1 | 0.0039 | 0.0039 | 0.0029 | 0.0028 | 0.0026 | 0.0016 | 0.0026 | 0.0031 | 0.0017 | -0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0046 | 0.0022 | 0.0065 | 0.0047 | 0.0035 | 0.0039 | 0.031 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.1.44 | 4-carboxymuconolactone decarboxylase. | 0.24 | 0.55 | 1 | 0.024 | 0.92 | 1 | 0.25 | 0.89 | 1 | 0.3 | 0.58 | 1 | 0.025 | 0.76 | 1 | 0.23 | 0.75 | 1 | 1e-04 | 3.4e-05 | 0 | 3e-05 | 0 | 5.2e-05 | 9.1e-05 | 0.00011 | 8.7e-05 | 1.6 | 12 / 36 (33%) | 3 / 7 (43%) | 5 / 7 (71%) | 0.429 | 0.714 | 8.8e-06 | 0 | 2.9e-05 | 2.7e-05 | 0 | 5.3e-05 | 1.6 | 1 / 11 (9.1%) | 3 / 11 (27%) | 0.0909 | 0.273 |
| 3.6.3.36 | Transferred entry: 7.6.2.7. | 0.56 | 0.78 | 1 | 0.26 | 1 | 1 | 0.79 | 1 | 1 | 0.78 | 0.92 | 1 | 0.24 | 0.88 | 1 | 0.3 | 0.78 | 1 | 0.00083 | 0.00058 | 0.00024 | 0.00052 | 0.00011 | 8e-04 | 0.00022 | 2.7e-05 | 0.00035 | -1.2 | 25 / 36 (69%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 0.00098 | 0.00029 | 0.0017 | 0.00044 | 3e-04 | 0.00064 | -1.2 | 7 / 11 (64%) | 9 / 11 (82%) | 0.636 | 0.818 |
| 2.1.3.6 | Putrescine carbamoyltransferase. | 0.088 | 0.49 | 1 | 0.56 | 1 | 1 | 0.26 | 0.89 | 1 | 0.11 | 0.47 | 1 | 0.68 | 0.95 | 1 | 0.37 | 0.82 | 1 | 0.00039 | 0.00025 | 0.00012 | 9.4e-05 | 4.8e-05 | 0.00015 | 0.00021 | 5.4e-05 | 0.00043 | 1.2 | 23 / 36 (64%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.00038 | 2e-04 | 0.00039 | 0.00025 | 0.00014 | 0.00031 | -0.6 | 8 / 11 (73%) | 7 / 11 (64%) | 0.727 | 0.636 |
| 1.14.99.46 | Pyrimidine monooxygenase. | 0.12 | 0.5 | 1 | 0.29 | 1 | 1 | 0.12 | 0.82 | 1 | 0.095 | 0.45 | 1 | 0.66 | 0.95 | 1 | 0.33 | 0.8 | 1 | 0.00024 | 2.6e-05 | 0 | 7e-05 | 0 | 0.00018 | 5.8e-05 | 0 | 0.00015 | -0.27 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 7.1.2.2 | H(+)-transporting two-sector ATPase. | 0.34 | 0.62 | 1 | 0.55 | 1 | 1 | 0.54 | 0.95 | 1 | 0.27 | 0.56 | 1 | 0.046 | 0.81 | 1 | 0.032 | 0.62 | 1 | 0.099 | 0.099 | 0.1 | 0.094 | 0.09 | 0.017 | 0.098 | 0.1 | 0.028 | 0.06 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.1 | 0.1 | 0.013 | 0.1 | 0.099 | 0.016 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.5.1.50 | Dihydromonapterin reductase. | 0.18 | 0.52 | 1 | 0.63 | 1 | 1 | 0.24 | 0.89 | 1 | 0.2 | 0.5 | 1 | 0.77 | 0.97 | 1 | 0.62 | 0.92 | 1 | 0.00023 | 1.9e-05 | 0 | 5e-05 | 0 | 0.00013 | 3.9e-05 | 0 | 1e-04 | -0.36 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 6.2.1.26 | o-succinylbenzoate--CoA ligase. | 0.6 | 0.81 | 1 | 0.22 | 1 | 1 | 0.15 | 0.83 | 1 | 0.71 | 0.89 | 1 | 0.17 | 0.87 | 1 | 0.34 | 0.8 | 1 | 0.0014 | 0.0013 | 0.00082 | 0.0014 | 0.00089 | 0.0013 | 8e-04 | 0.00071 | 0.00064 | -0.81 | 34 / 36 (94%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.0011 | 0.00078 | 0.0011 | 0.0019 | 0.00091 | 0.0026 | 0.79 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 1.7.99.1 | Hydroxylamine reductase. | 0.79 | 0.9 | 1 | 0.81 | 1 | 1 | 0.89 | 1 | 1 | 0.75 | 0.9 | 1 | 0.23 | 0.88 | 1 | 0.47 | 0.87 | 1 | 0.019 | 0.019 | 0.019 | 0.018 | 0.018 | 0.0028 | 0.019 | 0.02 | 0.0041 | 0.078 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.018 | 0.019 | 0.0048 | 0.019 | 0.019 | 0.0022 | 0.078 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.4.1.13 | Glutamate synthase (NADPH). | 0.21 | 0.53 | 1 | 0.78 | 1 | 1 | 0.73 | 0.99 | 1 | 0.18 | 0.5 | 1 | 0.33 | 0.9 | 1 | 0.4 | 0.83 | 1 | 0.1 | 0.1 | 0.11 | 0.095 | 0.09 | 0.015 | 0.093 | 0.099 | 0.02 | -0.031 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.11 | 0.11 | 0.026 | 0.11 | 0.11 | 0.012 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.1.1.7 | Alanine--tRNA ligase. | 0.043 | 0.49 | 1 | 0.63 | 1 | 1 | 0.3 | 0.89 | 1 | 0.13 | 0.47 | 1 | 0.42 | 0.91 | 1 | 0.048 | 0.65 | 1 | 0.046 | 0.046 | 0.046 | 0.043 | 0.043 | 0.0059 | 0.043 | 0.045 | 0.0093 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.05 | 0.052 | 0.0073 | 0.046 | 0.046 | 0.0049 | -0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.139 | Alpha-glucuronidase. | 0.44 | 0.69 | 1 | 0.96 | 1 | 1 | 0.24 | 0.89 | 1 | 0.48 | 0.73 | 1 | 0.82 | 0.98 | 1 | 0.6 | 0.91 | 1 | 0.0012 | 8e-04 | 0.00014 | 0.00079 | 0.00022 | 0.0012 | 0.00061 | 0.00095 | 0.00057 | -0.37 | 24 / 36 (67%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.00037 | 0.00014 | 0.00075 | 0.0014 | 1e-04 | 0.0022 | 1.9 | 6 / 11 (55%) | 8 / 11 (73%) | 0.545 | 0.727 |
| 2.5.1.3 | Thiamine phosphate synthase. | 0.37 | 0.64 | 1 | 0.87 | 1 | 1 | 0.55 | 0.95 | 1 | 0.22 | 0.52 | 1 | 0.19 | 0.87 | 1 | 0.75 | 0.95 | 1 | 0.017 | 0.017 | 0.017 | 0.015 | 0.012 | 0.0041 | 0.015 | 0.018 | 0.0042 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.018 | 0.016 | 0.0087 | 0.019 | 0.017 | 0.0047 | 0.078 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.1.84 | Cocaine esterase. | 0.67 | 0.84 | 1 | 0.73 | 1 | 1 | 1 | 1 | 1 | 0.5 | 0.74 | 1 | 0.82 | 0.98 | 1 | 0.97 | 0.99 | 1 | 0.0029 | 0.0029 | 0.0026 | 0.0025 | 0.0025 | 0.001 | 0.0031 | 0.0025 | 0.0024 | 0.31 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0029 | 0.0031 | 0.0014 | 0.0032 | 0.0027 | 0.0017 | 0.14 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.1.54 | Undecaprenyl-phosphate mannosyltransferase. | 0.55 | 0.77 | 1 | 0.72 | 1 | 1 | 0.43 | 0.93 | 1 | 0.25 | 0.55 | 1 | 0.86 | 0.99 | 1 | 0.75 | 0.95 | 1 | 0.00087 | 0.00072 | 3e-04 | 0.00063 | 0.00035 | 6e-04 | 5e-04 | 0.00025 | 0.00049 | -0.33 | 30 / 36 (83%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00057 | 0.00026 | 0.00084 | 0.0011 | 2e-04 | 0.0019 | 0.95 | 8 / 11 (73%) | 8 / 11 (73%) | 0.727 | 0.727 |
| 7.2.4.1 | Carboxybiotin decarboxylase. | 0.85 | 0.94 | 1 | 0.068 | 1 | 1 | 0.73 | 0.99 | 1 | 0.68 | 0.87 | 1 | 0.15 | 0.87 | 1 | 0.91 | 0.98 | 1 | 0.00027 | 8.3e-05 | 0 | 8.3e-05 | 0 | 0.00011 | 3.1e-05 | 0 | 4.8e-05 | -1.4 | 11 / 36 (31%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 0.00013 | 0 | 0.00035 | 6.6e-05 | 0 | 0.00017 | -0.98 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 5.1.3.9 | N-acylglucosamine-6-phosphate 2-epimerase. | 0.78 | 0.89 | 1 | 0.84 | 1 | 1 | 0.86 | 1 | 1 | 0.62 | 0.83 | 1 | 0.41 | 0.9 | 1 | 0.38 | 0.83 | 1 | 0.0049 | 0.0049 | 0.0043 | 0.0053 | 0.0037 | 0.0033 | 0.005 | 0.0041 | 0.0023 | -0.084 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0047 | 0.0042 | 0.0015 | 0.0048 | 0.0046 | 0.0016 | 0.03 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.1.105 | L-tryptophan decarboxylase. | 0.03 | 0.49 | 1 | 0.00043 | 0.34 | 0.99 | 0.007 | 0.61 | 1 | 0.034 | 0.43 | 1 | 0.0017 | 0.62 | 1 | 0.019 | 0.55 | 1 | 0.00027 | 6.8e-05 | 0 | 0.00022 | 5.7e-05 | 0.00043 | 7.7e-05 | 0 | 2e-04 | -1.5 | 9 / 36 (25%) | 4 / 7 (57%) | 1 / 7 (14%) | 0.571 | 0.143 | 2.3e-05 | 0 | 6.5e-05 | 9.4e-06 | 0 | 2.3e-05 | -1.3 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 1.5.1.2 | Pyrroline-5-carboxylate reductase. | 0.01 | 0.49 | 1 | 0.28 | 1 | 1 | 0.21 | 0.87 | 1 | 0.026 | 0.42 | 1 | 0.039 | 0.8 | 1 | 0.03 | 0.61 | 1 | 0.013 | 0.013 | 0.013 | 0.011 | 0.011 | 0.0017 | 0.012 | 0.013 | 0.0031 | 0.13 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.014 | 0.014 | 0.0041 | 0.014 | 0.014 | 0.0015 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.4.2.7 | Phosphopentomutase. | 0.77 | 0.89 | 1 | 0.49 | 1 | 1 | 0.77 | 0.99 | 1 | 0.85 | 0.95 | 1 | 0.93 | 0.99 | 1 | 0.23 | 0.74 | 1 | 0.013 | 0.013 | 0.013 | 0.014 | 0.013 | 0.0033 | 0.013 | 0.014 | 0.0036 | -0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.013 | 0.014 | 0.0039 | 0.012 | 0.012 | 0.0038 | -0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.5.-.- | 0.18 | 0.52 | 1 | 0.27 | 1 | 1 | 0.76 | 0.99 | 1 | 0.21 | 0.52 | 1 | 0.25 | 0.88 | 1 | 0.68 | 0.93 | 1 | 0.00034 | 7.5e-05 | 0 | 0.00015 | 0 | 0.00035 | 0.00019 | 0 | 0.00038 | 0.34 | 8 / 36 (22%) | 2 / 7 (29%) | 3 / 7 (43%) | 0.286 | 0.429 | 6e-06 | 0 | 2e-05 | 2.6e-05 | 0 | 5.9e-05 | 2.1 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 | |
| 4.2.2.5 | Chondroitin AC lyase. | 0.021 | 0.49 | 1 | 0.76 | 1 | 1 | 0.44 | 0.93 | 1 | 0.019 | 0.42 | 1 | 0.31 | 0.9 | 1 | 0.47 | 0.87 | 1 | 0.0017 | 0.001 | 1e-04 | 0.0018 | 0.00069 | 0.002 | 0.0022 | 0.00022 | 0.0037 | 0.29 | 21 / 36 (58%) | 6 / 7 (86%) | 5 / 7 (71%) | 0.857 | 0.714 | 0.00015 | 8.5e-05 | 0.00025 | 0.00064 | 0 | 0.0015 | 2.1 | 6 / 11 (55%) | 4 / 11 (36%) | 0.545 | 0.364 |
| 5.1.1.13 | Aspartate racemase. | 0.16 | 0.5 | 1 | 0.67 | 1 | 1 | 0.46 | 0.93 | 1 | 0.15 | 0.48 | 1 | 0.25 | 0.88 | 1 | 0.21 | 0.74 | 1 | 0.0022 | 0.0022 | 0.0016 | 0.0014 | 0.0011 | 0.0011 | 0.0016 | 0.0016 | 0.0011 | 0.19 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.003 | 0.0017 | 0.0041 | 0.0023 | 0.0022 | 0.0017 | -0.38 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.3.7.9 | 4-hydroxybenzoyl-CoA reductase. | 0.94 | 0.97 | 1 | 0.23 | 1 | 1 | 0.22 | 0.88 | 1 | 0.68 | 0.87 | 1 | 0.52 | 0.93 | 1 | 0.56 | 0.9 | 1 | 0.025 | 0.025 | 0.025 | 0.026 | 0.022 | 0.01 | 0.023 | 0.028 | 0.011 | -0.18 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.025 | 0.022 | 0.0069 | 0.026 | 0.025 | 0.0071 | 0.057 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.1.59 | 3-hydroxyacyl-[acyl-carrier-protein] dehydratase. | 0.34 | 0.61 | 1 | 0.26 | 1 | 1 | 0.93 | 1 | 1 | 0.087 | 0.45 | 1 | 0.054 | 0.81 | 1 | 0.52 | 0.89 | 1 | 0.0099 | 0.0099 | 0.0092 | 0.0083 | 0.0082 | 0.001 | 0.0098 | 0.0086 | 0.0028 | 0.24 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0096 | 0.0092 | 0.0034 | 0.011 | 0.011 | 0.0034 | 0.2 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.2.17 | Inulin fructotransferase (DFA-I-forming). | 0.23 | 0.54 | 1 | 0.87 | 1 | 1 | 0.53 | 0.95 | 1 | 0.17 | 0.49 | 1 | 0.72 | 0.95 | 1 | 0.4 | 0.83 | 1 | 0.00066 | 0.00052 | 0.00016 | 0.00064 | 0.00037 | 0.00069 | 0.00083 | 0.00016 | 0.0015 | 0.38 | 28 / 36 (78%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00034 | 0.00012 | 0.00049 | 0.00041 | 0.00014 | 0.00057 | 0.27 | 7 / 11 (64%) | 9 / 11 (82%) | 0.636 | 0.818 |
| 3.5.4.- | 0.23 | 0.54 | 1 | 0.33 | 1 | 1 | 0.77 | 0.99 | 1 | 0.16 | 0.48 | 1 | 0.054 | 0.81 | 1 | 0.45 | 0.86 | 1 | 0.0062 | 0.0062 | 0.0059 | 0.0052 | 0.0058 | 0.0013 | 0.0058 | 0.0058 | 0.0014 | 0.16 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0063 | 0.0058 | 0.0022 | 0.007 | 0.007 | 0.0019 | 0.15 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 2.7.7.94 | Transferred entry: 6.2.1.51. | 0.7 | 0.86 | 1 | 0.17 | 1 | 1 | 0.35 | 0.9 | 1 | 0.42 | 0.69 | 1 | 0.099 | 0.86 | 1 | 0.22 | 0.74 | 1 | 1e-04 | 8.6e-06 | 0 | 0 | 0 | 0 | 2.9e-05 | 0 | 7.7e-05 | Inf | 3 / 36 (8.3%) | 0 / 7 (0%) | 1 / 7 (14%) | 0 | 0.143 | 3e-06 | 0 | 9.9e-06 | 6.8e-06 | 0 | 2.3e-05 | 1.2 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.1.1.n11 | Methylphosphotriester-DNA--[protein]-cysteine S-methyltransferase. | 0.19 | 0.53 | 1 | 0.38 | 1 | 1 | 0.47 | 0.93 | 1 | 0.21 | 0.52 | 1 | 0.41 | 0.9 | 1 | 0.59 | 0.91 | 1 | 8.9e-05 | 1.7e-05 | 0 | 3.1e-05 | 0 | 4.9e-05 | 2.9e-05 | 0 | 7.7e-05 | -0.096 | 7 / 36 (19%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 5.8e-06 | 0 | 1.3e-05 | 1.3e-05 | 0 | 4.3e-05 | 1.2 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 2.1.1.187 | 23S rRNA (guanine(745)-N(1))-methyltransferase. | 0.028 | 0.49 | 1 | 0.3 | 1 | 1 | 0.12 | 0.82 | 1 | 0.023 | 0.42 | 1 | 0.36 | 0.9 | 1 | 0.15 | 0.71 | 1 | 0.0021 | 0.0021 | 0.0022 | 0.0015 | 0.0015 | 0.00064 | 0.0013 | 0.0013 | 8e-04 | -0.21 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0024 | 0.0027 | 0.001 | 0.0026 | 0.0025 | 8e-04 | 0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.2.27 | Uracil-DNA glycosylase. | 0.062 | 0.49 | 1 | 0.6 | 1 | 1 | 0.63 | 0.96 | 1 | 0.057 | 0.43 | 1 | 0.27 | 0.88 | 1 | 0.31 | 0.79 | 1 | 0.0084 | 0.0084 | 0.0082 | 0.0071 | 0.0067 | 0.0017 | 0.0078 | 0.0078 | 0.0031 | 0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0092 | 0.0078 | 0.0032 | 0.0089 | 0.009 | 0.0012 | -0.048 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.1.80 | Ceramide glucosyltransferase. | 0.97 | 0.99 | 1 | 0.37 | 1 | 1 | 0.8 | 1 | 1 | 0.9 | 0.97 | 1 | 0.39 | 0.9 | 1 | 0.77 | 0.95 | 1 | 0.00017 | 2.4e-05 | 0 | 4.6e-06 | 0 | 1.2e-05 | 6.1e-05 | 0 | 0.00016 | 3.7 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 5.7e-06 | 0 | 1.9e-05 | 3e-05 | 0 | 9e-05 | 2.4 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 6.2.1.4 | Succinate--CoA ligase (GDP-forming). | 0.28 | 0.56 | 1 | 0.37 | 1 | 1 | 0.99 | 1 | 1 | 0.26 | 0.56 | 1 | 0.23 | 0.88 | 1 | 0.76 | 0.95 | 1 | 0.00012 | 9.7e-06 | 0 | 1.7e-05 | 0 | 4.5e-05 | 2.9e-05 | 0 | 7.7e-05 | 0.77 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.1.1.218 | Morphine 6-dehydrogenase. | 0.29 | 0.57 | 1 | 0.17 | 1 | 1 | 0.55 | 0.95 | 1 | 0.055 | 0.43 | 1 | 0.019 | 0.76 | 1 | 0.076 | 0.66 | 1 | 0.00096 | 0.00093 | 0.00065 | 0.00062 | 0.00028 | 0.00091 | 0.00094 | 0.00084 | 0.00093 | 0.6 | 35 / 36 (97%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.00089 | 0.00057 | 0.00087 | 0.0012 | 0.00086 | 0.0011 | 0.43 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.6.6.4 | Transferred entry: 1.7.1.4. | 0.18 | 0.52 | 1 | 0.23 | 1 | 1 | 0.19 | 0.87 | 1 | 0.38 | 0.65 | 1 | 0.76 | 0.97 | 1 | 0.59 | 0.91 | 1 | 0.00099 | 0.00019 | 0 | 0.00059 | 0 | 0.0016 | 0.00032 | 0 | 0.00084 | -0.88 | 7 / 36 (19%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 1.1e-05 | 0 | 2.9e-05 | 3.7e-05 | 0 | 8.3e-05 | 1.8 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 1.5.99.8 | Transferred entry: 1.5.5.2. | 0.34 | 0.62 | 1 | 0.062 | 1 | 1 | 0.22 | 0.88 | 1 | 0.83 | 0.94 | 1 | 0.26 | 0.88 | 1 | 0.67 | 0.93 | 1 | 0.00049 | 2e-04 | 0 | 5e-04 | 0 | 0.0011 | 0.00023 | 0 | 0.00058 | -1.1 | 15 / 36 (42%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 1e-04 | 3.1e-05 | 0.00015 | 1e-04 | 0 | 0.00016 | 0 | 6 / 11 (55%) | 4 / 11 (36%) | 0.545 | 0.364 |
| 3.2.1.67 | Galacturonan 1.4-alpha-galacturonidase. | 0.66 | 0.84 | 1 | 0.26 | 1 | 1 | 0.26 | 0.89 | 1 | 0.78 | 0.92 | 1 | 0.23 | 0.88 | 1 | 0.18 | 0.71 | 1 | 0.00017 | 1.9e-05 | 0 | 1.6e-05 | 0 | 4.2e-05 | 7.2e-05 | 0 | 0.00018 | 2.2 | 4 / 36 (11%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 5.1e-06 | 0 | 1.7e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.4.1.212 | Hyaluronan synthase. | 0.31 | 0.59 | 1 | 0.14 | 1 | 1 | 0.24 | 0.88 | 1 | 0.22 | 0.52 | 1 | 0.58 | 0.93 | 1 | 0.82 | 0.96 | 1 | 0.034 | 0.034 | 0.034 | 0.032 | 0.034 | 0.0085 | 0.029 | 0.024 | 0.01 | -0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.037 | 0.04 | 0.011 | 0.037 | 0.037 | 0.0079 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.1.3.15 | Glucose-6-phosphate 1-epimerase. | 0.9 | 0.96 | 1 | 0.53 | 1 | 1 | 0.54 | 0.95 | 1 | 0.4 | 0.66 | 1 | 0.32 | 0.9 | 1 | 0.43 | 0.85 | 1 | 0.0013 | 0.0012 | 0.0011 | 0.0013 | 0.0015 | 0.00091 | 0.0011 | 0.001 | 0.001 | -0.24 | 33 / 36 (92%) | 6 / 7 (86%) | 5 / 7 (71%) | 0.857 | 0.714 | 0.0012 | 0.00088 | 0.0011 | 0.0012 | 0.001 | 0.00092 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.7.87 | L-threonylcarbamoyladenylate synthase. | 0.15 | 0.5 | 1 | 0.72 | 1 | 1 | 0.44 | 0.93 | 1 | 0.64 | 0.85 | 1 | 0.66 | 0.95 | 1 | 0.92 | 0.98 | 1 | 0.012 | 0.012 | 0.012 | 0.012 | 0.011 | 0.0024 | 0.012 | 0.012 | 0.0024 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.012 | 0.003 | 0.011 | 0.011 | 0.0015 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.4.- | 0.39 | 0.65 | 1 | 0.21 | 1 | 1 | 0.45 | 0.93 | 1 | 0.28 | 0.57 | 1 | 0.5 | 0.93 | 1 | 0.92 | 0.98 | 1 | 0.018 | 0.018 | 0.018 | 0.017 | 0.017 | 0.0031 | 0.016 | 0.017 | 0.0054 | -0.087 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.019 | 0.018 | 0.0042 | 0.018 | 0.019 | 0.0022 | -0.078 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 2.7.1.100 | S-methyl-5-thioribose kinase. | 0.065 | 0.49 | 1 | 0.63 | 1 | 1 | 0.65 | 0.97 | 1 | 0.2 | 0.5 | 1 | 0.54 | 0.93 | 1 | 0.58 | 0.91 | 1 | 0.0012 | 0.0012 | 0.001 | 0.0014 | 0.0014 | 0.00065 | 0.0016 | 0.0015 | 0.00067 | 0.19 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00098 | 0.00082 | 0.00061 | 0.00092 | 0.00085 | 0.00039 | -0.091 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.5.1.84 | All-trans-nonaprenyl-diphosphate synthase (geranyl-diphosphate specific). | 0.058 | 0.49 | 1 | 0.079 | 1 | 1 | 0.13 | 0.82 | 1 | 0.11 | 0.47 | 1 | 0.16 | 0.87 | 1 | 0.25 | 0.76 | 1 | 0.00031 | 8.7e-05 | 0 | 0.00024 | 1.6e-05 | 0.00044 | 7.6e-05 | 0 | 0.00013 | -1.7 | 10 / 36 (28%) | 4 / 7 (57%) | 2 / 7 (29%) | 0.571 | 0.286 | 3.2e-05 | 0 | 8.1e-05 | 5.3e-05 | 0 | 0.00015 | 0.73 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 2.4.2.3 | Uridine phosphorylase. | 0.82 | 0.92 | 1 | 0.71 | 1 | 1 | 0.31 | 0.89 | 1 | 0.73 | 0.9 | 1 | 0.14 | 0.87 | 1 | 0.031 | 0.62 | 1 | 0.0072 | 0.0072 | 0.0066 | 0.0076 | 0.0077 | 0.0032 | 0.0079 | 0.008 | 0.0033 | 0.056 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0072 | 0.0069 | 0.0021 | 0.0064 | 0.0065 | 0.0014 | -0.17 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.188 | Prostaglandin-F synthase. | 0.05 | 0.49 | 1 | 0.48 | 1 | 1 | 0.48 | 0.93 | 1 | 0.048 | 0.43 | 1 | 0.088 | 0.86 | 1 | 0.071 | 0.66 | 1 | 0.0025 | 0.002 | 0.00077 | 0.00036 | 0.00028 | 0.00034 | 0.00072 | 0.00064 | 0.00067 | 1 | 30 / 36 (83%) | 5 / 7 (71%) | 6 / 7 (86%) | 0.714 | 0.857 | 0.0034 | 0.0014 | 0.0068 | 0.0026 | 0.0011 | 0.003 | -0.39 | 10 / 11 (91%) | 9 / 11 (82%) | 0.909 | 0.818 |
| 1.1.5.- | 0.25 | 0.56 | 1 | 0.61 | 1 | 1 | 0.4 | 0.92 | 1 | 0.43 | 0.69 | 1 | 0.9 | 0.99 | 1 | 0.75 | 0.95 | 1 | 5e-04 | 6.9e-05 | 0 | 0.00019 | 0 | 0.00049 | 0.00014 | 0 | 0.00036 | -0.44 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 7.3e-06 | 0 | 2.4e-05 | 1.6e-05 | 0 | 3.7e-05 | 1.1 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 | |
| 6.3.1.2 | Glutamine synthetase. | 0.55 | 0.77 | 1 | 0.84 | 1 | 1 | 0.52 | 0.95 | 1 | 0.34 | 0.62 | 1 | 0.3 | 0.9 | 1 | 0.65 | 0.92 | 1 | 0.053 | 0.053 | 0.056 | 0.051 | 0.051 | 0.01 | 0.05 | 0.055 | 0.011 | -0.029 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.054 | 0.058 | 0.013 | 0.056 | 0.056 | 0.0044 | 0.052 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.49 | Glucose-6-phosphate dehydrogenase (NADP(+)). | 0.27 | 0.56 | 1 | 0.4 | 1 | 1 | 0.45 | 0.93 | 1 | 0.31 | 0.59 | 1 | 0.21 | 0.88 | 1 | 0.34 | 0.8 | 1 | 0.0034 | 0.0034 | 0.0023 | 0.0019 | 0.0016 | 0.0013 | 0.0027 | 0.0023 | 0.0017 | 0.51 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0045 | 0.0025 | 0.0071 | 0.0037 | 0.0032 | 0.0032 | -0.28 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.5.1.- | 0.072 | 0.49 | 1 | 0.57 | 1 | 1 | 0.31 | 0.89 | 1 | 0.036 | 0.43 | 1 | 0.1 | 0.86 | 1 | 0.041 | 0.63 | 1 | 0.015 | 0.015 | 0.016 | 0.014 | 0.014 | 0.0019 | 0.015 | 0.016 | 0.003 | 0.1 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.017 | 0.016 | 0.0043 | 0.016 | 0.016 | 0.0018 | -0.087 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 3.2.1.98 | Glucan 1.4-alpha-maltohexaosidase. | 0.37 | 0.65 | 1 | 0.52 | 1 | 1 | 0.72 | 0.98 | 1 | 0.53 | 0.77 | 1 | 0.97 | 0.99 | 1 | 0.35 | 0.81 | 1 | 0.00049 | 0.00044 | 0.00025 | 0.00065 | 0.00045 | 0.00073 | 5e-04 | 0.00025 | 0.00065 | -0.38 | 32 / 36 (89%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00041 | 0.00023 | 0.00046 | 0.00029 | 0.00017 | 0.00032 | -0.5 | 10 / 11 (91%) | 8 / 11 (73%) | 0.909 | 0.727 |
| 2.1.1.35 | tRNA (uracil(54)-C(5))-methyltransferase. | 0.49 | 0.73 | 1 | 0.15 | 1 | 1 | 0.14 | 0.83 | 1 | 0.32 | 0.59 | 1 | 0.1 | 0.86 | 1 | 0.26 | 0.76 | 1 | 0.00097 | 0.00092 | 0.00069 | 0.00078 | 0.00024 | 0.00093 | 0.0013 | 0.00048 | 0.0016 | 0.74 | 34 / 36 (94%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.001 | 0.00078 | 0.001 | 0.00068 | 0.00064 | 0.00038 | -0.56 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 2.8.4.2 | Arsenate-mycothiol transferase. | 0.29 | 0.57 | 1 | 0.96 | 1 | 1 | 0.73 | 0.99 | 1 | 0.26 | 0.56 | 1 | 0.93 | 0.99 | 1 | 0.71 | 0.94 | 1 | 0.00013 | 5.1e-05 | 0 | 3.3e-05 | 0 | 7.4e-05 | 3e-05 | 0 | 6.9e-05 | -0.14 | 14 / 36 (39%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 8.2e-05 | 0 | 0.00013 | 4.6e-05 | 0 | 7.1e-05 | -0.83 | 5 / 11 (45%) | 5 / 11 (45%) | 0.455 | 0.455 |
| 4.2.1.60 | Transferred entry: 4.2.1.59. | 0.031 | 0.49 | 1 | 0.0041 | 0.67 | 1 | 0.019 | 0.71 | 1 | 0.012 | 0.42 | 1 | 0.047 | 0.81 | 1 | 0.14 | 0.69 | 1 | 0.00027 | 3e-05 | 0 | 9.8e-05 | 0 | 0.00022 | 5.8e-05 | 0 | 0.00015 | -0.76 | 4 / 36 (11%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 2.1.1.178 | 16S rRNA (cytosine(1407)-C(5))-methyltransferase. | 0.35 | 0.63 | 1 | 0.86 | 1 | 1 | 0.85 | 1 | 1 | 0.27 | 0.56 | 1 | 0.38 | 0.9 | 1 | 0.38 | 0.82 | 1 | 0.011 | 0.011 | 0.01 | 0.0098 | 0.01 | 0.0036 | 0.01 | 0.012 | 0.0049 | 0.029 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.011 | 0.0032 | 0.011 | 0.01 | 0.0021 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.283 | Methylglyoxal reductase (NADPH). | 0.33 | 0.6 | 1 | 0.11 | 1 | 1 | 0.056 | 0.71 | 1 | 0.21 | 0.52 | 1 | 0.075 | 0.86 | 1 | 0.019 | 0.55 | 1 | 0.00038 | 0.00035 | 0.00025 | 0.00025 | 0.00017 | 0.00021 | 4e-04 | 0.00028 | 0.00025 | 0.68 | 33 / 36 (92%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.00038 | 0.00026 | 0.00035 | 0.00034 | 0.00024 | 0.00044 | -0.16 | 11 / 11 (100%) | 9 / 11 (82%) | 1 | 0.818 |
| 3.6.3.3 | Transferred entry: 7.2.2.21. | 0.73 | 0.87 | 1 | 0.98 | 1 | 1 | 0.74 | 0.99 | 1 | 0.36 | 0.63 | 1 | 0.41 | 0.9 | 1 | 0.62 | 0.92 | 1 | 0.045 | 0.045 | 0.046 | 0.044 | 0.046 | 0.01 | 0.044 | 0.046 | 0.016 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.045 | 0.045 | 0.0095 | 0.047 | 0.046 | 0.0054 | 0.063 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.4.1.23 | 2-hydroxypropyl-CoM lyase. | 0.81 | 0.91 | 1 | 0.16 | 1 | 1 | 0.51 | 0.95 | 1 | 0.82 | 0.93 | 1 | 0.21 | 0.88 | 1 | 0.52 | 0.89 | 1 | 0.00023 | 0.00013 | 7.3e-05 | 9.2e-05 | 8.1e-05 | 1e-04 | 0.00021 | 0.00022 | 2e-04 | 1.2 | 20 / 36 (56%) | 4 / 7 (57%) | 5 / 7 (71%) | 0.571 | 0.714 | 9e-05 | 0 | 0.00013 | 0.00014 | 6.6e-05 | 2e-04 | 0.64 | 5 / 11 (45%) | 6 / 11 (55%) | 0.455 | 0.545 |
| 3.6.3.53 | Transferred entry: 7.2.2.15. | 0.61 | 0.81 | 1 | 0.48 | 1 | 1 | 0.9 | 1 | 1 | 0.72 | 0.89 | 1 | 0.39 | 0.9 | 1 | 0.62 | 0.92 | 1 | 0.0019 | 0.0017 | 0.00064 | 0.0014 | 0.0013 | 0.0014 | 0.0012 | 0.00048 | 0.0018 | -0.22 | 32 / 36 (89%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.002 | 6e-04 | 0.0021 | 0.0018 | 0.00094 | 0.0026 | -0.15 | 11 / 11 (100%) | 8 / 11 (73%) | 1 | 0.727 |
| 2.7.1.36 | Mevalonate kinase. | 0.36 | 0.63 | 1 | 0.13 | 1 | 1 | 0.23 | 0.88 | 1 | 0.44 | 0.69 | 1 | 0.077 | 0.86 | 1 | 0.15 | 0.71 | 1 | 0.00032 | 0.00021 | 0.00014 | 0.00011 | 8e-05 | 0.00013 | 0.00024 | 0.00013 | 0.00022 | 1.1 | 24 / 36 (67%) | 4 / 7 (57%) | 6 / 7 (86%) | 0.571 | 0.857 | 0.00024 | 2e-04 | 0.00029 | 0.00023 | 0.00022 | 0.00026 | -0.061 | 7 / 11 (64%) | 7 / 11 (64%) | 0.636 | 0.636 |
| 3.4.24.13 | IgA-specific metalloendopeptidase. | 0.28 | 0.56 | 1 | 0.67 | 1 | 1 | 0.21 | 0.87 | 1 | 0.55 | 0.78 | 1 | 0.59 | 0.93 | 1 | 0.6 | 0.91 | 1 | 0.00096 | 0.00094 | 0.00043 | 0.00092 | 0.00039 | 0.0011 | 0.00079 | 0.00046 | 0.00089 | -0.22 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0015 | 0.0018 | 0.001 | 0.00052 | 0.00036 | 0.00064 | -1.5 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 3.2.2.20 | DNA-3-methyladenine glycosylase I. | 0.99 | 1 | 1 | 0.89 | 1 | 1 | 0.94 | 1 | 1 | 0.53 | 0.76 | 1 | 0.54 | 0.93 | 1 | 0.56 | 0.9 | 1 | 0.0027 | 0.0027 | 0.0026 | 0.0027 | 0.0025 | 0.0013 | 0.0028 | 0.0034 | 0.0011 | 0.052 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0027 | 0.0025 | 0.001 | 0.0027 | 0.0026 | 0.001 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.4.1.6 | Acetone carboxylase. | 0.69 | 0.86 | 1 | 0.21 | 1 | 1 | 0.76 | 0.99 | 1 | 0.82 | 0.93 | 1 | 0.24 | 0.88 | 1 | 0.87 | 0.97 | 1 | 9.9e-05 | 2.7e-05 | 0 | 4.2e-05 | 0 | 5.6e-05 | 1.9e-05 | 0 | 5.1e-05 | -1.1 | 10 / 36 (28%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 3.4e-05 | 0 | 5.8e-05 | 1.6e-05 | 0 | 4.5e-05 | -1.1 | 4 / 11 (36%) | 2 / 11 (18%) | 0.364 | 0.182 |
| 2.1.1.133 | Precorrin-4 C(11)-methyltransferase. | 0.16 | 0.5 | 1 | 0.61 | 1 | 1 | 0.72 | 0.98 | 1 | 0.57 | 0.8 | 1 | 0.15 | 0.87 | 1 | 0.58 | 0.91 | 1 | 0.0076 | 0.0076 | 0.0077 | 0.0082 | 0.0076 | 0.0025 | 0.0086 | 0.0092 | 0.0024 | 0.069 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0068 | 0.0077 | 0.0019 | 0.0074 | 0.0075 | 0.0021 | 0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.8.11 | CDP-diacylglycerol--inositol 3-phosphatidyltransferase. | 0.44 | 0.69 | 1 | 0.67 | 1 | 1 | 0.74 | 0.99 | 1 | 0.8 | 0.93 | 1 | 0.37 | 0.9 | 1 | 0.87 | 0.97 | 1 | 0.0012 | 0.001 | 0.00057 | 0.00047 | 0.00055 | 0.00027 | 7e-04 | 0.00027 | 0.00088 | 0.57 | 31 / 36 (86%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.0015 | 4e-04 | 0.0034 | 0.001 | 0.00076 | 0.001 | -0.58 | 9 / 11 (82%) | 9 / 11 (82%) | 0.818 | 0.818 |
| 4.1.2.22 | Fructose-6-phosphate phosphoketolase. | 0.12 | 0.5 | 1 | 0.52 | 1 | 1 | 0.32 | 0.89 | 1 | 0.23 | 0.53 | 1 | 0.66 | 0.95 | 1 | 0.83 | 0.96 | 1 | 0.0029 | 0.0027 | 0.0015 | 0.0012 | 9e-04 | 0.0013 | 0.0017 | 0.0019 | 0.0012 | 0.5 | 34 / 36 (94%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.0044 | 0.0016 | 0.0082 | 0.0026 | 0.0017 | 0.0024 | -0.76 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 2.5.1.97 | Pseudaminic acid synthase. | 0.0017 | 0.49 | 1 | 0.36 | 1 | 1 | 0.25 | 0.89 | 1 | 0.0032 | 0.36 | 1 | 0.3 | 0.89 | 1 | 0.22 | 0.74 | 1 | 0.00041 | 0.00034 | 0.00017 | 0.00066 | 0.00056 | 0.00056 | 0.00064 | 0.00019 | 0.00085 | -0.044 | 30 / 36 (83%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00011 | 8.7e-05 | 9.6e-05 | 0.00017 | 0.00011 | 0.00017 | 0.63 | 8 / 11 (73%) | 9 / 11 (82%) | 0.727 | 0.818 |
| 1.5.1.11 | D-octopine dehydrogenase. | 0.84 | 0.93 | 1 | 0.6 | 1 | 1 | 0.52 | 0.95 | 1 | 0.89 | 0.96 | 1 | 0.51 | 0.93 | 1 | 0.53 | 0.9 | 1 | 0.00046 | 0.00015 | 0 | 1e-04 | 0 | 2e-04 | 5.7e-05 | 0 | 0.00015 | -0.81 | 12 / 36 (33%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 0.00016 | 0 | 0.00034 | 0.00024 | 0 | 0.00052 | 0.58 | 4 / 11 (36%) | 4 / 11 (36%) | 0.364 | 0.364 |
| 2.8.3.6 | 3-oxoadipate CoA-transferase. | 1 | 1 | 1 | 0.035 | 0.97 | 1 | 0.17 | 0.85 | 1 | 0.7 | 0.88 | 1 | 0.0065 | 0.7 | 1 | 0.064 | 0.66 | 1 | 8.8e-05 | 7.3e-06 | 0 | 0 | 0 | 0 | 3.3e-05 | 0 | 7.6e-05 | Inf | 3 / 36 (8.3%) | 0 / 7 (0%) | 2 / 7 (29%) | 0 | 0.286 | 0 | 0 | 0 | 3e-06 | 0 | 1e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 5.4.99.24 | 23S rRNA pseudouridine(955/2504/2580) synthase. | 0.9 | 0.96 | 1 | 0.48 | 1 | 1 | 0.069 | 0.73 | 1 | 0.69 | 0.88 | 1 | 0.92 | 0.99 | 1 | 0.26 | 0.76 | 1 | 0.0072 | 0.0072 | 0.007 | 0.007 | 0.006 | 0.0019 | 0.0066 | 0.0065 | 0.0023 | -0.085 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0069 | 0.0068 | 0.0019 | 0.0081 | 0.0075 | 0.0025 | 0.23 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.8.20 | Phosphatidylglycerol--membrane-oligosaccharide glycerophosphotransferase. | 0.77 | 0.89 | 1 | 0.33 | 1 | 1 | 0.091 | 0.79 | 1 | 0.85 | 0.95 | 1 | 0.12 | 0.87 | 1 | 0.037 | 0.63 | 1 | 0.00064 | 0.00047 | 0.00016 | 0.00076 | 0.00014 | 0.0015 | 0.00069 | 0.00059 | 0.00061 | -0.14 | 26 / 36 (72%) | 5 / 7 (71%) | 6 / 7 (86%) | 0.714 | 0.857 | 0.00044 | 0.00017 | 0.00061 | 0.00017 | 0.00011 | 0.00023 | -1.4 | 8 / 11 (73%) | 7 / 11 (64%) | 0.727 | 0.636 |
| 1.6.6.9 | Deleted entry. | 0.079 | 0.49 | 1 | 0.034 | 0.97 | 1 | 0.023 | 0.71 | 1 | 0.053 | 0.43 | 1 | 0.078 | 0.86 | 1 | 0.055 | 0.66 | 1 | 0.00092 | 0.00018 | 0 | 0.00048 | 0 | 0.0011 | 0.00037 | 0 | 0.00097 | -0.38 | 7 / 36 (19%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 8.9e-06 | 0 | 3e-05 | 3.4e-05 | 0 | 7.8e-05 | 1.9 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 1.17.5.3 | Formate dehydrogenase-N. | 0.022 | 0.49 | 1 | 0.00055 | 0.34 | 1 | 0.00073 | 0.51 | 1 | 0.0034 | 0.36 | 1 | 0.0046 | 0.7 | 1 | 0.0033 | 0.48 | 1 | 0.00052 | 0.00016 | 0 | 5e-04 | 0.00012 | 0.0011 | 0.00026 | 0 | 0.00069 | -0.94 | 11 / 36 (31%) | 5 / 7 (71%) | 1 / 7 (14%) | 0.714 | 0.143 | 1e-05 | 0 | 3.4e-05 | 2.2e-05 | 0 | 3.8e-05 | 1.1 | 1 / 11 (9.1%) | 4 / 11 (36%) | 0.0909 | 0.364 |
| 7.2.2.7 | ABC-type Fe(3+) transporter. | 0.032 | 0.49 | 1 | 0.96 | 1 | 1 | 0.12 | 0.82 | 1 | 0.055 | 0.43 | 1 | 0.82 | 0.98 | 1 | 0.071 | 0.66 | 1 | 0.0012 | 0.0011 | 0.0011 | 0.00072 | 0.00079 | 0.00058 | 0.00071 | 0.00057 | 0.00061 | -0.02 | 32 / 36 (89%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.0015 | 0.0015 | 0.00076 | 0.0011 | 0.001 | 0.00078 | -0.45 | 11 / 11 (100%) | 9 / 11 (82%) | 1 | 0.818 |
| 2.4.1.8 | Maltose phosphorylase. | 0.17 | 0.5 | 1 | 0.89 | 1 | 1 | 0.71 | 0.98 | 1 | 0.32 | 0.59 | 1 | 0.62 | 0.95 | 1 | 0.68 | 0.93 | 1 | 0.00045 | 0.00035 | 0.00013 | 0.00051 | 0.00037 | 0.00064 | 0.00047 | 0.00038 | 0.00042 | -0.12 | 28 / 36 (78%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00017 | 9.5e-05 | 2e-04 | 0.00034 | 0.00011 | 0.00049 | 1 | 9 / 11 (82%) | 7 / 11 (64%) | 0.818 | 0.636 |
| 2.7.1.207 | Protein-N(pi)-phosphohistidine--lactose phosphotransferase. | 0.36 | 0.64 | 1 | 0.18 | 1 | 1 | 0.5 | 0.95 | 1 | 0.21 | 0.51 | 1 | 0.28 | 0.88 | 1 | 0.65 | 0.92 | 1 | 0.00021 | 6.4e-05 | 0 | 2.3e-06 | 0 | 6.1e-06 | 0.00017 | 0 | 0.00044 | 6.2 | 11 / 36 (31%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3.8e-05 | 0 | 5.9e-05 | 6.6e-05 | 0 | 9.7e-05 | 0.8 | 4 / 11 (36%) | 5 / 11 (45%) | 0.364 | 0.455 |
| 5.4.2.2 | Phosphoglucomutase (alpha-D-glucose-1.6-bisphosphate-dependent). | 0.16 | 0.5 | 1 | 0.71 | 1 | 1 | 0.77 | 0.99 | 1 | 0.16 | 0.48 | 1 | 0.05 | 0.81 | 1 | 0.069 | 0.66 | 1 | 0.034 | 0.034 | 0.034 | 0.031 | 0.033 | 0.0095 | 0.032 | 0.032 | 0.009 | 0.046 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.036 | 0.036 | 0.0096 | 0.036 | 0.037 | 0.0063 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.12 | Gluconokinase. | 0.11 | 0.5 | 1 | 0.19 | 1 | 1 | 0.44 | 0.93 | 1 | 0.0063 | 0.37 | 1 | 0.0056 | 0.7 | 1 | 0.046 | 0.65 | 1 | 0.0014 | 0.0012 | 0.00044 | 0.00045 | 5.7e-05 | 0.00095 | 0.00069 | 0.00032 | 8e-04 | 0.62 | 31 / 36 (86%) | 4 / 7 (57%) | 6 / 7 (86%) | 0.571 | 0.857 | 0.0017 | 0.00057 | 0.004 | 0.0014 | 0.00083 | 0.0014 | -0.28 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 3.6.3.34 | Transferred entry: 7.2.2.16. | 0.8 | 0.9 | 1 | 0.87 | 1 | 1 | 0.98 | 1 | 1 | 0.86 | 0.95 | 1 | 0.67 | 0.95 | 1 | 0.7 | 0.94 | 1 | 0.0069 | 0.0069 | 0.0064 | 0.0069 | 0.0063 | 0.0018 | 0.0075 | 0.0088 | 0.0036 | 0.12 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0067 | 0.0065 | 0.0022 | 0.0069 | 0.0061 | 0.0024 | 0.042 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.2.1.2 | Transferred entry: 1.17.1.9. | 0.14 | 0.5 | 1 | 0.29 | 1 | 1 | 0.87 | 1 | 1 | 0.33 | 0.6 | 1 | 0.3 | 0.89 | 1 | 0.98 | 1 | 1 | 0.0065 | 0.0065 | 0.0057 | 0.0085 | 0.0071 | 0.0038 | 0.0075 | 0.0053 | 0.0049 | -0.18 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.006 | 0.0064 | 0.0024 | 0.0051 | 0.0041 | 0.0026 | -0.23 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.11.18 | [Myosin light-chain] kinase. | 0.43 | 0.69 | 1 | 0.64 | 1 | 1 | 0.57 | 0.96 | 1 | 0.54 | 0.77 | 1 | 0.57 | 0.93 | 1 | 0.8 | 0.96 | 1 | 0.00075 | 0.00015 | 0 | 3.2e-05 | 0 | 8.6e-05 | 9.7e-05 | 0 | 0.00026 | 1.6 | 7 / 36 (19%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0.00027 | 0 | 0.00076 | 0.00013 | 0 | 0.00026 | -1.1 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 2.4.1.320 | 1.4-beta-mannosyl-N-acetylglucosamine phosphorylase. | 0.19 | 0.53 | 1 | 0.21 | 1 | 1 | 0.053 | 0.71 | 1 | 0.16 | 0.49 | 1 | 0.19 | 0.87 | 1 | 0.075 | 0.66 | 1 | 3e-04 | 6.6e-05 | 0 | 9.2e-05 | 0 | 0.00014 | 6.3e-05 | 0 | 0.00014 | -0.55 | 8 / 36 (22%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 2.4e-05 | 0 | 7.9e-05 | 9.5e-05 | 0 | 0.00028 | 2 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 3.6.3.29 | Transferred entry: 7.3.2.5. | 0.2 | 0.53 | 1 | 0.56 | 1 | 1 | 0.61 | 0.96 | 1 | 0.34 | 0.62 | 1 | 0.75 | 0.97 | 1 | 0.96 | 0.99 | 1 | 0.00033 | 0.00017 | 6.7e-05 | 0.00035 | 0.00011 | 0.00073 | 0.00024 | 0.00011 | 0.00044 | -0.54 | 19 / 36 (53%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 7.5e-05 | 6.2e-05 | 9.3e-05 | 0.00012 | 0 | 0.00018 | 0.68 | 6 / 11 (55%) | 4 / 11 (36%) | 0.545 | 0.364 |
| 2.3.1.181 | Lipoyl(octanoyl) transferase. | 0.23 | 0.54 | 1 | 0.9 | 1 | 1 | 0.81 | 1 | 1 | 0.47 | 0.72 | 1 | 0.52 | 0.93 | 1 | 0.49 | 0.88 | 1 | 0.0011 | 0.00097 | 0.00065 | 0.0013 | 0.0014 | 9e-04 | 0.0011 | 0.0011 | 0.00068 | -0.24 | 33 / 36 (92%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.00073 | 0.00029 | 0.00084 | 0.00094 | 0.00033 | 0.0014 | 0.36 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 4.4.1.2 | Homocysteine desulfhydrase. | 0.038 | 0.49 | 1 | 0.11 | 1 | 1 | 0.13 | 0.82 | 1 | 0.053 | 0.43 | 1 | 0.19 | 0.87 | 1 | 0.17 | 0.71 | 1 | 0.0012 | 0.00013 | 0 | 4e-04 | 0 | 0.00072 | 0.00025 | 0 | 0.00067 | -0.68 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 9e-06 | 0 | 3e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.17.4.1 | Ribonucleoside-diphosphate reductase. | 0.75 | 0.88 | 1 | 0.54 | 1 | 1 | 0.3 | 0.89 | 1 | 0.89 | 0.97 | 1 | 0.87 | 0.99 | 1 | 0.56 | 0.9 | 1 | 0.014 | 0.014 | 0.013 | 0.014 | 0.012 | 0.0058 | 0.013 | 0.015 | 0.0063 | -0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.013 | 0.012 | 0.0057 | 0.015 | 0.013 | 0.007 | 0.21 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.1.182 | 16S rRNA (adenine(1518)-N(6)/adenine(1519)-N(6))-dimethyltransferase. | 0.095 | 0.49 | 1 | 0.83 | 1 | 1 | 0.4 | 0.92 | 1 | 0.12 | 0.47 | 1 | 0.24 | 0.88 | 1 | 0.026 | 0.59 | 1 | 0.017 | 0.017 | 0.018 | 0.016 | 0.016 | 0.0042 | 0.015 | 0.014 | 0.0048 | -0.093 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.019 | 0.019 | 0.0041 | 0.017 | 0.018 | 0.0022 | -0.16 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.3.4 | Phosphatidate phosphatase. | 0.14 | 0.5 | 1 | 0.27 | 1 | 1 | 0.35 | 0.9 | 1 | 0.14 | 0.47 | 1 | 0.055 | 0.81 | 1 | 0.096 | 0.66 | 1 | 0.0027 | 0.0024 | 0.0013 | 0.00081 | 0.00082 | 0.00072 | 0.0017 | 0.00096 | 0.0018 | 1.1 | 32 / 36 (89%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.0035 | 0.0016 | 0.0069 | 0.0028 | 0.0019 | 0.0031 | -0.32 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 4.2.1.79 | 2-methylcitrate dehydratase. | 0.08 | 0.49 | 1 | 0.13 | 1 | 1 | 0.31 | 0.89 | 1 | 0.1 | 0.46 | 1 | 0.34 | 0.9 | 1 | 0.65 | 0.92 | 1 | 0.00033 | 7.4e-05 | 0 | 0.00021 | 0 | 4e-04 | 0.00014 | 0 | 0.00036 | -0.58 | 8 / 36 (22%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 1.2e-05 | 0 | 3.2e-05 | 1e-05 | 0 | 3.4e-05 | -0.26 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 3.4.23.43 | Prepilin peptidase. | 0.42 | 0.68 | 1 | 0.81 | 1 | 1 | 0.96 | 1 | 1 | 0.51 | 0.75 | 1 | 0.7 | 0.95 | 1 | 0.87 | 0.97 | 1 | 0.0029 | 0.0029 | 0.0021 | 0.002 | 0.0012 | 0.0011 | 0.0022 | 0.0019 | 0.0016 | 0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0036 | 0.002 | 0.0057 | 0.0031 | 0.0033 | 0.0018 | -0.22 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.4.2 | Phosphomevalonate kinase. | 0.23 | 0.54 | 1 | 0.24 | 1 | 1 | 0.11 | 0.82 | 1 | 0.24 | 0.54 | 1 | 0.19 | 0.87 | 1 | 0.099 | 0.66 | 1 | 0.00036 | 2e-04 | 9e-05 | 0.00011 | 0 | 0.00017 | 0.00025 | 2e-04 | 0.00026 | 1.2 | 20 / 36 (56%) | 3 / 7 (43%) | 5 / 7 (71%) | 0.429 | 0.714 | 0.00027 | 2e-04 | 0.00037 | 0.00014 | 0 | 2e-04 | -0.95 | 7 / 11 (64%) | 5 / 11 (45%) | 0.636 | 0.455 |
| 6.2.1.14 | 6-carboxyhexanoate--CoA ligase. | 0.88 | 0.95 | 1 | 0.3 | 1 | 1 | 0.91 | 1 | 1 | 0.9 | 0.97 | 1 | 0.35 | 0.9 | 1 | 0.93 | 0.98 | 1 | 0.00051 | 0.00043 | 0.00029 | 0.00046 | 0.00019 | 0.00046 | 0.00033 | 0.00013 | 0.00039 | -0.48 | 30 / 36 (83%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00051 | 0.00026 | 0.00047 | 0.00038 | 0.00034 | 0.00035 | -0.42 | 9 / 11 (82%) | 9 / 11 (82%) | 0.818 | 0.818 |
| 3.1.3.90 | Maltose 6'-phosphate phosphatase. | 0.75 | 0.88 | 1 | 0.039 | 0.97 | 1 | 0.091 | 0.79 | 1 | 0.78 | 0.92 | 1 | 0.035 | 0.8 | 1 | 0.051 | 0.65 | 1 | 0.00024 | 1e-04 | 0 | 4e-05 | 0 | 5.5e-05 | 0.00018 | 6.8e-05 | 0.00026 | 2.2 | 15 / 36 (42%) | 3 / 7 (43%) | 5 / 7 (71%) | 0.429 | 0.714 | 8e-05 | 0 | 0.00013 | 0.00011 | 0 | 0.00024 | 0.46 | 4 / 11 (36%) | 3 / 11 (27%) | 0.364 | 0.273 |
| 1.21.98.- | 0.43 | 0.69 | 1 | 0.58 | 1 | 1 | 0.12 | 0.82 | 1 | 0.52 | 0.76 | 1 | 0.24 | 0.88 | 1 | 0.089 | 0.66 | 1 | 0.00087 | 0.00079 | 0.00054 | 0.00079 | 5e-04 | 0.00063 | 0.00076 | 0.00095 | 0.00062 | -0.056 | 33 / 36 (92%) | 7 / 7 (100%) | 5 / 7 (71%) | 1 | 0.714 | 0.00052 | 0.00058 | 0.00037 | 0.0011 | 0.00044 | 0.0015 | 1.1 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 | |
| 2.7.4.28 | ([Pyruvate. water dikinase] phosphate) phosphotransferase. | 0.11 | 0.5 | 1 | 0.15 | 1 | 1 | 0.058 | 0.71 | 1 | 0.12 | 0.47 | 1 | 0.26 | 0.88 | 1 | 0.12 | 0.66 | 1 | 2e-04 | 4.9e-05 | 0 | 0.00012 | 0 | 0.00026 | 7.7e-05 | 0 | 2e-04 | -0.64 | 9 / 36 (25%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 5.5e-06 | 0 | 1.2e-05 | 3.2e-05 | 0 | 6.7e-05 | 2.5 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 2.8.4.- | 0.08 | 0.49 | 1 | 0.48 | 1 | 1 | 0.71 | 0.98 | 1 | 0.19 | 0.5 | 1 | 0.45 | 0.92 | 1 | 0.64 | 0.92 | 1 | 7e-04 | 0.00056 | 0.00049 | 0.00041 | 0.00026 | 0.00048 | 0.00029 | 9.3e-05 | 0.00034 | -0.5 | 29 / 36 (81%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 0.00094 | 0.00072 | 0.00087 | 0.00045 | 0.00045 | 3e-04 | -1.1 | 9 / 11 (82%) | 11 / 11 (100%) | 0.818 | 1 | |
| 2.4.1.20 | Cellobiose phosphorylase. | 0.029 | 0.49 | 1 | 0.75 | 1 | 1 | 0.81 | 1 | 1 | 0.018 | 0.42 | 1 | 0.45 | 0.92 | 1 | 0.73 | 0.94 | 1 | 0.0072 | 0.0072 | 0.0061 | 0.0038 | 0.0029 | 0.0028 | 0.0046 | 0.0022 | 0.0047 | 0.28 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0097 | 0.0084 | 0.0099 | 0.0085 | 0.0082 | 0.0038 | -0.19 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.3.4 | 3-dehydroquinate synthase. | 0.1 | 0.49 | 1 | 0.82 | 1 | 1 | 0.66 | 0.97 | 1 | 0.073 | 0.43 | 1 | 0.29 | 0.89 | 1 | 0.25 | 0.76 | 1 | 0.018 | 0.018 | 0.017 | 0.015 | 0.014 | 0.0032 | 0.016 | 0.016 | 0.0042 | 0.093 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.02 | 0.016 | 0.01 | 0.019 | 0.019 | 0.0041 | -0.074 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.1.11 | Phosphopyruvate hydratase. | 0.87 | 0.94 | 1 | 0.75 | 1 | 1 | 0.97 | 1 | 1 | 0.44 | 0.69 | 1 | 0.13 | 0.87 | 1 | 0.21 | 0.74 | 1 | 0.026 | 0.026 | 0.026 | 0.025 | 0.026 | 0.0047 | 0.026 | 0.028 | 0.0058 | 0.057 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.025 | 0.026 | 0.0035 | 0.026 | 0.026 | 0.0042 | 0.057 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.26.4 | Ribonuclease H. | 0.011 | 0.49 | 1 | 0.82 | 1 | 1 | 0.17 | 0.85 | 1 | 0.022 | 0.42 | 1 | 0.17 | 0.87 | 1 | 0.018 | 0.55 | 1 | 0.022 | 0.022 | 0.021 | 0.018 | 0.018 | 0.0034 | 0.019 | 0.018 | 0.0041 | 0.078 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.025 | 0.023 | 0.0095 | 0.022 | 0.023 | 0.0036 | -0.18 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.1.118 | 3-dehydroshikimate dehydratase. | 0.79 | 0.9 | 1 | 0.27 | 1 | 1 | 0.03 | 0.71 | 1 | 0.09 | 0.45 | 1 | 0.53 | 0.93 | 1 | 0.39 | 0.83 | 1 | 0.00094 | 0.00076 | 4e-04 | 0.00079 | 0.00014 | 0.0014 | 0.00041 | 8.1e-05 | 9e-04 | -0.95 | 29 / 36 (81%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.00046 | 0.00042 | 0.00036 | 0.0013 | 9e-04 | 0.001 | 1.5 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 3.4.11.23 | PepB aminopeptidase. | 0.35 | 0.62 | 1 | 0.85 | 1 | 1 | 0.75 | 0.99 | 1 | 0.74 | 0.9 | 1 | 0.25 | 0.88 | 1 | 0.26 | 0.76 | 1 | 0.00019 | 6.2e-05 | 0 | 0.00017 | 0 | 0.00044 | 7.7e-05 | 2.7e-05 | 1e-04 | -1.1 | 12 / 36 (33%) | 2 / 7 (29%) | 4 / 7 (57%) | 0.286 | 0.571 | 2.8e-05 | 0 | 5.7e-05 | 1.7e-05 | 0 | 4e-05 | -0.72 | 3 / 11 (27%) | 3 / 11 (27%) | 0.273 | 0.273 |
| 7.6.2.8 | ABC-type vitamin B12 transporter. | 0.084 | 0.49 | 1 | 0.51 | 1 | 1 | 0.6 | 0.96 | 1 | 0.042 | 0.43 | 1 | 0.44 | 0.91 | 1 | 0.61 | 0.92 | 1 | 0.00024 | 2e-05 | 0 | 4.5e-05 | 0 | 8.9e-05 | 5.8e-05 | 0 | 0.00015 | 0.37 | 3 / 36 (8.3%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 3.4.22.- | 0.74 | 0.88 | 1 | 0.64 | 1 | 1 | 0.28 | 0.89 | 1 | 0.43 | 0.69 | 1 | 0.42 | 0.91 | 1 | 0.52 | 0.89 | 1 | 0.019 | 0.019 | 0.017 | 0.016 | 0.014 | 0.0067 | 0.018 | 0.017 | 0.0094 | 0.17 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.018 | 0.015 | 0.0096 | 0.022 | 0.018 | 0.0076 | 0.29 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 3.5.1.103 | N-acetyl-1-D-myo-inositol-2-amino-2-deoxy-alpha-D-glucopyranoside | 0.35 | 0.62 | 1 | 0.64 | 1 | 1 | 0.45 | 0.93 | 1 | 0.6 | 0.81 | 1 | 0.86 | 0.99 | 1 | 0.76 | 0.95 | 1 | 0.00022 | 7.3e-05 | 0 | 0.00014 | 0 | 0.00031 | 9.5e-05 | 0 | 2e-04 | -0.56 | 12 / 36 (33%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 2.4e-05 | 0 | 4.2e-05 | 6.6e-05 | 0 | 0.00011 | 1.5 | 4 / 11 (36%) | 3 / 11 (27%) | 0.364 | 0.273 |
| 6.1.1.22 | Asparagine--tRNA ligase. | 0.34 | 0.62 | 1 | 0.74 | 1 | 1 | 1 | 1 | 1 | 0.3 | 0.58 | 1 | 0.38 | 0.9 | 1 | 0.24 | 0.76 | 1 | 0.021 | 0.021 | 0.022 | 0.02 | 0.021 | 0.0044 | 0.02 | 0.02 | 0.0034 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.022 | 0.024 | 0.0044 | 0.022 | 0.02 | 0.0041 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.2.8 | Acetylglutamate kinase. | 0.044 | 0.49 | 1 | 0.85 | 1 | 1 | 0.78 | 1 | 1 | 0.062 | 0.43 | 1 | 0.29 | 0.89 | 1 | 0.51 | 0.89 | 1 | 0.016 | 0.016 | 0.016 | 0.014 | 0.014 | 0.0015 | 0.015 | 0.016 | 0.0044 | 0.1 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.017 | 0.017 | 0.0028 | 0.018 | 0.018 | 0.0021 | 0.082 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.5.1.3 | RNA ligase (ATP). | 0.81 | 0.91 | 1 | 0.63 | 1 | 1 | 0.89 | 1 | 1 | 0.97 | 0.99 | 1 | 0.45 | 0.92 | 1 | 0.71 | 0.94 | 1 | 0.00027 | 6.9e-05 | 0 | 8.2e-05 | 0 | 0.00021 | 8.7e-05 | 0 | 0.00018 | 0.085 | 9 / 36 (25%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 5.8e-05 | 0 | 0.00015 | 5.9e-05 | 0 | 0.00014 | 0.025 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 1.1.1.179 | D-xylose 1-dehydrogenase (NADP(+)). | 0.099 | 0.49 | 1 | 0.97 | 1 | 1 | 0.57 | 0.96 | 1 | 0.057 | 0.43 | 1 | 0.86 | 0.99 | 1 | 0.43 | 0.85 | 1 | 0.002 | 0.0015 | 0.00041 | 0.00049 | 8.1e-05 | 0.00077 | 0.00054 | 3.1e-05 | 0.001 | 0.14 | 27 / 36 (75%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.0027 | 0.00095 | 0.0059 | 0.0015 | 0.0012 | 0.0018 | -0.85 | 10 / 11 (91%) | 9 / 11 (82%) | 0.909 | 0.818 |
| 1.17.98.1 | Transferred entry: 1.1.1.395. | 0.16 | 0.5 | 1 | 0.52 | 1 | 1 | 0.075 | 0.74 | 1 | 0.11 | 0.47 | 1 | 0.35 | 0.9 | 1 | 0.029 | 0.61 | 1 | 0.0028 | 0.0028 | 0.0028 | 0.0025 | 0.0026 | 0.0013 | 0.0029 | 0.003 | 0.0017 | 0.21 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0033 | 0.0032 | 9e-04 | 0.0026 | 0.0024 | 0.0013 | -0.34 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.31.- | 0.21 | 0.54 | 1 | 0.71 | 1 | 1 | 0.28 | 0.89 | 1 | 0.13 | 0.47 | 1 | 0.95 | 0.99 | 1 | 0.55 | 0.9 | 1 | 0.00061 | 0.00049 | 3e-04 | 0.00054 | 0.00056 | 0.00031 | 0.00046 | 0.00021 | 0.00045 | -0.23 | 29 / 36 (81%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.00034 | 0.00016 | 0.00044 | 0.00063 | 0.00014 | 0.001 | 0.89 | 7 / 11 (64%) | 9 / 11 (82%) | 0.636 | 0.818 | |
| 2.8.1.- | 0.33 | 0.61 | 1 | 0.88 | 1 | 1 | 0.73 | 0.99 | 1 | 0.079 | 0.44 | 1 | 0.82 | 0.98 | 1 | 0.65 | 0.92 | 1 | 0.0015 | 0.0015 | 0.0012 | 0.0014 | 0.0011 | 0.0015 | 0.0013 | 0.00094 | 0.0015 | -0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0017 | 0.0013 | 0.0011 | 0.0014 | 0.0013 | 0.00058 | -0.28 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 2.3.1.79 | Maltose O-acetyltransferase. | 0.43 | 0.69 | 1 | 0.68 | 1 | 1 | 0.6 | 0.96 | 1 | 0.24 | 0.54 | 1 | 0.91 | 0.99 | 1 | 0.93 | 0.98 | 1 | 0.0045 | 0.0045 | 0.0041 | 0.0039 | 0.0045 | 0.0018 | 0.0037 | 0.0035 | 0.0019 | -0.076 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0049 | 0.004 | 0.0034 | 0.0049 | 0.0043 | 0.0024 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.13.- | 0.69 | 0.85 | 1 | 0.87 | 1 | 1 | 0.46 | 0.93 | 1 | 0.72 | 0.89 | 1 | 0.082 | 0.86 | 1 | 0.43 | 0.85 | 1 | 0.014 | 0.014 | 0.013 | 0.014 | 0.014 | 0.0062 | 0.014 | 0.013 | 0.0054 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.013 | 0.013 | 0.0032 | 0.014 | 0.014 | 0.0026 | 0.11 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 2.1.1.271 | Cobalt-precorrin-4 methyltransferase. | 0.3 | 0.58 | 1 | 0.43 | 1 | 1 | 0.064 | 0.72 | 1 | 0.18 | 0.5 | 1 | 0.41 | 0.9 | 1 | 0.11 | 0.66 | 1 | 0.00085 | 0.00073 | 0.00048 | 0.00076 | 0.00048 | 0.00065 | 0.00058 | 0.00059 | 0.00032 | -0.39 | 31 / 36 (86%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00053 | 0.00031 | 0.00085 | 0.001 | 0.00057 | 0.0015 | 0.92 | 9 / 11 (82%) | 9 / 11 (82%) | 0.818 | 0.818 |
| 5.3.4.1 | Protein disulfide-isomerase. | 0.1 | 0.49 | 1 | 0.89 | 1 | 1 | 0.35 | 0.9 | 1 | 0.095 | 0.45 | 1 | 0.85 | 0.98 | 1 | 0.54 | 0.9 | 1 | 3e-04 | 5e-05 | 0 | 0.00011 | 0 | 0.00026 | 9.3e-05 | 0 | 2e-04 | -0.24 | 6 / 36 (17%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0 | 0 | 0 | 3.3e-05 | 0 | 9.8e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 6.2.1.16 | Acetoacetate--CoA ligase. | 0.98 | 0.99 | 1 | 0.34 | 1 | 1 | 0.041 | 0.71 | 1 | 0.81 | 0.93 | 1 | 0.35 | 0.9 | 1 | 0.06 | 0.66 | 1 | 0.00057 | 4e-04 | 0.00012 | 0.00028 | 0.00014 | 0.00041 | 0.00014 | 0 | 0.00026 | -1 | 25 / 36 (69%) | 4 / 7 (57%) | 3 / 7 (43%) | 0.571 | 0.429 | 3e-04 | 9.7e-05 | 0.00062 | 0.00073 | 0.00015 | 0.0013 | 1.3 | 8 / 11 (73%) | 10 / 11 (91%) | 0.727 | 0.909 |
| 3.5.1.31 | Formylmethionine deformylase. | 0.25 | 0.56 | 1 | 0.62 | 1 | 1 | 0.52 | 0.95 | 1 | 0.38 | 0.65 | 1 | 0.88 | 0.99 | 1 | 0.99 | 1 | 1 | 0.00023 | 2.5e-05 | 0 | 6.7e-05 | 0 | 0.00018 | 4.8e-05 | 0 | 0.00013 | -0.48 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 6.8e-06 | 0 | 2.3e-05 | 1.4 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.2.1.45 | Glucosylceramidase. | 0.16 | 0.5 | 1 | 1 | 1 | 1 | 0.99 | 1 | 1 | 0.29 | 0.58 | 1 | 0.82 | 0.98 | 1 | 0.75 | 0.95 | 1 | 0.0016 | 0.0016 | 0.00069 | 0.00063 | 0.00047 | 0.00079 | 0.00059 | 0.00038 | 0.00054 | -0.095 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0025 | 0.0011 | 0.0049 | 0.0019 | 0.0016 | 0.0018 | -0.4 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 3.6.5.3 | Protein-synthesizing GTPase. | 0.31 | 0.59 | 1 | 0.76 | 1 | 1 | 0.93 | 1 | 1 | 0.22 | 0.52 | 1 | 0.43 | 0.91 | 1 | 0.2 | 0.73 | 1 | 0.027 | 0.027 | 0.027 | 0.025 | 0.026 | 0.0025 | 0.025 | 0.029 | 0.0063 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.028 | 0.029 | 0.0061 | 0.027 | 0.027 | 0.0059 | -0.052 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.7.2.4 | Nitrous-oxide reductase. | 0.65 | 0.84 | 1 | 0.21 | 1 | 1 | 0.13 | 0.82 | 1 | 0.76 | 0.91 | 1 | 0.25 | 0.88 | 1 | 0.098 | 0.66 | 1 | 2e-04 | 1.6e-05 | 0 | 1.7e-05 | 0 | 4.5e-05 | 5.8e-05 | 0 | 0.00015 | 1.8 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 6e-06 | 0 | 2e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 1.1.1.81 | Hydroxypyruvate reductase. | 0.84 | 0.93 | 1 | 0.68 | 1 | 1 | 0.46 | 0.93 | 1 | 0.53 | 0.77 | 1 | 0.19 | 0.87 | 1 | 0.58 | 0.91 | 1 | 0.0016 | 0.0014 | 0.0012 | 0.0013 | 0.0011 | 0.0013 | 0.0013 | 0.00048 | 0.0013 | 0 | 33 / 36 (92%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.0013 | 0.0012 | 0.00095 | 0.0017 | 0.0012 | 0.0011 | 0.39 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 4.2.1.36 | Homoaconitate hydratase. | 0.18 | 0.52 | 1 | 0.18 | 1 | 1 | 0.64 | 0.97 | 1 | 0.56 | 0.79 | 1 | 0.35 | 0.9 | 1 | 0.99 | 1 | 1 | 0.00067 | 0.00067 | 0.00051 | 0.00099 | 0.0011 | 0.00062 | 0.00077 | 4e-04 | 0.00091 | -0.36 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00061 | 0.00049 | 0.00037 | 0.00048 | 5e-04 | 0.00022 | -0.35 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.6.2.2 | DNA topoisomerase (ATP-hydrolyzing). | 0.019 | 0.49 | 1 | 0.82 | 1 | 1 | 0.53 | 0.95 | 1 | 0.067 | 0.43 | 1 | 0.32 | 0.9 | 1 | 0.075 | 0.66 | 1 | 0.16 | 0.16 | 0.16 | 0.14 | 0.14 | 0.026 | 0.14 | 0.14 | 0.036 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.17 | 0.17 | 0.036 | 0.16 | 0.16 | 0.02 | -0.087 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.3.46 | Fructose-2.6-bisphosphate 2-phosphatase. | 0.34 | 0.61 | 1 | 0.75 | 1 | 1 | 0.95 | 1 | 1 | 0.38 | 0.65 | 1 | 1 | 1 | 1 | 0.93 | 0.98 | 1 | 0.0016 | 0.0013 | 0.00071 | 0.00063 | 0.00031 | 0.00077 | 0.00082 | 0.00072 | 0.001 | 0.38 | 30 / 36 (83%) | 6 / 7 (86%) | 5 / 7 (71%) | 0.857 | 0.714 | 0.0019 | 0.00061 | 0.0042 | 0.0016 | 0.0011 | 0.0018 | -0.25 | 10 / 11 (91%) | 9 / 11 (82%) | 0.909 | 0.818 |
| 3.6.1.15 | Nucleoside-triphosphate phosphatase. | 0.2 | 0.53 | 1 | 0.52 | 1 | 1 | 0.39 | 0.91 | 1 | 0.025 | 0.42 | 1 | 0.042 | 0.8 | 1 | 0.072 | 0.66 | 1 | 0.0078 | 0.0078 | 0.0037 | 0.0052 | 0.00073 | 0.0078 | 0.0059 | 0.0037 | 0.007 | 0.18 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.0034 | 0.02 | 0.0074 | 0.0071 | 0.0063 | -0.57 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.6.2.3 | 0.99 | 1 | 1 | 0.43 | 1 | 1 | 0.94 | 1 | 1 | 0.42 | 0.68 | 1 | 0.44 | 0.91 | 1 | 0.95 | 0.99 | 1 | 0.0018 | 0.0016 | 0.001 | 0.0017 | 0.00096 | 0.0028 | 0.0022 | 0.0013 | 0.0034 | 0.37 | 32 / 36 (89%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.0012 | 0.00094 | 0.0012 | 0.0015 | 0.0012 | 0.0013 | 0.32 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 | |
| 4.3.1.4 | Formimidoyltetrahydrofolate cyclodeaminase. | 0.097 | 0.49 | 1 | 0.63 | 1 | 1 | 0.53 | 0.95 | 1 | 0.3 | 0.58 | 1 | 0.83 | 0.98 | 1 | 0.94 | 0.98 | 1 | 0.0016 | 0.0016 | 0.0015 | 0.0021 | 0.0019 | 0.0013 | 0.0019 | 0.0021 | 0.00091 | -0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0014 | 0.0013 | 0.00069 | 0.0014 | 0.0015 | 0.00064 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.306 | S-(hydroxymethyl)mycothiol dehydrogenase. | 0.93 | 0.97 | 1 | 0.59 | 1 | 1 | 0.68 | 0.98 | 1 | 0.91 | 0.97 | 1 | 0.71 | 0.95 | 1 | 0.81 | 0.96 | 1 | 9.9e-05 | 2.5e-05 | 0 | 3e-05 | 0 | 6.1e-05 | 1.7e-05 | 0 | 4.5e-05 | -0.82 | 9 / 36 (25%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-05 | 0 | 4.7e-05 | 2.5e-05 | 0 | 4.4e-05 | -0.057 | 3 / 11 (27%) | 3 / 11 (27%) | 0.273 | 0.273 |
| 6.3.2.1 | Pantoate--beta-alanine ligase (AMP-forming). | 0.84 | 0.93 | 1 | 0.11 | 1 | 1 | 0.47 | 0.93 | 1 | 0.52 | 0.76 | 1 | 0.2 | 0.87 | 1 | 0.76 | 0.95 | 1 | 0.0064 | 0.0064 | 0.0063 | 0.0067 | 0.0064 | 0.0025 | 0.0056 | 0.0055 | 0.0027 | -0.26 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0069 | 0.0069 | 0.0021 | 0.0062 | 0.0062 | 0.0016 | -0.15 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.3.2.10 | Soluble epoxide hydrolase. | 0.54 | 0.76 | 1 | 0.2 | 1 | 1 | 0.73 | 0.99 | 1 | 0.9 | 0.97 | 1 | 0.53 | 0.93 | 1 | 0.66 | 0.92 | 1 | 0.0016 | 0.0016 | 0.0012 | 0.0021 | 0.0018 | 0.0017 | 0.0015 | 0.0011 | 0.0011 | -0.49 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0016 | 0.0012 | 0.00081 | 0.0013 | 0.001 | 0.00077 | -0.3 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.3.1.2 | Dihydropyrimidine dehydrogenase (NADP(+)). | 0.21 | 0.53 | 1 | 0.57 | 1 | 1 | 0.92 | 1 | 1 | 0.31 | 0.59 | 1 | 0.082 | 0.86 | 1 | 0.25 | 0.76 | 1 | 0.0024 | 0.0017 | 0.00052 | 0.00077 | 0.00033 | 9e-04 | 0.00069 | 0 | 0.0011 | -0.16 | 26 / 36 (72%) | 5 / 7 (71%) | 3 / 7 (43%) | 0.714 | 0.429 | 0.003 | 9e-04 | 0.0058 | 0.0017 | 6e-04 | 0.0021 | -0.82 | 9 / 11 (82%) | 9 / 11 (82%) | 0.818 | 0.818 |
| 1.5.3.- | 0.9 | 0.96 | 1 | 0.27 | 1 | 1 | 0.15 | 0.84 | 1 | 0.69 | 0.87 | 1 | 0.38 | 0.9 | 1 | 0.15 | 0.71 | 1 | 0.00019 | 2.7e-05 | 0 | 1.7e-05 | 0 | 4.5e-05 | 8.7e-05 | 0 | 0.00023 | 2.4 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2e-05 | 0 | 5e-05 | 2.7e-06 | 0 | 8.9e-06 | -2.9 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 | |
| 3.2.1.8 | Endo-1.4-beta-xylanase. | 0.24 | 0.55 | 1 | 0.5 | 1 | 1 | 0.34 | 0.89 | 1 | 0.14 | 0.47 | 1 | 0.091 | 0.86 | 1 | 0.71 | 0.94 | 1 | 0.023 | 0.023 | 0.025 | 0.018 | 0.018 | 0.0079 | 0.02 | 0.02 | 0.0081 | 0.15 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.023 | 0.026 | 0.009 | 0.028 | 0.029 | 0.0058 | 0.28 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.39 | Malate dehydrogenase (decarboxylating). | 0.18 | 0.52 | 1 | 0.57 | 1 | 1 | 0.26 | 0.89 | 1 | 0.23 | 0.53 | 1 | 0.66 | 0.95 | 1 | 0.33 | 0.8 | 1 | 0.00027 | 7.5e-05 | 0 | 0.00015 | 0 | 0.00035 | 0.00015 | 0 | 0.00038 | 0 | 10 / 36 (28%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 1.5e-05 | 0 | 3.9e-05 | 3.6e-05 | 0 | 7.3e-05 | 1.3 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 2.4.2.45 | Decaprenyl-phosphate phosphoribosyltransferase. | 0.18 | 0.52 | 1 | 0.62 | 1 | 1 | 0.33 | 0.89 | 1 | 0.29 | 0.58 | 1 | 0.8 | 0.97 | 1 | 0.64 | 0.92 | 1 | 0.00067 | 0.00022 | 0 | 0.00038 | 0 | 0.00062 | 3e-04 | 0 | 0.00065 | -0.34 | 12 / 36 (33%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 4e-05 | 0 | 8.5e-05 | 0.00025 | 0 | 0.00078 | 2.6 | 3 / 11 (27%) | 3 / 11 (27%) | 0.273 | 0.273 |
| 2.3.1.81 | Aminoglycoside N(3)-acetyltransferase. | 0.58 | 0.79 | 1 | 0.62 | 1 | 1 | 0.95 | 1 | 1 | 0.79 | 0.92 | 1 | 0.46 | 0.92 | 1 | 0.68 | 0.93 | 1 | 0.00081 | 0.00081 | 0.00056 | 0.00079 | 0.00073 | 0.00055 | 0.00096 | 0.00072 | 7e-04 | 0.28 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00061 | 0.00049 | 0.00041 | 0.00091 | 0.00045 | 0.0011 | 0.58 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.1.24 | Choloylglycine hydrolase. | 0.066 | 0.49 | 1 | 0.76 | 1 | 1 | 0.33 | 0.89 | 1 | 0.085 | 0.44 | 1 | 0.096 | 0.86 | 1 | 0.017 | 0.55 | 1 | 0.015 | 0.015 | 0.014 | 0.013 | 0.013 | 0.0024 | 0.014 | 0.013 | 0.0038 | 0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.017 | 0.018 | 0.0045 | 0.015 | 0.015 | 0.0038 | -0.18 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.1.104 | Cyanase. | 0.23 | 0.54 | 1 | 0.83 | 1 | 1 | 0.91 | 1 | 1 | 0.19 | 0.5 | 1 | 0.94 | 0.99 | 1 | 0.71 | 0.94 | 1 | 0.00026 | 8.5e-05 | 0 | 4.6e-05 | 0 | 0.00012 | 3.1e-05 | 0 | 8.2e-05 | -0.57 | 12 / 36 (33%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0.00012 | 0 | 2e-04 | 0.00011 | 0 | 0.00019 | -0.13 | 5 / 11 (45%) | 5 / 11 (45%) | 0.455 | 0.455 |
| 3.4.21.19 | Glutamyl endopeptidase. | 0.12 | 0.5 | 1 | 1 | 1 | 1 | 0.39 | 0.91 | 1 | 0.067 | 0.43 | 1 | 0.81 | 0.98 | 1 | 0.29 | 0.78 | 1 | 0.00012 | 1e-05 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 3 / 36 (8.3%) | 0 / 7 (0%) | 0 / 7 (0%) | 0 | 0 | 3.1e-05 | 0 | 7.9e-05 | 3e-06 | 0 | 1e-05 | -3.4 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 2.2.1.6 | Acetolactate synthase. | 0.52 | 0.75 | 1 | 0.78 | 1 | 1 | 0.96 | 1 | 1 | 0.36 | 0.63 | 1 | 0.4 | 0.9 | 1 | 0.23 | 0.75 | 1 | 0.056 | 0.056 | 0.059 | 0.055 | 0.059 | 0.0089 | 0.054 | 0.059 | 0.012 | -0.026 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.058 | 0.061 | 0.012 | 0.056 | 0.055 | 0.0085 | -0.051 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.1.51 | Prephenate dehydratase. | 0.055 | 0.49 | 1 | 0.81 | 1 | 1 | 0.95 | 1 | 1 | 0.033 | 0.43 | 1 | 0.72 | 0.95 | 1 | 0.58 | 0.91 | 1 | 0.015 | 0.015 | 0.014 | 0.012 | 0.011 | 0.002 | 0.012 | 0.012 | 0.0042 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.017 | 0.015 | 0.0087 | 0.016 | 0.015 | 0.003 | -0.087 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.5.1.29 | Transferred entry: 1.5.1.38. 1.5.1.39 and 1.5.1.41. | 0.31 | 0.59 | 1 | 0.86 | 1 | 1 | 0.89 | 1 | 1 | 0.49 | 0.74 | 1 | 0.83 | 0.98 | 1 | 0.81 | 0.96 | 1 | 0.00056 | 6.2e-05 | 0 | 0.00015 | 0 | 4e-04 | 0.00014 | 0 | 0.00036 | -0.1 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 1.1e-05 | 0 | 3.6e-05 | 1.1e-05 | 0 | 3.6e-05 | 0 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.3.1.46 | Homoserine O-succinyltransferase. | 0.84 | 0.93 | 1 | 0.68 | 1 | 1 | 0.52 | 0.95 | 1 | 0.89 | 0.96 | 1 | 0.65 | 0.95 | 1 | 0.83 | 0.96 | 1 | 0.0011 | 0.0011 | 0.00083 | 0.00087 | 0.00065 | 0.00086 | 0.00097 | 0.00084 | 0.00074 | 0.16 | 34 / 36 (94%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00098 | 0.00077 | 0.00095 | 0.0014 | 0.0012 | 0.001 | 0.51 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 3.6.1.25 | Triphosphatase. | 0.058 | 0.49 | 1 | 0.018 | 0.92 | 1 | 0.042 | 0.71 | 1 | 0.055 | 0.43 | 1 | 0.078 | 0.86 | 1 | 0.14 | 0.69 | 1 | 2e-04 | 4.5e-05 | 0 | 0.00014 | 0 | 0.00031 | 6.8e-05 | 0 | 0.00018 | -1 | 8 / 36 (22%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 5.5e-06 | 0 | 1.2e-05 | 9.2e-06 | 0 | 2.2e-05 | 0.74 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 4.1.1.101 | Malolactic enzyme. | 0.26 | 0.56 | 1 | 0.41 | 1 | 1 | 0.32 | 0.89 | 1 | 0.18 | 0.5 | 1 | 0.23 | 0.88 | 1 | 0.14 | 0.7 | 1 | 0.00013 | 1.4e-05 | 0 | 0 | 0 | 0 | 7.7e-06 | 0 | 2e-05 | Inf | 4 / 36 (11%) | 0 / 7 (0%) | 1 / 7 (14%) | 0 | 0.143 | 2.1e-05 | 0 | 4.6e-05 | 2.1e-05 | 0 | 6.8e-05 | 0 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 6.3.5.3 | Phosphoribosylformylglycinamidine synthase. | 0.3 | 0.58 | 1 | 0.9 | 1 | 1 | 0.85 | 1 | 1 | 0.24 | 0.53 | 1 | 0.21 | 0.88 | 1 | 0.2 | 0.73 | 1 | 0.052 | 0.052 | 0.052 | 0.049 | 0.047 | 0.0078 | 0.05 | 0.053 | 0.012 | 0.029 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.054 | 0.055 | 0.014 | 0.053 | 0.052 | 0.0063 | -0.027 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.6.1.12 | dCTP diphosphatase. | 0.27 | 0.56 | 1 | 0.27 | 1 | 1 | 0.8 | 1 | 1 | 0.24 | 0.53 | 1 | 0.23 | 0.88 | 1 | 0.75 | 0.95 | 1 | 0.00022 | 1.9e-05 | 0 | 3.4e-05 | 0 | 8.9e-05 | 5.8e-05 | 0 | 0.00015 | 0.77 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 3.1.2.28 | 1.4-dihydroxy-2-naphthoyl-CoA hydrolase. | 0.053 | 0.49 | 1 | 0.83 | 1 | 1 | 0.69 | 0.98 | 1 | 0.05 | 0.43 | 1 | 0.86 | 0.98 | 1 | 0.71 | 0.94 | 1 | 8.8e-05 | 1.9e-05 | 0 | 4.1e-05 | 0 | 6e-05 | 4.7e-05 | 0 | 8.3e-05 | 0.2 | 8 / 36 (22%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 2.6e-06 | 0 | 8.5e-06 | 5.7e-06 | 0 | 1.3e-05 | 1.1 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 5.1.3.22 | L-ribulose-5-phosphate 3-epimerase. | 0.81 | 0.91 | 1 | 0.57 | 1 | 1 | 0.72 | 0.98 | 1 | 0.83 | 0.94 | 1 | 0.26 | 0.88 | 1 | 0.29 | 0.78 | 1 | 0.0013 | 0.0013 | 0.0011 | 0.0013 | 0.0013 | 0.00067 | 0.0013 | 0.0011 | 0.00099 | 0 | 35 / 36 (97%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.0013 | 0.0012 | 0.00099 | 0.0012 | 0.00098 | 0.00066 | -0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 7.6.2.10 | ABC-type glycerol 3-phosphate transporter. | 0.42 | 0.68 | 1 | 0.14 | 1 | 1 | 0.43 | 0.93 | 1 | 0.44 | 0.69 | 1 | 0.14 | 0.87 | 1 | 0.61 | 0.91 | 1 | 0.00027 | 3.7e-05 | 0 | 5e-05 | 0 | 0.00013 | 0.00013 | 0 | 0.00033 | 1.4 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 5.7e-06 | 0 | 1.3e-05 | 1.1 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 1.18.-.- | 0.15 | 0.5 | 1 | 0.81 | 1 | 1 | 0.96 | 1 | 1 | 0.13 | 0.47 | 1 | 0.58 | 0.93 | 1 | 0.71 | 0.94 | 1 | 0.0012 | 0.0011 | 0.00058 | 0.00056 | 0.00034 | 5e-04 | 0.00067 | 0.00057 | 0.00066 | 0.26 | 34 / 36 (94%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.0013 | 0.00097 | 0.0013 | 0.0016 | 0.0011 | 0.0019 | 0.3 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 2.7.1.113 | Deoxyguanosine kinase. | 0.43 | 0.69 | 1 | 0.21 | 1 | 1 | 0.17 | 0.85 | 1 | 0.24 | 0.53 | 1 | 0.2 | 0.87 | 1 | 0.078 | 0.66 | 1 | 0.00017 | 7.1e-05 | 0 | 4.4e-05 | 0 | 1e-04 | 0.00011 | 0 | 0.00018 | 1.3 | 15 / 36 (42%) | 2 / 7 (29%) | 3 / 7 (43%) | 0.286 | 0.429 | 5.6e-05 | 3.3e-05 | 5.8e-05 | 7.8e-05 | 0 | 0.00017 | 0.48 | 7 / 11 (64%) | 3 / 11 (27%) | 0.636 | 0.273 |
| 2.7.7.2 | FAD synthase. | 0.16 | 0.5 | 1 | 0.65 | 1 | 1 | 0.87 | 1 | 1 | 0.18 | 0.5 | 1 | 0.069 | 0.86 | 1 | 0.11 | 0.66 | 1 | 0.013 | 0.013 | 0.013 | 0.012 | 0.012 | 0.002 | 0.012 | 0.014 | 0.0027 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.013 | 0.013 | 0.0023 | 0.013 | 0.013 | 0.0015 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.1.105 | Chitin disaccharide deacetylase. | 0.82 | 0.92 | 1 | 0.36 | 1 | 1 | 0.69 | 0.98 | 1 | 0.97 | 0.99 | 1 | 0.23 | 0.88 | 1 | 0.36 | 0.81 | 1 | 0.00029 | 1e-04 | 0 | 9.1e-05 | 0 | 0.00018 | 0.00014 | 3.1e-05 | 0.00017 | 0.62 | 13 / 36 (36%) | 2 / 7 (29%) | 4 / 7 (57%) | 0.286 | 0.571 | 5.1e-05 | 0 | 1e-04 | 0.00014 | 0 | 0.00037 | 1.5 | 4 / 11 (36%) | 3 / 11 (27%) | 0.364 | 0.273 |
| 6.2.1.27 | 4-hydroxybenzoate--CoA ligase. | 0.24 | 0.55 | 1 | 0.81 | 1 | 1 | 0.57 | 0.96 | 1 | 0.27 | 0.56 | 1 | 0.86 | 0.99 | 1 | 0.44 | 0.85 | 1 | 0.00028 | 0.00019 | 8.4e-05 | 0.00022 | 0.00014 | 0.00025 | 0.00031 | 6.2e-05 | 0.00045 | 0.49 | 24 / 36 (67%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 9.8e-05 | 0 | 0.00015 | 0.00017 | 0.00011 | 0.00028 | 0.79 | 5 / 11 (45%) | 9 / 11 (82%) | 0.455 | 0.818 |
| 1.1.1.317 | Perakine reductase. | 0.087 | 0.49 | 1 | 0.45 | 1 | 1 | 0.45 | 0.93 | 1 | 0.098 | 0.46 | 1 | 0.16 | 0.87 | 1 | 0.18 | 0.72 | 1 | 0.0066 | 0.0055 | 0.0032 | 0.0018 | 0.0012 | 0.0019 | 0.0031 | 0.0019 | 0.0031 | 0.78 | 30 / 36 (83%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.0084 | 0.0046 | 0.015 | 0.0065 | 0.0042 | 0.0062 | -0.37 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 1.16.1.1 | Mercury(II) reductase. | 0.91 | 0.96 | 1 | 0.059 | 1 | 1 | 0.47 | 0.93 | 1 | 0.75 | 0.9 | 1 | 0.039 | 0.8 | 1 | 0.15 | 0.71 | 1 | 0.0024 | 0.0024 | 0.0017 | 0.0021 | 0.0013 | 0.0016 | 0.003 | 0.0022 | 0.0025 | 0.51 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.002 | 0.0015 | 0.0017 | 0.0025 | 0.0024 | 0.002 | 0.32 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.272 | D-2-hydroxyacid dehydrogenase (NADP(+)). | 0.73 | 0.87 | 1 | 0.43 | 1 | 1 | 0.42 | 0.92 | 1 | 0.73 | 0.89 | 1 | 0.35 | 0.9 | 1 | 0.27 | 0.77 | 1 | 0.0021 | 0.0021 | 0.0015 | 0.0019 | 0.0014 | 0.0012 | 0.0017 | 0.0012 | 0.0013 | -0.16 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0023 | 0.0025 | 0.0015 | 0.0023 | 0.0016 | 0.0013 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.53 | L-xylulokinase. | 0.81 | 0.91 | 1 | 0.85 | 1 | 1 | 0.58 | 0.96 | 1 | 0.91 | 0.97 | 1 | 0.76 | 0.97 | 1 | 0.31 | 0.79 | 1 | 0.00074 | 0.00068 | 0.00054 | 0.00065 | 6e-04 | 0.00056 | 0.00073 | 0.00063 | 0.00069 | 0.17 | 33 / 36 (92%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00062 | 0.00023 | 0.00065 | 0.00071 | 0.00047 | 0.00051 | 0.2 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 4.3.1.1 | Aspartate ammonia-lyase. | 0.52 | 0.75 | 1 | 0.87 | 1 | 1 | 0.98 | 1 | 1 | 0.94 | 0.98 | 1 | 0.57 | 0.93 | 1 | 0.58 | 0.91 | 1 | 0.0077 | 0.0077 | 0.0077 | 0.0082 | 0.0086 | 0.0027 | 0.0086 | 0.009 | 0.0046 | 0.069 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0072 | 0.0072 | 0.0014 | 0.0074 | 0.0067 | 0.0018 | 0.04 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.6.1.1 | Magnesium chelatase. | 0.29 | 0.57 | 1 | 0.82 | 1 | 1 | 0.86 | 1 | 1 | 0.59 | 0.81 | 1 | 0.98 | 0.99 | 1 | 0.36 | 0.81 | 1 | 0.0018 | 0.0018 | 0.0011 | 0.00091 | 0.00045 | 9e-04 | 0.00078 | 0.00043 | 0.00078 | -0.22 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0027 | 0.0011 | 0.0051 | 0.002 | 0.0012 | 0.002 | -0.43 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 3.5.4.10 | IMP cyclohydrolase. | 0.14 | 0.5 | 1 | 0.46 | 1 | 1 | 0.5 | 0.95 | 1 | 0.077 | 0.44 | 1 | 0.96 | 0.99 | 1 | 0.98 | 1 | 1 | 0.015 | 0.015 | 0.014 | 0.013 | 0.012 | 0.0028 | 0.012 | 0.012 | 0.0038 | -0.12 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.016 | 0.014 | 0.0069 | 0.016 | 0.016 | 0.0034 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.8.4.14 | L-methionine (R)-S-oxide reductase. | 0.43 | 0.69 | 1 | 0.8 | 1 | 1 | 0.85 | 1 | 1 | 0.54 | 0.77 | 1 | 0.44 | 0.91 | 1 | 0.71 | 0.94 | 1 | 0.00072 | 7e-04 | 0.00054 | 0.00076 | 0.00069 | 0.00036 | 0.00083 | 0.00084 | 0.00064 | 0.13 | 35 / 36 (97%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 6e-04 | 0.00036 | 0.00047 | 0.00067 | 0.00043 | 0.00077 | 0.16 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.6.1.51 | Serine--pyruvate transaminase. | 0.31 | 0.59 | 1 | 1 | 1 | 1 | 0.58 | 0.96 | 1 | 0.21 | 0.52 | 1 | 0.83 | 0.98 | 1 | 0.74 | 0.94 | 1 | 9.4e-05 | 1.3e-05 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 5 / 36 (14%) | 0 / 7 (0%) | 0 / 7 (0%) | 0 | 0 | 1.5e-05 | 0 | 3.9e-05 | 2.8e-05 | 0 | 5e-05 | 0.9 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 4.2.1.2 | Fumarate hydratase. | 0.41 | 0.67 | 1 | 0.94 | 1 | 1 | 0.41 | 0.92 | 1 | 0.2 | 0.5 | 1 | 0.3 | 0.9 | 1 | 0.72 | 0.94 | 1 | 0.018 | 0.018 | 0.018 | 0.017 | 0.017 | 0.003 | 0.017 | 0.016 | 0.0042 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.018 | 0.018 | 0.0039 | 0.019 | 0.02 | 0.0022 | 0.078 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.3.1.14 | Diphthine--ammonia ligase. | 0.9 | 0.96 | 1 | 0.77 | 1 | 1 | 0.41 | 0.92 | 1 | 0.72 | 0.89 | 1 | 0.84 | 0.98 | 1 | 0.34 | 0.8 | 1 | 0.00018 | 3.4e-05 | 0 | 3.6e-05 | 0 | 5.7e-05 | 3.1e-05 | 0 | 5.5e-05 | -0.22 | 7 / 36 (19%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 6.9e-05 | 0 | 0.00018 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 2.1.1.342 | Anaerobilin synthase. | 0.056 | 0.49 | 1 | 0.021 | 0.92 | 1 | 0.026 | 0.71 | 1 | 0.02 | 0.42 | 1 | 0.1 | 0.86 | 1 | 0.11 | 0.66 | 1 | 0.00038 | 5.2e-05 | 0 | 0.00015 | 0 | 0.00035 | 0.00012 | 0 | 0.00031 | -0.32 | 5 / 36 (14%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 4.1.99.2 | Tyrosine phenol-lyase. | 0.12 | 0.5 | 1 | 0.53 | 1 | 1 | 0.52 | 0.95 | 1 | 0.073 | 0.43 | 1 | 0.89 | 0.99 | 1 | 0.21 | 0.74 | 1 | 0.00069 | 5e-04 | 0.00022 | 0.00049 | 0.00069 | 0.00035 | 0.001 | 0.00051 | 0.0017 | 1 | 26 / 36 (72%) | 6 / 7 (86%) | 5 / 7 (71%) | 0.857 | 0.714 | 0.00018 | 2.8e-05 | 0.00028 | 5e-04 | 0.00022 | 0.00064 | 1.5 | 6 / 11 (55%) | 9 / 11 (82%) | 0.545 | 0.818 |
| 6.3.5.7 | Glutaminyl-tRNA synthase (glutamine-hydrolyzing). | 0.066 | 0.49 | 1 | 0.3 | 1 | 1 | 0.21 | 0.87 | 1 | 0.1 | 0.46 | 1 | 0.096 | 0.86 | 1 | 0.063 | 0.66 | 1 | 0.019 | 0.019 | 0.019 | 0.016 | 0.016 | 0.0031 | 0.018 | 0.019 | 0.0056 | 0.17 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.02 | 0.021 | 0.0064 | 0.019 | 0.02 | 0.0046 | -0.074 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 7.2.2.16 | ABC-type ferric hydroxamate transporter. | 0.12 | 0.5 | 1 | 0.29 | 1 | 1 | 0.061 | 0.71 | 1 | 0.12 | 0.47 | 1 | 0.67 | 0.95 | 1 | 0.16 | 0.71 | 1 | 0.00027 | 3.8e-05 | 0 | 1e-04 | 0 | 0.00027 | 6.8e-05 | 0 | 0.00018 | -0.56 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 1.6e-05 | 0 | 3.7e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 2.1.1.210 | Demethylspheroidene O-methyltransferase. | 0.54 | 0.76 | 1 | 0.36 | 1 | 1 | 0.52 | 0.95 | 1 | 0.19 | 0.5 | 1 | 0.22 | 0.88 | 1 | 0.24 | 0.76 | 1 | 0.00038 | 0.00017 | 0 | 0.00017 | 0.00014 | 0.00018 | 8.3e-05 | 0 | 0.00016 | -1 | 16 / 36 (44%) | 5 / 7 (71%) | 3 / 7 (43%) | 0.714 | 0.429 | 0.00024 | 0 | 0.00061 | 0.00015 | 0 | 0.00037 | -0.68 | 3 / 11 (27%) | 5 / 11 (45%) | 0.273 | 0.455 |
| 2.4.1.227 | Undecaprenyldiphospho-muramoylpentapeptide beta-N- | 0.049 | 0.49 | 1 | 0.63 | 1 | 1 | 0.27 | 0.89 | 1 | 0.084 | 0.44 | 1 | 0.49 | 0.92 | 1 | 0.049 | 0.65 | 1 | 0.018 | 0.018 | 0.017 | 0.017 | 0.016 | 0.0037 | 0.016 | 0.016 | 0.0039 | -0.087 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.02 | 0.02 | 0.0044 | 0.018 | 0.017 | 0.0023 | -0.15 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.2.1.36 | Retinal dehydrogenase. | 0.19 | 0.53 | 1 | 0.95 | 1 | 1 | 0.38 | 0.9 | 1 | 0.19 | 0.5 | 1 | 0.5 | 0.93 | 1 | 0.88 | 0.97 | 1 | 0.00056 | 4.6e-05 | 0 | 0.00012 | 0 | 0.00031 | 0.00012 | 0 | 0.00031 | 0 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.7.1.172 | Protein-ribulosamine 3-kinase. | 0.19 | 0.53 | 1 | 0.83 | 1 | 1 | 0.99 | 1 | 1 | 0.15 | 0.47 | 1 | 0.57 | 0.93 | 1 | 0.56 | 0.9 | 1 | 0.0018 | 0.0014 | 0.00066 | 7e-04 | 2e-04 | 0.00088 | 0.00047 | 0.00054 | 0.00047 | -0.57 | 27 / 36 (75%) | 4 / 7 (57%) | 5 / 7 (71%) | 0.571 | 0.714 | 0.0021 | 0.00097 | 0.0041 | 0.0016 | 9e-04 | 0.0017 | -0.39 | 9 / 11 (82%) | 9 / 11 (82%) | 0.818 | 0.818 |
| 6.3.4.15 | Biotin--[biotin carboxyl-carrier protein] ligase. | 0.57 | 0.78 | 1 | 0.81 | 1 | 1 | 0.99 | 1 | 1 | 0.4 | 0.66 | 1 | 0.19 | 0.87 | 1 | 0.32 | 0.8 | 1 | 0.018 | 0.018 | 0.018 | 0.017 | 0.016 | 0.0025 | 0.017 | 0.016 | 0.0062 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.018 | 0.019 | 0.0047 | 0.018 | 0.018 | 0.0026 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.1.186 | 23S rRNA (cytidine(2498)-2'-O)-methyltransferase. | 0.33 | 0.6 | 1 | 0.28 | 1 | 1 | 0.95 | 1 | 1 | 0.3 | 0.58 | 1 | 0.43 | 0.91 | 1 | 0.63 | 0.92 | 1 | 0.00033 | 3.6e-05 | 0 | 5e-05 | 0 | 0.00013 | 0.00012 | 0 | 0.00031 | 1.3 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 1.3e-05 | 0 | 3.5e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 1.3.1.14 | Dihydroorotate dehydrogenase (NAD(+)). | 0.1 | 0.49 | 1 | 0.58 | 1 | 1 | 0.89 | 1 | 1 | 0.13 | 0.47 | 1 | 0.32 | 0.9 | 1 | 0.8 | 0.96 | 1 | 0.0026 | 0.0025 | 0.0014 | 0.00084 | 0.0011 | 0.00051 | 0.0013 | 0.0011 | 0.0013 | 0.63 | 34 / 36 (94%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.0035 | 0.0015 | 0.0064 | 0.0033 | 0.0031 | 0.0021 | -0.085 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 3.4.24.11 | Neprilysin. | 0.16 | 0.5 | 1 | 0.73 | 1 | 1 | 0.73 | 0.99 | 1 | 0.19 | 0.5 | 1 | 0.22 | 0.88 | 1 | 0.63 | 0.92 | 1 | 0.0047 | 0.0042 | 0.0024 | 0.0019 | 0.00091 | 0.0021 | 0.0023 | 0.0016 | 0.0026 | 0.28 | 32 / 36 (89%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.0065 | 0.0031 | 0.013 | 0.0045 | 0.0036 | 0.004 | -0.53 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 4.1.2.29 | 5-dehydro-2-deoxyphosphogluconate aldolase. | 0.37 | 0.64 | 1 | 0.48 | 1 | 1 | 0.069 | 0.73 | 1 | 0.31 | 0.58 | 1 | 0.63 | 0.95 | 1 | 0.076 | 0.66 | 1 | 0.00046 | 0.00046 | 0.00029 | 4e-04 | 0.00028 | 0.00029 | 0.00062 | 3e-04 | 0.00086 | 0.63 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00056 | 0.00035 | 0.00038 | 0.00031 | 0.00027 | 0.00019 | -0.85 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.1.28 | Chloramphenicol O-acetyltransferase. | 0.91 | 0.96 | 1 | 0.56 | 1 | 1 | 0.39 | 0.91 | 1 | 0.92 | 0.97 | 1 | 0.62 | 0.95 | 1 | 0.67 | 0.93 | 1 | 0.0025 | 0.0025 | 0.0019 | 0.0024 | 0.0026 | 0.00093 | 0.0021 | 0.0017 | 0.0012 | -0.19 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0024 | 0.0018 | 0.0014 | 0.003 | 0.0017 | 0.0031 | 0.32 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.1.96 | Carboxynorspermidine decarboxylase. | 0.57 | 0.78 | 1 | 0.67 | 1 | 1 | 0.27 | 0.89 | 1 | 0.99 | 1 | 1 | 0.91 | 0.99 | 1 | 0.49 | 0.88 | 1 | 0.007 | 0.007 | 0.0065 | 0.0072 | 0.0061 | 0.0027 | 0.0068 | 0.0058 | 0.0027 | -0.082 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0066 | 0.0059 | 0.0025 | 0.0075 | 0.0076 | 0.0017 | 0.18 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.6.1.57 | UDP-2.4-diacetamido-2.4.6-trideoxy-beta-L-altropyranose hydrolase. | 0.0056 | 0.49 | 1 | 0.34 | 1 | 1 | 0.1 | 0.82 | 1 | 0.0051 | 0.36 | 1 | 0.19 | 0.87 | 1 | 0.08 | 0.66 | 1 | 0.00049 | 0.00037 | 0.00023 | 0.00064 | 0.00037 | 0.00056 | 0.00069 | 0.00024 | 0.00099 | 0.11 | 27 / 36 (75%) | 7 / 7 (100%) | 5 / 7 (71%) | 1 | 0.714 | 0.00011 | 7.2e-05 | 0.00014 | 0.00026 | 0.00023 | 0.00024 | 1.2 | 7 / 11 (64%) | 8 / 11 (73%) | 0.636 | 0.727 |
| 2.7.7.80 | Molybdopterin-synthase adenylyltransferase. | 0.035 | 0.49 | 1 | 0.47 | 1 | 1 | 0.26 | 0.89 | 1 | 0.029 | 0.43 | 1 | 0.33 | 0.9 | 1 | 0.19 | 0.73 | 1 | 0.0043 | 0.0043 | 0.0037 | 0.0032 | 0.0028 | 0.0014 | 0.0038 | 0.0031 | 0.0023 | 0.25 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0052 | 0.0044 | 0.0032 | 0.0044 | 0.0046 | 0.0013 | -0.24 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.21.62 | Subtilisin. | 0.67 | 0.85 | 1 | 0.31 | 1 | 1 | 0.3 | 0.89 | 1 | 0.91 | 0.97 | 1 | 0.4 | 0.9 | 1 | 0.33 | 0.8 | 1 | 0.0062 | 0.0062 | 0.0048 | 0.0072 | 0.0046 | 0.0069 | 0.0054 | 0.0048 | 0.0039 | -0.42 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0061 | 0.0048 | 0.0045 | 0.0063 | 0.0054 | 0.0034 | 0.047 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.13.12.16 | Nitronate monooxygenase. | 0.25 | 0.55 | 1 | 0.44 | 1 | 1 | 0.72 | 0.98 | 1 | 0.27 | 0.56 | 1 | 0.5 | 0.93 | 1 | 0.81 | 0.96 | 1 | 0.011 | 0.011 | 0.011 | 0.01 | 0.01 | 0.0022 | 0.0094 | 0.0086 | 0.005 | -0.089 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.013 | 0.013 | 0.0051 | 0.012 | 0.012 | 0.0028 | -0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.1.24 | Porphobilinogen synthase. | 0.11 | 0.5 | 1 | 0.6 | 1 | 1 | 0.2 | 0.87 | 1 | 0.38 | 0.65 | 1 | 0.75 | 0.97 | 1 | 0.56 | 0.9 | 1 | 0.0065 | 0.0065 | 0.0069 | 0.0072 | 0.007 | 0.0017 | 0.0069 | 0.007 | 0.0019 | -0.061 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0058 | 0.0059 | 0.0022 | 0.0064 | 0.0068 | 0.0019 | 0.14 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.3.99.2 | Transferred entry: 1.3.8.1. | 0.66 | 0.84 | 1 | 0.83 | 1 | 1 | 0.24 | 0.89 | 1 | 0.94 | 0.98 | 1 | 0.93 | 0.99 | 1 | 0.36 | 0.81 | 1 | 0.0097 | 0.0097 | 0.009 | 0.0093 | 0.0085 | 0.0027 | 0.0095 | 0.007 | 0.0056 | 0.031 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0087 | 0.0087 | 0.0033 | 0.011 | 0.01 | 0.0045 | 0.34 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.1.104 | Caffeoyl-CoA O-methyltransferase. | 0.0076 | 0.49 | 1 | 0.03 | 0.97 | 1 | 0.0084 | 0.61 | 1 | 0.005 | 0.36 | 1 | 0.008 | 0.71 | 1 | 0.0024 | 0.48 | 1 | 0.00021 | 0.00011 | 4.7e-05 | 9.2e-06 | 0 | 2.4e-05 | 0.00013 | 6.8e-05 | 0.00017 | 3.8 | 19 / 36 (53%) | 1 / 7 (14%) | 4 / 7 (57%) | 0.143 | 0.571 | 0.00019 | 9.8e-05 | 0.00026 | 8e-05 | 2.9e-05 | 9.2e-05 | -1.2 | 8 / 11 (73%) | 6 / 11 (55%) | 0.727 | 0.545 |
| 4.2.99.20 | 2-succinyl-6-hydroxy-2.4-cyclohexadiene-1-carboxylate synthase. | 0.29 | 0.58 | 1 | 0.15 | 1 | 1 | 0.13 | 0.82 | 1 | 0.36 | 0.63 | 1 | 0.26 | 0.88 | 1 | 0.3 | 0.78 | 1 | 0.00015 | 9e-05 | 5.1e-05 | 0.00013 | 0.00011 | 0.00016 | 5.3e-05 | 5.4e-05 | 5.7e-05 | -1.3 | 21 / 36 (58%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 6.4e-05 | 3.3e-05 | 8.7e-05 | 0.00011 | 2.9e-05 | 0.00021 | 0.78 | 6 / 11 (55%) | 6 / 11 (55%) | 0.545 | 0.545 |
| 1.1.1.264 | L-idonate 5-dehydrogenase (NAD(P)(+)). | 0.41 | 0.67 | 1 | 0.045 | 0.97 | 1 | 0.057 | 0.71 | 1 | 0.76 | 0.91 | 1 | 0.028 | 0.76 | 1 | 0.031 | 0.62 | 1 | 0.00047 | 9.1e-05 | 0 | 0.00019 | 0 | 0.00049 | 0.00023 | 0 | 0.00053 | 0.28 | 7 / 36 (19%) | 1 / 7 (14%) | 3 / 7 (43%) | 0.143 | 0.429 | 1.8e-05 | 0 | 4.2e-05 | 1.3e-05 | 0 | 4.5e-05 | -0.47 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 2.7.1.90 | Diphosphate--fructose-6-phosphate 1-phosphotransferase. | 0.12 | 0.5 | 1 | 0.35 | 1 | 1 | 0.17 | 0.85 | 1 | 0.66 | 0.86 | 1 | 0.85 | 0.98 | 1 | 0.49 | 0.88 | 1 | 0.021 | 0.021 | 0.021 | 0.022 | 0.023 | 0.0043 | 0.02 | 0.02 | 0.0057 | -0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.019 | 0.021 | 0.0042 | 0.021 | 0.02 | 0.0039 | 0.14 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.6 | Galactokinase. | 0.97 | 0.99 | 1 | 0.89 | 1 | 1 | 0.32 | 0.89 | 1 | 0.69 | 0.88 | 1 | 0.66 | 0.95 | 1 | 0.77 | 0.95 | 1 | 0.017 | 0.017 | 0.018 | 0.016 | 0.017 | 0.0034 | 0.016 | 0.017 | 0.006 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.016 | 0.017 | 0.0044 | 0.019 | 0.019 | 0.0053 | 0.25 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.3.11 | N-succinylornithine carbamoyltransferase. | 0.058 | 0.49 | 1 | 0.43 | 1 | 1 | 0.72 | 0.98 | 1 | 0.013 | 0.42 | 1 | 0.99 | 1 | 1 | 0.44 | 0.85 | 1 | 0.00049 | 0.00025 | 1.4e-05 | 0.00024 | 0.00024 | 0.00023 | 0.00057 | 0.00019 | 9e-04 | 1.2 | 18 / 36 (50%) | 6 / 7 (86%) | 5 / 7 (71%) | 0.857 | 0.714 | 4.1e-05 | 0 | 9.9e-05 | 0.00025 | 0 | 0.00047 | 2.6 | 3 / 11 (27%) | 4 / 11 (36%) | 0.273 | 0.364 |
| 5.99.1.3 | Transferred entry: 5.6.2.2. | 0.91 | 0.96 | 1 | 0.26 | 1 | 1 | 0.12 | 0.82 | 1 | 0.83 | 0.94 | 1 | 0.22 | 0.88 | 1 | 0.11 | 0.66 | 1 | 0.011 | 0.011 | 0.0087 | 0.01 | 0.012 | 0.004 | 0.0079 | 0.007 | 0.0048 | -0.34 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.012 | 0.0086 | 0.011 | 0.014 | 0.012 | 0.008 | 0.22 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.2.5.1 | Pyruvate dehydrogenase (quinone). | 0.08 | 0.49 | 1 | 0.057 | 1 | 1 | 0.034 | 0.71 | 1 | 0.025 | 0.42 | 1 | 0.039 | 0.8 | 1 | 0.056 | 0.66 | 1 | 0.00037 | 0.00021 | 6.2e-05 | 0.00029 | 0.00016 | 0.00026 | 0.00021 | 2.7e-05 | 0.00038 | -0.47 | 21 / 36 (58%) | 7 / 7 (100%) | 4 / 7 (57%) | 1 | 0.571 | 0.00011 | 0 | 0.00026 | 0.00026 | 0 | 0.00065 | 1.2 | 5 / 11 (45%) | 5 / 11 (45%) | 0.455 | 0.455 |
| 2.7.4.25 | (d)CMP kinase. | 0.066 | 0.49 | 1 | 0.91 | 1 | 1 | 0.97 | 1 | 1 | 0.035 | 0.43 | 1 | 0.74 | 0.96 | 1 | 0.74 | 0.94 | 1 | 0.014 | 0.014 | 0.013 | 0.011 | 0.011 | 0.0021 | 0.011 | 0.011 | 0.0039 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.016 | 0.013 | 0.0082 | 0.015 | 0.016 | 0.0028 | -0.093 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.3.1.44 | Trans-2-enoyl-CoA reductase (NAD(+)). | 0.06 | 0.49 | 1 | 0.49 | 1 | 1 | 0.35 | 0.9 | 1 | 0.2 | 0.5 | 1 | 0.39 | 0.9 | 1 | 0.27 | 0.77 | 1 | 0.0024 | 0.0024 | 0.0021 | 0.0032 | 0.0027 | 0.0021 | 0.0036 | 0.0031 | 0.0023 | 0.17 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0019 | 0.0019 | 0.00081 | 0.0017 | 0.0016 | 0.00077 | -0.16 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.6.1.22 | (S)-3-amino-2-methylpropionate transaminase. | 0.06 | 0.49 | 1 | 0.032 | 0.97 | 1 | 0.036 | 0.71 | 1 | 0.021 | 0.42 | 1 | 0.11 | 0.87 | 1 | 0.11 | 0.66 | 1 | 0.00039 | 5.4e-05 | 0 | 0.00015 | 0 | 0.00035 | 0.00013 | 0 | 0.00033 | -0.21 | 5 / 36 (14%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 3.1.21.2 | Deoxyribonuclease IV. | 0.51 | 0.74 | 1 | 0.72 | 1 | 1 | 0.87 | 1 | 1 | 0.38 | 0.65 | 1 | 0.59 | 0.93 | 1 | 0.44 | 0.85 | 1 | 0.012 | 0.012 | 0.012 | 0.011 | 0.011 | 0.002 | 0.011 | 0.012 | 0.0037 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.012 | 0.012 | 0.0026 | 0.012 | 0.012 | 0.0015 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.6.3.28 | Transferred entry: 7.3.2.2. | 0.13 | 0.5 | 1 | 0.58 | 1 | 1 | 0.52 | 0.95 | 1 | 0.068 | 0.43 | 1 | 0.072 | 0.86 | 1 | 0.1 | 0.66 | 1 | 0.017 | 0.017 | 0.011 | 0.0076 | 0.004 | 0.0075 | 0.0092 | 0.0085 | 0.0075 | 0.28 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.025 | 0.012 | 0.046 | 0.019 | 0.014 | 0.017 | -0.4 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.6.1.45 | UDP-sugar diphosphatase. | 0.13 | 0.5 | 1 | 0.19 | 1 | 1 | 0.031 | 0.71 | 1 | 0.11 | 0.47 | 1 | 0.25 | 0.88 | 1 | 0.073 | 0.66 | 1 | 0.00094 | 0.00065 | 0.00021 | 0.001 | 0.00045 | 0.0018 | 0.00064 | 0.00024 | 0.0011 | -0.64 | 25 / 36 (69%) | 6 / 7 (86%) | 5 / 7 (71%) | 0.857 | 0.714 | 0.00029 | 5.7e-05 | 5e-04 | 0.00077 | 0.00011 | 0.0012 | 1.4 | 6 / 11 (55%) | 8 / 11 (73%) | 0.545 | 0.727 |
| 5.3.99.11 | 2-keto-myo-inositol isomerase. | 0.18 | 0.53 | 1 | 0.74 | 1 | 1 | 0.27 | 0.89 | 1 | 0.38 | 0.65 | 1 | 0.6 | 0.94 | 1 | 0.58 | 0.91 | 1 | 0.00028 | 0.00024 | 0.00012 | 0.00034 | 0.00012 | 4e-04 | 0.00022 | 0.00013 | 0.00017 | -0.63 | 31 / 36 (86%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.00016 | 0.00011 | 0.00019 | 0.00026 | 2e-04 | 0.00025 | 0.7 | 8 / 11 (73%) | 10 / 11 (91%) | 0.727 | 0.909 |
| 1.14.13.7 | Phenol 2-monooxygenase (NADPH). | 0.1 | 0.49 | 1 | 0.78 | 1 | 1 | 0.8 | 1 | 1 | 0.38 | 0.65 | 1 | 0.43 | 0.91 | 1 | 0.86 | 0.96 | 1 | 0.0016 | 0.0016 | 0.0013 | 0.002 | 0.002 | 0.0014 | 0.0021 | 0.0019 | 0.0015 | 0.07 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0011 | 0.0011 | 0.00056 | 0.0013 | 0.0013 | 6e-04 | 0.24 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.1.175 | Glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N- | 0.097 | 0.49 | 1 | 0.33 | 1 | 1 | 0.31 | 0.89 | 1 | 0.035 | 0.43 | 1 | 0.06 | 0.85 | 1 | 0.066 | 0.66 | 1 | 0.0045 | 0.004 | 0.0026 | 0.0017 | 0.00054 | 0.0021 | 0.0028 | 0.0031 | 0.0027 | 0.72 | 32 / 36 (89%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.006 | 0.0034 | 0.011 | 0.0042 | 0.0029 | 0.0038 | -0.51 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.1.226 | 23S rRNA (cytidine(1920)-2'-O)-methyltransferase. | 0.13 | 0.5 | 1 | 0.91 | 1 | 1 | 0.97 | 1 | 1 | 0.14 | 0.47 | 1 | 0.64 | 0.95 | 1 | 0.54 | 0.9 | 1 | 0.01 | 0.01 | 0.0097 | 0.0086 | 0.0086 | 0.0021 | 0.0088 | 0.0076 | 0.004 | 0.033 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.011 | 0.0041 | 0.011 | 0.011 | 0.003 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.1.292 | GalNAc-alpha-(1->4)-GalNAc-alpha-(1->3)-diNAcBac-PP-undecaprenol alpha- | 0.19 | 0.53 | 1 | 0.32 | 1 | 1 | 0.091 | 0.79 | 1 | 0.19 | 0.5 | 1 | 0.32 | 0.9 | 1 | 0.096 | 0.66 | 1 | 0.00018 | 6.5e-05 | 0 | 8.7e-05 | 2.8e-05 | 0.00015 | 3.5e-05 | 0 | 8.1e-05 | -1.3 | 13 / 36 (36%) | 4 / 7 (57%) | 2 / 7 (29%) | 0.571 | 0.286 | 2.4e-05 | 0 | 6e-05 | 0.00011 | 0 | 0.00023 | 2.2 | 2 / 11 (18%) | 5 / 11 (45%) | 0.182 | 0.455 |
| 3.4.11.2 | Membrane alanyl aminopeptidase. | 0.087 | 0.49 | 1 | 0.66 | 1 | 1 | 0.58 | 0.96 | 1 | 0.018 | 0.42 | 1 | 0.25 | 0.88 | 1 | 0.24 | 0.76 | 1 | 0.0056 | 0.0056 | 0.0036 | 0.0027 | 0.0029 | 0.0021 | 0.0032 | 0.0028 | 0.0023 | 0.25 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0079 | 0.0037 | 0.012 | 0.0066 | 0.0057 | 0.0055 | -0.26 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.4.37 | 2'.3'-cyclic-nucleotide 3'-phosphodiesterase. | 0.73 | 0.87 | 1 | 0.62 | 1 | 1 | 0.83 | 1 | 1 | 0.56 | 0.78 | 1 | 0.94 | 0.99 | 1 | 0.73 | 0.94 | 1 | 0.00051 | 0.00014 | 0 | 6.1e-05 | 0 | 8.8e-05 | 0.00021 | 0 | 0.00047 | 1.8 | 10 / 36 (28%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 6.4e-05 | 0 | 0.00018 | 0.00023 | 0 | 0.00068 | 1.8 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 2.1.1.31 | Transferred entry: 2.1.1.221 and 2.1.1.228. | 0.094 | 0.49 | 1 | 0.055 | 1 | 1 | 0.22 | 0.88 | 1 | 0.14 | 0.47 | 1 | 0.17 | 0.87 | 1 | 0.73 | 0.94 | 1 | 0.00053 | 5.8e-05 | 0 | 0.00024 | 0 | 0.00062 | 4.8e-05 | 0 | 0.00013 | -2.3 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 7.3e-06 | 0 | 2.4e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 5.3.1.- | 0.072 | 0.49 | 1 | 0.1 | 1 | 1 | 0.3 | 0.89 | 1 | 0.35 | 0.62 | 1 | 0.47 | 0.92 | 1 | 0.85 | 0.96 | 1 | 0.0038 | 0.0038 | 0.0036 | 0.0047 | 0.0042 | 0.0017 | 0.0039 | 0.0035 | 0.0013 | -0.27 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0036 | 0.0039 | 0.0011 | 0.0034 | 0.0034 | 7e-04 | -0.082 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 2.4.1.58 | Lipopolysaccharide glucosyltransferase I. | 0.53 | 0.75 | 1 | 0.2 | 1 | 1 | 0.13 | 0.82 | 1 | 0.97 | 0.99 | 1 | 0.12 | 0.87 | 1 | 0.075 | 0.66 | 1 | 0.00033 | 0.00013 | 0 | 0.00026 | 0 | 0.00062 | 0.00026 | 2.7e-05 | 0.00045 | 0 | 14 / 36 (39%) | 2 / 7 (29%) | 4 / 7 (57%) | 0.286 | 0.571 | 5.9e-05 | 0 | 0.00012 | 3.6e-05 | 0 | 7.6e-05 | -0.71 | 5 / 11 (45%) | 3 / 11 (27%) | 0.455 | 0.273 |
| 6.5.-.- | 0.81 | 0.91 | 1 | 0.41 | 1 | 1 | 0.3 | 0.89 | 1 | 0.92 | 0.97 | 1 | 0.63 | 0.95 | 1 | 0.4 | 0.83 | 1 | 0.00019 | 2.6e-05 | 0 | 3.2e-05 | 0 | 8.6e-05 | 4.4e-06 | 0 | 1.2e-05 | -2.9 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.9e-05 | 0 | 9.5e-05 | 3.4e-05 | 0 | 7.7e-05 | 0.23 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 | |
| 1.10.3.- | 0.045 | 0.49 | 1 | 0.47 | 1 | 1 | 0.08 | 0.76 | 1 | 0.021 | 0.42 | 1 | 0.41 | 0.9 | 1 | 0.24 | 0.75 | 1 | 0.0023 | 0.0023 | 0.00086 | 0.0031 | 0.0017 | 0.0029 | 0.0029 | 0.0011 | 0.0034 | -0.096 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00093 | 0.00039 | 0.0014 | 0.0027 | 0.00085 | 0.0041 | 1.5 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 | |
| 4.1.1.22 | Histidine decarboxylase. | 0.86 | 0.94 | 1 | 0.81 | 1 | 1 | 0.5 | 0.95 | 1 | 0.77 | 0.91 | 1 | 0.97 | 0.99 | 1 | 0.94 | 0.98 | 1 | 0.00037 | 0.00015 | 0 | 9e-05 | 0 | 0.00016 | 5.3e-05 | 0 | 7.3e-05 | -0.76 | 15 / 36 (42%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 0.00011 | 0 | 2e-04 | 3e-04 | 0 | 0.00056 | 1.4 | 5 / 11 (45%) | 4 / 11 (36%) | 0.455 | 0.364 |
| 2.7.1.189 | Autoinducer-2 kinase. | 0.11 | 0.5 | 1 | 0.42 | 1 | 1 | 0.24 | 0.88 | 1 | 0.26 | 0.55 | 1 | 0.71 | 0.95 | 1 | 0.67 | 0.93 | 1 | 0.00027 | 0.00011 | 0 | 0.00033 | 0 | 0.00074 | 0.00014 | 2.7e-05 | 0.00027 | -1.2 | 15 / 36 (42%) | 3 / 7 (43%) | 4 / 7 (57%) | 0.429 | 0.571 | 2.6e-05 | 0 | 4.7e-05 | 4.3e-05 | 0 | 5.1e-05 | 0.73 | 3 / 11 (27%) | 5 / 11 (45%) | 0.273 | 0.455 |
| 1.1.1.336 | UDP-N-acetyl-D-mannosamine dehydrogenase. | 0.23 | 0.54 | 1 | 0.84 | 1 | 1 | 0.88 | 1 | 1 | 0.044 | 0.43 | 1 | 0.32 | 0.9 | 1 | 0.16 | 0.71 | 1 | 6e-04 | 0.00036 | 0.00012 | 0.00034 | 0.00016 | 0.00035 | 0.00079 | 8.1e-05 | 0.0013 | 1.2 | 22 / 36 (61%) | 7 / 7 (100%) | 4 / 7 (57%) | 1 | 0.571 | 0.00024 | 0 | 0.00048 | 0.00023 | 1e-04 | 0.00029 | -0.061 | 4 / 11 (36%) | 7 / 11 (64%) | 0.364 | 0.636 |
| 4.2.1.47 | GDP-mannose 4.6-dehydratase. | 0.13 | 0.5 | 1 | 0.79 | 1 | 1 | 0.61 | 0.96 | 1 | 0.24 | 0.53 | 1 | 0.68 | 0.95 | 1 | 0.66 | 0.92 | 1 | 0.0039 | 0.0039 | 0.0038 | 0.0043 | 0.0049 | 0.0014 | 0.0047 | 0.0043 | 0.0027 | 0.13 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0032 | 0.0033 | 0.0016 | 0.0037 | 0.0033 | 0.0016 | 0.21 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.5.8 | Quinate dehydrogenase (quinone). | 0.08 | 0.49 | 1 | 0.98 | 1 | 1 | 0.75 | 0.99 | 1 | 0.086 | 0.44 | 1 | 0.54 | 0.93 | 1 | 0.84 | 0.96 | 1 | 0.00013 | 1.8e-05 | 0 | 5.3e-05 | 0 | 0.00013 | 3.6e-05 | 0 | 6.2e-05 | -0.56 | 5 / 36 (14%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 3.4.14.11 | Xaa-Pro dipeptidyl-peptidase. | 0.023 | 0.49 | 1 | 0.053 | 1 | 1 | 0.012 | 0.61 | 1 | 0.02 | 0.42 | 1 | 0.012 | 0.76 | 1 | 0.0068 | 0.52 | 1 | 0.0016 | 0.0016 | 0.0014 | 0.00097 | 0.00082 | 0.00083 | 0.0018 | 0.0017 | 0.0015 | 0.89 | 35 / 36 (97%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.0021 | 0.0019 | 0.0013 | 0.0013 | 0.0013 | 0.00074 | -0.69 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.8.3.12 | Glutaconate CoA-transferase. | 0.97 | 0.99 | 1 | 1 | 1 | 1 | 0.59 | 0.96 | 1 | 0.99 | 0.99 | 1 | 0.83 | 0.98 | 1 | 0.83 | 0.96 | 1 | 2e-04 | 0.00013 | 0.00011 | 1e-04 | 0.00011 | 0.00011 | 9.1e-05 | 0.00011 | 6.9e-05 | -0.14 | 24 / 36 (67%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.00011 | 7.2e-05 | 0.00013 | 0.00019 | 0.00015 | 0.00025 | 0.79 | 7 / 11 (64%) | 7 / 11 (64%) | 0.636 | 0.636 |
| 4.1.99.17 | Phosphomethylpyrimidine synthase. | 0.92 | 0.96 | 1 | 0.23 | 1 | 1 | 0.14 | 0.83 | 1 | 0.62 | 0.83 | 1 | 0.74 | 0.96 | 1 | 0.54 | 0.9 | 1 | 0.02 | 0.02 | 0.02 | 0.02 | 0.022 | 0.0049 | 0.018 | 0.02 | 0.0053 | -0.15 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.02 | 0.019 | 0.005 | 0.021 | 0.022 | 0.0036 | 0.07 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.3.1.14 | L-rhamnose isomerase. | 0.87 | 0.94 | 1 | 0.2 | 1 | 1 | 0.063 | 0.71 | 1 | 0.9 | 0.97 | 1 | 0.38 | 0.9 | 1 | 0.18 | 0.72 | 1 | 0.003 | 0.003 | 0.0025 | 0.003 | 0.0026 | 0.0018 | 0.002 | 0.0016 | 0.00099 | -0.58 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0028 | 0.003 | 0.0014 | 0.0039 | 0.0028 | 0.0026 | 0.48 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.3.25 | Inositol-phosphate phosphatase. | 0.29 | 0.58 | 1 | 0.4 | 1 | 1 | 0.69 | 0.98 | 1 | 0.23 | 0.53 | 1 | 0.57 | 0.93 | 1 | 0.8 | 0.96 | 1 | 0.0035 | 0.0035 | 0.003 | 0.003 | 0.0028 | 0.00094 | 0.0025 | 0.0027 | 0.00089 | -0.26 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0042 | 0.0034 | 0.0033 | 0.0038 | 0.0033 | 0.0016 | -0.14 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.14.13.9 | Kynurenine 3-monooxygenase. | 0.63 | 0.82 | 1 | 0.68 | 1 | 1 | 0.87 | 1 | 1 | 0.3 | 0.58 | 1 | 0.71 | 0.95 | 1 | 0.45 | 0.86 | 1 | 0.00072 | 0.00022 | 0 | 0.00026 | 0 | 0.00068 | 8.2e-05 | 0 | 0.00018 | -1.7 | 11 / 36 (31%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 0.00022 | 0 | 4e-04 | 0.00028 | 0 | 0.00059 | 0.35 | 5 / 11 (45%) | 3 / 11 (27%) | 0.455 | 0.273 |
| 3.1.1.32 | Phospholipase A(1). | 0.47 | 0.72 | 1 | 0.11 | 1 | 1 | 0.28 | 0.89 | 1 | 0.5 | 0.74 | 1 | 0.14 | 0.87 | 1 | 0.21 | 0.74 | 1 | 0.00028 | 6.9e-05 | 0 | 0.00011 | 0 | 0.00026 | 9.7e-05 | 0 | 0.00026 | -0.18 | 9 / 36 (25%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 5.9e-05 | 0 | 0.00016 | 3.2e-05 | 0 | 6.7e-05 | -0.88 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 1.5.1.- | 0.34 | 0.61 | 1 | 0.89 | 1 | 1 | 0.16 | 0.85 | 1 | 0.55 | 0.78 | 1 | 0.91 | 0.99 | 1 | 0.26 | 0.76 | 1 | 0.00055 | 0.00011 | 0 | 2e-04 | 0 | 0.00053 | 0.00017 | 0 | 0.00046 | -0.23 | 7 / 36 (19%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 1.1e-05 | 0 | 3.6e-05 | 1e-04 | 0 | 0.00018 | 3.2 | 1 / 11 (9.1%) | 4 / 11 (36%) | 0.0909 | 0.364 | |
| 4.2.1.130 | D-lactate dehydratase. | 0.89 | 0.96 | 1 | 0.48 | 1 | 1 | 0.61 | 0.96 | 1 | 0.59 | 0.81 | 1 | 0.8 | 0.98 | 1 | 0.43 | 0.85 | 1 | 0.00023 | 8.2e-05 | 0 | 0.00014 | 0 | 0.00035 | 6.2e-05 | 0 | 0.00013 | -1.2 | 13 / 36 (36%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 0.00011 | 0 | 0.00019 | 2.7e-05 | 0 | 4.7e-05 | -2 | 5 / 11 (45%) | 3 / 11 (27%) | 0.455 | 0.273 |
| 3.11.1.1 | Phosphonoacetaldehyde hydrolase. | 0.069 | 0.49 | 1 | 0.9 | 1 | 1 | 0.42 | 0.93 | 1 | 0.19 | 0.5 | 1 | 0.66 | 0.95 | 1 | 0.84 | 0.96 | 1 | 0.00091 | 0.00083 | 0.00058 | 0.0011 | 0.0012 | 6e-04 | 0.0012 | 0.00086 | 0.0015 | 0.13 | 33 / 36 (92%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.00042 | 0.00017 | 0.00052 | 0.00083 | 5e-04 | 0.001 | 0.98 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 2.1.1.298 | Ribosomal protein L3 N(5)-glutamine methyltransferase. | 0.19 | 0.53 | 1 | 0.1 | 1 | 1 | 0.17 | 0.85 | 1 | 0.19 | 0.5 | 1 | 0.52 | 0.93 | 1 | 0.71 | 0.94 | 1 | 2e-04 | 2.8e-05 | 0 | 7e-05 | 0 | 0.00018 | 6.8e-05 | 0 | 0.00018 | -0.042 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 3e-06 | 0 | 1e-05 | 0.21 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.-.-.- | 0.47 | 0.71 | 1 | 0.77 | 1 | 1 | 0.64 | 0.97 | 1 | 0.28 | 0.57 | 1 | 0.31 | 0.9 | 1 | 0.25 | 0.76 | 1 | 0.06 | 0.06 | 0.061 | 0.056 | 0.059 | 0.013 | 0.059 | 0.056 | 0.021 | 0.075 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.062 | 0.061 | 0.02 | 0.06 | 0.061 | 0.01 | -0.047 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 1.11.1.19 | Dye decolorizing peroxidase. | 0.059 | 0.49 | 1 | 0.023 | 0.92 | 1 | 0.035 | 0.71 | 1 | 0.038 | 0.43 | 1 | 0.16 | 0.87 | 1 | 0.15 | 0.71 | 1 | 5e-04 | 5.6e-05 | 0 | 2e-04 | 0 | 0.00049 | 8.7e-05 | 0 | 0.00023 | -1.2 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 3.5.1.9 | Arylformamidase. | 0.64 | 0.83 | 1 | 0.59 | 1 | 1 | 0.42 | 0.93 | 1 | 0.66 | 0.85 | 1 | 0.87 | 0.99 | 1 | 0.62 | 0.92 | 1 | 0.0013 | 0.0013 | 0.00098 | 0.0013 | 0.00094 | 0.00074 | 0.0011 | 0.001 | 6e-04 | -0.24 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0012 | 0.00076 | 0.001 | 0.0013 | 0.0011 | 0.001 | 0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.1.11 | Penicillin amidase. | 0.36 | 0.64 | 1 | 0.011 | 0.81 | 1 | 0.12 | 0.82 | 1 | 0.51 | 0.75 | 1 | 0.0095 | 0.73 | 1 | 0.12 | 0.66 | 1 | 0.0026 | 0.0026 | 0.0021 | 0.0022 | 0.0021 | 0.00068 | 0.0011 | 0.0011 | 0.00088 | -1 | 35 / 36 (97%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.0032 | 0.0028 | 0.0018 | 0.003 | 0.0013 | 0.0027 | -0.093 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.130 | 3-dehydro-L-gulonate 2-dehydrogenase. | 0.0016 | 0.49 | 1 | 0.75 | 1 | 1 | 0.67 | 0.98 | 1 | 0.018 | 0.42 | 1 | 0.84 | 0.98 | 1 | 0.47 | 0.87 | 1 | 5e-04 | 0.00047 | 0.00033 | 0.00089 | 0.00098 | 0.00058 | 0.00078 | 0.00067 | 0.00042 | -0.19 | 34 / 36 (94%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 3e-04 | 2e-04 | 0.00026 | 0.00019 | 0.00015 | 0.00014 | -0.66 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 6.1.1.10 | Methionine--tRNA ligase. | 0.063 | 0.49 | 1 | 0.84 | 1 | 1 | 0.15 | 0.84 | 1 | 0.15 | 0.48 | 1 | 0.12 | 0.87 | 1 | 0.007 | 0.52 | 1 | 0.048 | 0.048 | 0.05 | 0.045 | 0.047 | 0.0082 | 0.046 | 0.048 | 0.012 | 0.032 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.052 | 0.054 | 0.0078 | 0.047 | 0.05 | 0.006 | -0.15 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.2.11 | Glutamate 5-kinase. | 0.0048 | 0.49 | 1 | 0.73 | 1 | 1 | 0.57 | 0.96 | 1 | 0.014 | 0.42 | 1 | 0.24 | 0.88 | 1 | 0.17 | 0.71 | 1 | 0.012 | 0.012 | 0.013 | 0.01 | 0.01 | 0.0016 | 0.011 | 0.01 | 0.0029 | 0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.014 | 0.013 | 0.0032 | 0.013 | 0.013 | 0.0015 | -0.11 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.7.7.1 | Ferredoxin--nitrite reductase. | 0.76 | 0.89 | 1 | 0.85 | 1 | 1 | 0.21 | 0.87 | 1 | 0.54 | 0.77 | 1 | 0.98 | 0.99 | 1 | 0.29 | 0.78 | 1 | 0.00065 | 0.00061 | 5e-04 | 0.00047 | 0.00036 | 0.00043 | 0.00058 | 4e-04 | 0.00067 | 0.3 | 34 / 36 (94%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 5e-04 | 0.00052 | 0.00038 | 0.00083 | 0.00073 | 0.00046 | 0.73 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.2.99.4 | Transferred entry: 1.2.98.1. | 0.12 | 0.5 | 1 | 0.56 | 1 | 1 | 0.049 | 0.71 | 1 | 0.13 | 0.47 | 1 | 0.51 | 0.93 | 1 | 0.037 | 0.63 | 1 | 0.00017 | 8.2e-05 | 0 | 3.6e-05 | 0 | 5.4e-05 | 8.2e-05 | 0 | 0.00016 | 1.2 | 17 / 36 (47%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 0.00016 | 9.7e-05 | 0.00027 | 3.1e-05 | 0 | 4.8e-05 | -2.4 | 7 / 11 (64%) | 4 / 11 (36%) | 0.636 | 0.364 |
| 1.1.99.14 | Glycolate dehydrogenase. | 0.44 | 0.69 | 1 | 0.96 | 1 | 1 | 0.44 | 0.93 | 1 | 0.85 | 0.95 | 1 | 0.92 | 0.99 | 1 | 0.49 | 0.87 | 1 | 0.00062 | 0.00014 | 0 | 0.00029 | 0 | 0.00076 | 0.00035 | 0 | 0.00092 | 0.27 | 8 / 36 (22%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 3.5e-05 | 0 | 7.4e-05 | 9.2e-06 | 0 | 2.2e-05 | -1.9 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 2.7.1.4 | Fructokinase. | 0.075 | 0.49 | 1 | 0.79 | 1 | 1 | 0.92 | 1 | 1 | 0.072 | 0.43 | 1 | 0.88 | 0.99 | 1 | 0.93 | 0.98 | 1 | 0.013 | 0.013 | 0.012 | 0.01 | 0.011 | 0.0032 | 0.01 | 0.0072 | 0.0065 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.016 | 0.013 | 0.0099 | 0.014 | 0.015 | 0.0031 | -0.19 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.180 | FAD:protein FMN transferase. | 0.6 | 0.8 | 1 | 0.7 | 1 | 1 | 0.75 | 0.99 | 1 | 0.3 | 0.58 | 1 | 0.4 | 0.9 | 1 | 0.12 | 0.66 | 1 | 0.021 | 0.021 | 0.02 | 0.021 | 0.021 | 0.0063 | 0.02 | 0.02 | 0.0056 | -0.07 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.022 | 0.02 | 0.0045 | 0.02 | 0.019 | 0.0027 | -0.14 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.6.1.3 | Adenosinetriphosphatase. | 0.028 | 0.49 | 1 | 0.47 | 1 | 1 | 0.44 | 0.93 | 1 | 0.27 | 0.56 | 1 | 0.22 | 0.88 | 1 | 0.18 | 0.71 | 1 | 0.021 | 0.021 | 0.02 | 0.024 | 0.021 | 0.0055 | 0.026 | 0.026 | 0.0089 | 0.12 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.018 | 0.019 | 0.0038 | 0.018 | 0.018 | 0.0038 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.99.2 | Transferred entry: 4.2.3.1. | 0.17 | 0.51 | 1 | 0.41 | 1 | 1 | 0.33 | 0.89 | 1 | 0.2 | 0.51 | 1 | 0.77 | 0.97 | 1 | 0.66 | 0.92 | 1 | 0.00047 | 6.5e-05 | 0 | 0.00017 | 0 | 0.00044 | 0.00015 | 0 | 0.00038 | -0.18 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1e-05 | 0 | 3.4e-05 | 1.9 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.3.1.31 | Homoserine O-acetyltransferase. | 0.028 | 0.49 | 1 | 0.55 | 1 | 1 | 0.97 | 1 | 1 | 0.039 | 0.43 | 1 | 0.13 | 0.87 | 1 | 0.37 | 0.82 | 1 | 0.016 | 0.016 | 0.016 | 0.013 | 0.012 | 0.0025 | 0.014 | 0.015 | 0.0042 | 0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.017 | 0.016 | 0.005 | 0.018 | 0.018 | 0.0025 | 0.082 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.2.2 | Carbamate kinase. | 0.39 | 0.65 | 1 | 0.55 | 1 | 1 | 0.2 | 0.87 | 1 | 0.11 | 0.47 | 1 | 0.54 | 0.93 | 1 | 0.22 | 0.74 | 1 | 0.00078 | 0.00071 | 0.00049 | 7e-04 | 4e-04 | 0.001 | 0.00081 | 0.00065 | 0.001 | 0.21 | 33 / 36 (92%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 8e-04 | 0.00057 | 0.00072 | 0.00056 | 0.00044 | 0.00045 | -0.51 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 3.2.2.10 | Pyrimidine-5'-nucleotide nucleosidase. | 0.2 | 0.53 | 1 | 0.97 | 1 | 1 | 0.39 | 0.91 | 1 | 0.21 | 0.51 | 1 | 0.67 | 0.95 | 1 | 0.68 | 0.93 | 1 | 0.00058 | 4.8e-05 | 0 | 0.00012 | 0 | 0.00031 | 0.00012 | 0 | 0.00031 | 0 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 8.1e-06 | 0 | 2.7e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 3.1.30.- | 0.95 | 0.98 | 1 | 0.052 | 1 | 1 | 0.67 | 0.98 | 1 | 0.81 | 0.93 | 1 | 0.044 | 0.8 | 1 | 0.63 | 0.92 | 1 | 0.0045 | 0.0045 | 0.0049 | 0.004 | 0.0044 | 0.0015 | 0.0056 | 0.0067 | 0.0027 | 0.49 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.004 | 0.0043 | 0.0017 | 0.0048 | 0.0051 | 0.0011 | 0.26 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 4.3.1.32 | 7.8-didemethyl-8-hydroxy-5-deazariboflavin synthase. | 0.88 | 0.95 | 1 | 0.42 | 1 | 1 | 0.24 | 0.88 | 1 | 0.94 | 0.98 | 1 | 0.62 | 0.95 | 1 | 0.33 | 0.8 | 1 | 0.00027 | 3.7e-05 | 0 | 3.6e-05 | 0 | 9.7e-05 | 4.4e-06 | 0 | 1.2e-05 | -3 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3.8e-05 | 0 | 0.00013 | 5.7e-05 | 0 | 0.00013 | 0.58 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 2.6.1.90 | dTDP-3-amino-3.6-dideoxy-alpha-D-galactopyranose transaminase. | 0.29 | 0.57 | 1 | 0.85 | 1 | 1 | 0.49 | 0.94 | 1 | 0.31 | 0.59 | 1 | 0.7 | 0.95 | 1 | 0.48 | 0.87 | 1 | 0.00015 | 2.9e-05 | 0 | 4.7e-05 | 0 | 6.6e-05 | 6.9e-05 | 0 | 0.00016 | 0.55 | 7 / 36 (19%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 2.2e-05 | 0 | 5.3e-05 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 3.7.1.5 | Acylpyruvate hydrolase. | 0.51 | 0.74 | 1 | 0.2 | 1 | 1 | 0.42 | 0.93 | 1 | 0.24 | 0.53 | 1 | 0.16 | 0.87 | 1 | 0.53 | 0.9 | 1 | 0.0011 | 0.00088 | 0.00052 | 0.00062 | 0.00059 | 0.00079 | 0.0011 | 0.0012 | 0.0014 | 0.83 | 28 / 36 (78%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.00091 | 0.00039 | 0.0014 | 0.00086 | 0.00053 | 0.00084 | -0.082 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 2.7.8.12 | Teichoic acid poly(glycerol phosphate) polymerase. | 0.22 | 0.54 | 1 | 0.93 | 1 | 1 | 0.19 | 0.87 | 1 | 0.57 | 0.8 | 1 | 0.69 | 0.95 | 1 | 0.27 | 0.77 | 1 | 0.015 | 0.015 | 0.014 | 0.017 | 0.011 | 0.011 | 0.017 | 0.014 | 0.011 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.012 | 0.012 | 0.0045 | 0.015 | 0.015 | 0.0035 | 0.32 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.1.267 | [Ribosomal protein S5]-alanine N-acetyltransferase. | 0.27 | 0.56 | 1 | 0.32 | 1 | 1 | 0.74 | 0.99 | 1 | 0.21 | 0.52 | 1 | 0.26 | 0.88 | 1 | 0.6 | 0.91 | 1 | 0.00025 | 0.00013 | 3.1e-05 | 0.00021 | 0.00011 | 0.00029 | 0.00017 | 0 | 0.00027 | -0.3 | 19 / 36 (53%) | 6 / 7 (86%) | 3 / 7 (43%) | 0.857 | 0.429 | 0.00013 | 0 | 0.00019 | 6.8e-05 | 0 | 0.00013 | -0.93 | 5 / 11 (45%) | 5 / 11 (45%) | 0.455 | 0.455 |
| 1.2.1.22 | Lactaldehyde dehydrogenase. | 0.19 | 0.53 | 1 | 0.056 | 1 | 1 | 0.0091 | 0.61 | 1 | 0.14 | 0.47 | 1 | 0.092 | 0.86 | 1 | 0.028 | 0.61 | 1 | 0.00074 | 0.00056 | 0.00026 | 0.00071 | 0.00042 | 0.00079 | 0.00032 | 3e-04 | 0.00035 | -1.1 | 27 / 36 (75%) | 6 / 7 (86%) | 4 / 7 (57%) | 0.857 | 0.571 | 0.00036 | 8.7e-05 | 0.00068 | 0.00081 | 0.00028 | 0.0014 | 1.2 | 8 / 11 (73%) | 9 / 11 (82%) | 0.727 | 0.818 |
| 3.1.26.5 | Ribonuclease P. | 0.92 | 0.96 | 1 | 0.13 | 1 | 1 | 0.15 | 0.83 | 1 | 0.58 | 0.8 | 1 | 0.37 | 0.9 | 1 | 0.41 | 0.84 | 1 | 0.0072 | 0.0072 | 0.0073 | 0.0073 | 0.0066 | 0.0017 | 0.0065 | 0.006 | 0.0026 | -0.17 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0074 | 0.008 | 0.0018 | 0.0076 | 0.0075 | 0.0011 | 0.038 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.4.4.2 | Isochorismate synthase. | 0.11 | 0.5 | 1 | 0.073 | 1 | 1 | 0.0036 | 0.55 | 1 | 0.04 | 0.43 | 1 | 0.29 | 0.89 | 1 | 0.077 | 0.66 | 1 | 8e-04 | 0.00075 | 0.00034 | 9e-04 | 0.00081 | 0.00064 | 0.00048 | 0.00038 | 0.00042 | -0.91 | 34 / 36 (94%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00045 | 0.00019 | 0.00074 | 0.0011 | 4e-04 | 0.0016 | 1.3 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 2.5.1.56 | N-acetylneuraminate synthase. | 0.47 | 0.71 | 1 | 0.85 | 1 | 1 | 0.25 | 0.89 | 1 | 0.66 | 0.86 | 1 | 0.69 | 0.95 | 1 | 0.78 | 0.95 | 1 | 0.00061 | 0.00057 | 0.00025 | 0.00061 | 0.00034 | 0.00066 | 0.00054 | 0.00025 | 0.00071 | -0.18 | 34 / 36 (94%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.00034 | 0.00022 | 0.00043 | 0.00081 | 0.00023 | 0.0017 | 1.3 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 1.1.1.163 | Cyclopentanol dehydrogenase. | 0.0092 | 0.49 | 1 | 0.12 | 1 | 1 | 0.089 | 0.79 | 1 | 0.013 | 0.42 | 1 | 0.18 | 0.87 | 1 | 0.14 | 0.68 | 1 | 0.00013 | 6.8e-05 | 2.9e-05 | 0.00014 | 8.5e-05 | 0.00013 | 8e-05 | 2.7e-05 | 1e-04 | -0.81 | 19 / 36 (53%) | 6 / 7 (86%) | 4 / 7 (57%) | 0.857 | 0.571 | 3.1e-05 | 0 | 6.4e-05 | 5.3e-05 | 0 | 7.5e-05 | 0.77 | 4 / 11 (36%) | 5 / 11 (45%) | 0.364 | 0.455 |
| 5.3.1.35 | 2-dehydrotetronate isomerase. | 0.59 | 0.79 | 1 | 0.34 | 1 | 1 | 0.84 | 1 | 1 | 0.66 | 0.85 | 1 | 0.42 | 0.91 | 1 | 0.92 | 0.98 | 1 | 0.00014 | 1.9e-05 | 0 | 1.7e-05 | 0 | 4.5e-05 | 5.8e-05 | 0 | 0.00015 | 1.8 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1.3e-05 | 0 | 3.5e-05 | 2.3 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 6.3.2.6 | Phosphoribosylaminoimidazolesuccinocarboxamide synthase. | 0.04 | 0.49 | 1 | 0.85 | 1 | 1 | 0.85 | 1 | 1 | 0.085 | 0.44 | 1 | 0.24 | 0.88 | 1 | 0.12 | 0.66 | 1 | 0.014 | 0.014 | 0.014 | 0.013 | 0.013 | 0.0024 | 0.013 | 0.013 | 0.0028 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.015 | 0.015 | 0.0036 | 0.015 | 0.014 | 0.002 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.1.151 | Cobalt-factor II C(20)-methyltransferase. | 0.6 | 0.8 | 1 | 0.87 | 1 | 1 | 0.54 | 0.95 | 1 | 0.44 | 0.69 | 1 | 0.37 | 0.9 | 1 | 0.16 | 0.71 | 1 | 0.0047 | 0.0047 | 0.0047 | 0.0046 | 0.0048 | 0.0011 | 0.0047 | 0.0056 | 0.0016 | 0.031 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0049 | 0.0043 | 0.0013 | 0.0046 | 0.0046 | 0.0012 | -0.091 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.19.1.1 | Flavodoxin--NADP(+) reductase. | 0.0078 | 0.49 | 1 | 0.04 | 0.97 | 1 | 0.062 | 0.71 | 1 | 0.0012 | 0.31 | 1 | 0.016 | 0.76 | 1 | 0.032 | 0.62 | 1 | 0.00022 | 3.6e-05 | 0 | 9.5e-05 | 3.2e-05 | 0.00013 | 8.7e-05 | 0 | 0.00023 | -0.13 | 6 / 36 (17%) | 4 / 7 (57%) | 1 / 7 (14%) | 0.571 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.2.1.72 | Erythrose-4-phosphate dehydrogenase. | 0.27 | 0.56 | 1 | 0.64 | 1 | 1 | 0.55 | 0.95 | 1 | 0.42 | 0.69 | 1 | 0.29 | 0.89 | 1 | 0.26 | 0.76 | 1 | 0.00043 | 6e-05 | 0 | 0.00015 | 0 | 4e-04 | 0.00014 | 0 | 0.00036 | -0.1 | 5 / 36 (14%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 7.3e-06 | 0 | 2.4e-05 | 2.7e-06 | 0 | 8.9e-06 | -1.4 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 1.4.1.2 | Glutamate dehydrogenase. | 0.064 | 0.49 | 1 | 0.13 | 1 | 1 | 0.012 | 0.61 | 1 | 0.11 | 0.47 | 1 | 0.21 | 0.88 | 1 | 0.052 | 0.65 | 1 | 0.0029 | 0.0029 | 0.0027 | 0.0033 | 0.0034 | 0.00079 | 0.0023 | 0.0025 | 0.0016 | -0.52 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.002 | 0.002 | 0.0012 | 0.0038 | 0.0038 | 0.0022 | 0.93 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.37 | Transferred entry: 2.7.11.1. 2.7.11.8. 2.7.11.9. 2.7.11.10. 2.7.11.11. | 0.25 | 0.56 | 1 | 0.16 | 1 | 1 | 0.47 | 0.93 | 1 | 0.12 | 0.47 | 1 | 0.18 | 0.87 | 1 | 0.34 | 0.8 | 1 | 0.0014 | 0.0011 | 0.00077 | 0.00056 | 0.00056 | 0.00061 | 0.001 | 0.00096 | 0.0011 | 0.84 | 28 / 36 (78%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.0012 | 0.00067 | 0.0017 | 0.0013 | 0.00087 | 0.0011 | 0.12 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 2.7.1.19 | Phosphoribulokinase. | 0.31 | 0.59 | 1 | 0.41 | 1 | 1 | 0.73 | 0.99 | 1 | 0.44 | 0.69 | 1 | 0.38 | 0.9 | 1 | 0.6 | 0.91 | 1 | 0.00024 | 6.7e-05 | 0 | 0.00014 | 0 | 0.00024 | 0.00013 | 0 | 0.00033 | -0.11 | 10 / 36 (28%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 4.6e-05 | 0 | 0.00011 | 2.7e-06 | 0 | 8.9e-06 | -4.1 | 4 / 11 (36%) | 1 / 11 (9.1%) | 0.364 | 0.0909 |
| 4.1.3.18 | Transferred entry: 2.2.1.6. | 0.55 | 0.77 | 1 | 0.69 | 1 | 1 | 0.33 | 0.89 | 1 | 0.7 | 0.88 | 1 | 0.58 | 0.93 | 1 | 0.46 | 0.87 | 1 | 0.00036 | 0.00025 | 0.00011 | 0.00022 | 6.4e-05 | 0.00032 | 0.00026 | 0.00024 | 0.00026 | 0.24 | 25 / 36 (69%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.00038 | 0.00011 | 0.00055 | 0.00014 | 1e-04 | 0.00016 | -1.4 | 7 / 11 (64%) | 8 / 11 (73%) | 0.636 | 0.727 |
| 3.6.5.- | 0.067 | 0.49 | 1 | 0.96 | 1 | 1 | 0.81 | 1 | 1 | 0.023 | 0.42 | 1 | 0.44 | 0.91 | 1 | 0.41 | 0.84 | 1 | 0.035 | 0.035 | 0.031 | 0.028 | 0.027 | 0.0049 | 0.029 | 0.028 | 0.0077 | 0.051 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.041 | 0.031 | 0.024 | 0.038 | 0.037 | 0.01 | -0.11 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 1.4.3.19 | Glycine oxidase. | 1 | 1 | 1 | 0.13 | 1 | 1 | 0.28 | 0.89 | 1 | 0.86 | 0.95 | 1 | 0.2 | 0.87 | 1 | 0.3 | 0.78 | 1 | 0.00077 | 0.00056 | 3e-04 | 0.00069 | 0.00055 | 0.00089 | 0.00027 | 8.1e-05 | 0.00038 | -1.4 | 26 / 36 (72%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 0.00064 | 0.00052 | 7e-04 | 0.00058 | 0.00036 | 0.00068 | -0.14 | 8 / 11 (73%) | 9 / 11 (82%) | 0.727 | 0.818 |
| 3.5.3.7 | Guanidinobutyrase. | 0.59 | 0.79 | 1 | 0.65 | 1 | 1 | 0.25 | 0.89 | 1 | 0.88 | 0.96 | 1 | 0.43 | 0.91 | 1 | 0.27 | 0.77 | 1 | 0.0013 | 0.0011 | 0.00035 | 0.00045 | 0.00024 | 0.00041 | 0.00036 | 0.00012 | 0.00049 | -0.32 | 31 / 36 (86%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.0011 | 0.00044 | 0.0016 | 0.0021 | 0.001 | 0.0028 | 0.93 | 9 / 11 (82%) | 9 / 11 (82%) | 0.818 | 0.818 |
| 2.7.1.23 | NAD(+) kinase. | 0.037 | 0.49 | 1 | 0.94 | 1 | 1 | 0.46 | 0.93 | 1 | 0.063 | 0.43 | 1 | 0.17 | 0.87 | 1 | 0.039 | 0.63 | 1 | 0.013 | 0.013 | 0.013 | 0.012 | 0.012 | 0.0026 | 0.012 | 0.012 | 0.0028 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.015 | 0.013 | 0.0035 | 0.014 | 0.014 | 0.0026 | -0.1 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.3.-.- | 0.056 | 0.49 | 1 | 0.51 | 1 | 1 | 0.65 | 0.97 | 1 | 0.078 | 0.44 | 1 | 0.88 | 0.99 | 1 | 0.17 | 0.71 | 1 | 0.015 | 0.015 | 0.016 | 0.014 | 0.014 | 0.0022 | 0.014 | 0.014 | 0.0042 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.017 | 0.017 | 0.0033 | 0.015 | 0.016 | 0.0015 | -0.18 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 2.5.1.89 | Tritrans.polycis-undecaprenyl-diphosphate synthase (geranylgeranyl- | 0.68 | 0.85 | 1 | 0.4 | 1 | 1 | 0.81 | 1 | 1 | 0.62 | 0.83 | 1 | 0.44 | 0.91 | 1 | 0.93 | 0.98 | 1 | 0.00026 | 2.9e-05 | 0 | 2.4e-05 | 0 | 6.4e-05 | 0 | 0 | 0 | -Inf | 4 / 36 (11%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 4.9e-05 | 0 | 0.00013 | 3.1e-05 | 0 | 1e-04 | -0.66 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 1.18.99.1 | Transferred entry: 1.12.7.2. | 0.028 | 0.49 | 1 | 0.0028 | 0.63 | 1 | 0.0069 | 0.61 | 1 | 0.0043 | 0.36 | 1 | 0.016 | 0.76 | 1 | 0.037 | 0.63 | 1 | 0.00071 | 0.00024 | 0 | 0.00072 | 0.00012 | 0.0017 | 0.00041 | 0 | 0.0011 | -0.81 | 12 / 36 (33%) | 6 / 7 (86%) | 2 / 7 (29%) | 0.857 | 0.286 | 1.6e-05 | 0 | 3.8e-05 | 4.3e-05 | 0 | 0.00011 | 1.4 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 6.3.5.11 | Cobyrinate a.c-diamide synthase (glutamine-hydrolyzing). | 0.2 | 0.53 | 1 | 1 | 1 | 1 | 0.56 | 0.96 | 1 | 0.59 | 0.81 | 1 | 0.29 | 0.89 | 1 | 0.83 | 0.96 | 1 | 0.0079 | 0.0079 | 0.0079 | 0.0086 | 0.0089 | 0.0025 | 0.0086 | 0.0085 | 0.0025 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0072 | 0.0064 | 0.0026 | 0.0077 | 0.0077 | 0.0022 | 0.097 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 7.6.2.11 | ABC-type polyamine transporter. | 0.19 | 0.53 | 1 | 0.47 | 1 | 1 | 0.42 | 0.92 | 1 | 0.68 | 0.87 | 1 | 0.9 | 0.99 | 1 | 0.96 | 0.99 | 1 | 0.017 | 0.017 | 0.017 | 0.018 | 0.019 | 0.0026 | 0.017 | 0.018 | 0.0023 | -0.082 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.016 | 0.016 | 0.0044 | 0.017 | 0.016 | 0.0041 | 0.087 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.4.3.6 | Tyrosine 2.3-aminomutase. | 0.74 | 0.88 | 1 | 0.48 | 1 | 1 | 0.13 | 0.82 | 1 | 0.78 | 0.92 | 1 | 0.52 | 0.93 | 1 | 0.29 | 0.78 | 1 | 0.00066 | 0.00037 | 8.9e-05 | 0.00029 | 0.00011 | 0.00038 | 0.00019 | 0 | 0.00032 | -0.61 | 20 / 36 (56%) | 4 / 7 (57%) | 3 / 7 (43%) | 0.571 | 0.429 | 0.00024 | 8e-05 | 0.00052 | 0.00065 | 0.00014 | 0.0013 | 1.4 | 6 / 11 (55%) | 7 / 11 (64%) | 0.545 | 0.636 |
| 4.2.1.35 | (R)-2-methylmalate dehydratase. | 0.74 | 0.88 | 1 | 0.74 | 1 | 1 | 0.24 | 0.89 | 1 | 0.57 | 0.8 | 1 | 0.6 | 0.94 | 1 | 0.16 | 0.71 | 1 | 0.00021 | 5.2e-05 | 0 | 6.2e-05 | 0 | 0.00016 | 8e-05 | 0 | 0.00016 | 0.37 | 9 / 36 (25%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 7.7e-05 | 0 | 0.00019 | 3e-06 | 0 | 1e-05 | -4.7 | 4 / 11 (36%) | 1 / 11 (9.1%) | 0.364 | 0.0909 |
| 2.3.3.1 | Citrate (Si)-synthase. | 0.29 | 0.57 | 1 | 0.23 | 1 | 1 | 0.6 | 0.96 | 1 | 0.31 | 0.59 | 1 | 0.26 | 0.88 | 1 | 0.87 | 0.97 | 1 | 0.0033 | 0.0033 | 0.0028 | 0.0029 | 0.0029 | 0.001 | 0.0023 | 0.0015 | 0.0016 | -0.33 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0039 | 0.0041 | 0.0016 | 0.0037 | 0.0026 | 0.0025 | -0.076 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.3.1.6 | Ribose-5-phosphate isomerase. | 0.21 | 0.53 | 1 | 0.3 | 1 | 1 | 0.35 | 0.9 | 1 | 0.13 | 0.47 | 1 | 0.027 | 0.76 | 1 | 0.035 | 0.63 | 1 | 0.01 | 0.01 | 0.01 | 0.0094 | 0.0086 | 0.0026 | 0.01 | 0.01 | 0.0029 | 0.089 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.011 | 0.01 | 0.003 | 0.011 | 0.011 | 0.0018 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.11.10 | Bacterial leucyl aminopeptidase. | 0.92 | 0.96 | 1 | 0.67 | 1 | 1 | 0.68 | 0.98 | 1 | 0.49 | 0.74 | 1 | 0.52 | 0.93 | 1 | 0.53 | 0.9 | 1 | 0.0012 | 0.0012 | 9e-04 | 0.0013 | 0.0013 | 0.00076 | 0.00098 | 0.00096 | 0.00059 | -0.41 | 35 / 36 (97%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.0012 | 0.00064 | 0.0013 | 0.0013 | 0.00075 | 0.0015 | 0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.1.149 | Crotonobetainyl-CoA hydratase. | 0.17 | 0.5 | 1 | 0.33 | 1 | 1 | 0.45 | 0.93 | 1 | 0.17 | 0.49 | 1 | 0.043 | 0.8 | 1 | 0.075 | 0.66 | 1 | 0.00045 | 3.7e-05 | 0 | 1e-04 | 0 | 0.00027 | 9.1e-05 | 0 | 0.00023 | -0.14 | 3 / 36 (8.3%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 5.4.99.2 | Methylmalonyl-CoA mutase. | 0.27 | 0.56 | 1 | 0.29 | 1 | 1 | 0.18 | 0.86 | 1 | 0.11 | 0.47 | 1 | 0.2 | 0.87 | 1 | 0.11 | 0.66 | 1 | 0.0039 | 0.0039 | 0.0022 | 0.0044 | 0.004 | 0.0026 | 0.0036 | 0.0015 | 0.0041 | -0.29 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0033 | 0.0017 | 0.0048 | 0.0044 | 0.0017 | 0.0061 | 0.42 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.3.9 | Malate synthase. | 0.2 | 0.53 | 1 | 0.091 | 1 | 1 | 0.68 | 0.98 | 1 | 0.28 | 0.57 | 1 | 0.23 | 0.88 | 1 | 0.99 | 1 | 1 | 0.00084 | 0.00019 | 0 | 0.00049 | 0 | 0.0012 | 0.00044 | 0 | 0.0012 | -0.16 | 8 / 36 (22%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 1.9e-05 | 0 | 3.9e-05 | 5.4e-06 | 0 | 1.8e-05 | -1.8 | 3 / 11 (27%) | 1 / 11 (9.1%) | 0.273 | 0.0909 |
| 5.3.1.30 | 5-deoxy-glucuronate isomerase. | 0.61 | 0.81 | 1 | 0.23 | 1 | 1 | 0.41 | 0.92 | 1 | 0.7 | 0.88 | 1 | 0.34 | 0.9 | 1 | 0.43 | 0.85 | 1 | 0.00021 | 4.7e-05 | 0 | 8.4e-05 | 0 | 0.00016 | 1.3e-05 | 0 | 3.5e-05 | -2.7 | 8 / 36 (22%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 6.1e-05 | 0 | 0.00015 | 3e-05 | 0 | 5.5e-05 | -1 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 2.7.7.33 | Glucose-1-phosphate cytidylyltransferase. | 0.7 | 0.86 | 1 | 0.54 | 1 | 1 | 0.85 | 1 | 1 | 0.64 | 0.85 | 1 | 0.77 | 0.97 | 1 | 0.44 | 0.85 | 1 | 0.0023 | 0.0022 | 0.0017 | 0.002 | 0.0017 | 0.001 | 0.0018 | 0.0013 | 0.0013 | -0.15 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0027 | 0.0022 | 0.0022 | 0.0021 | 0.0016 | 0.0014 | -0.36 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 7.2.2.- | 0.094 | 0.49 | 1 | 0.14 | 1 | 1 | 0.13 | 0.82 | 1 | 0.047 | 0.43 | 1 | 0.22 | 0.88 | 1 | 0.19 | 0.72 | 1 | 0.00075 | 8.3e-05 | 0 | 0.00022 | 0 | 0.00053 | 2e-04 | 0 | 0.00054 | -0.14 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 | |
| 3.1.1.4 | Phospholipase A(2). | 0.11 | 0.5 | 1 | 0.11 | 1 | 1 | 0.053 | 0.71 | 1 | 0.13 | 0.47 | 1 | 0.33 | 0.9 | 1 | 0.16 | 0.71 | 1 | 0.00024 | 4.1e-05 | 0 | 0.00011 | 0 | 0.00026 | 6.8e-05 | 0 | 0.00018 | -0.69 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 3e-06 | 0 | 9.9e-06 | 1.6e-05 | 0 | 3.7e-05 | 2.4 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 1.13.11.29 | Stizolobate synthase. | 0.049 | 0.49 | 1 | 0.042 | 0.97 | 1 | 0.027 | 0.71 | 1 | 0.034 | 0.43 | 1 | 0.18 | 0.87 | 1 | 0.086 | 0.66 | 1 | 0.00026 | 3.6e-05 | 0 | 0.00012 | 0 | 0.00026 | 5.8e-05 | 0 | 0.00015 | -1 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.2e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 2.4.1.83 | Dolichyl-phosphate beta-D-mannosyltransferase. | 0.55 | 0.77 | 1 | 0.41 | 1 | 1 | 0.74 | 0.99 | 1 | 0.15 | 0.47 | 1 | 0.8 | 0.97 | 1 | 0.45 | 0.86 | 1 | 0.00069 | 0.00069 | 0.00048 | 0.00074 | 0.00082 | 0.00073 | 4e-04 | 0.00047 | 0.00022 | -0.89 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00078 | 0.00065 | 0.00053 | 0.00074 | 0.00034 | 0.00083 | -0.076 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.1.171 | 16S rRNA (guanine(966)-N(2))-methyltransferase. | 0.37 | 0.64 | 1 | 0.8 | 1 | 1 | 0.5 | 0.95 | 1 | 0.17 | 0.49 | 1 | 0.62 | 0.95 | 1 | 0.93 | 0.98 | 1 | 0.0077 | 0.0077 | 0.008 | 0.0071 | 0.0079 | 0.0014 | 0.0069 | 0.0077 | 0.0018 | -0.041 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.008 | 0.0086 | 0.0031 | 0.0084 | 0.0085 | 0.00087 | 0.07 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.37 | Malate dehydrogenase. | 0.16 | 0.5 | 1 | 0.47 | 1 | 1 | 0.26 | 0.89 | 1 | 0.19 | 0.5 | 1 | 0.6 | 0.95 | 1 | 0.45 | 0.86 | 1 | 0.003 | 0.003 | 0.0024 | 0.0036 | 0.0031 | 0.0017 | 0.003 | 0.0027 | 0.002 | -0.26 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0023 | 0.0015 | 0.0019 | 0.0032 | 0.0013 | 0.0034 | 0.48 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 7.3.2.3 | ABC-type sulfate transporter. | 0.27 | 0.56 | 1 | 0.73 | 1 | 1 | 0.57 | 0.96 | 1 | 0.27 | 0.56 | 1 | 0.38 | 0.9 | 1 | 0.34 | 0.8 | 1 | 0.0065 | 0.0065 | 0.0062 | 0.0056 | 0.0059 | 0.0022 | 0.006 | 0.006 | 0.0026 | 0.1 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0071 | 0.0081 | 0.0032 | 0.0066 | 0.0064 | 0.0024 | -0.11 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.5.12 | D-lactate dehydrogenase (quinone). | 0.78 | 0.9 | 1 | 0.15 | 1 | 1 | 0.32 | 0.89 | 1 | 0.8 | 0.92 | 1 | 0.11 | 0.87 | 1 | 0.28 | 0.78 | 1 | 0.00034 | 1e-04 | 0 | 0.00013 | 0 | 0.00036 | 0.00026 | 0 | 0.00044 | 1 | 11 / 36 (31%) | 1 / 7 (14%) | 3 / 7 (43%) | 0.143 | 0.429 | 4.2e-05 | 0 | 8.4e-05 | 4.5e-05 | 0 | 7.4e-05 | 0.1 | 3 / 11 (27%) | 4 / 11 (36%) | 0.273 | 0.364 |
| 4.2.1.- | 0.33 | 0.61 | 1 | 0.16 | 1 | 1 | 0.083 | 0.77 | 1 | 0.13 | 0.47 | 1 | 0.039 | 0.8 | 1 | 0.014 | 0.55 | 1 | 0.012 | 0.012 | 0.011 | 0.011 | 0.01 | 0.0019 | 0.012 | 0.012 | 0.0022 | 0.13 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.012 | 0.012 | 0.0036 | 0.011 | 0.011 | 0.0035 | -0.13 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 5.3.1.4 | L-arabinose isomerase. | 0.86 | 0.94 | 1 | 0.77 | 1 | 1 | 0.7 | 0.98 | 1 | 0.5 | 0.74 | 1 | 0.67 | 0.95 | 1 | 0.28 | 0.78 | 1 | 0.017 | 0.017 | 0.017 | 0.018 | 0.017 | 0.005 | 0.017 | 0.019 | 0.0049 | -0.082 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.018 | 0.016 | 0.0066 | 0.016 | 0.016 | 0.0024 | -0.17 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.22.71 | Sortase B. | 0.95 | 0.98 | 1 | 0.82 | 1 | 1 | 0.99 | 1 | 1 | 0.76 | 0.91 | 1 | 1 | 1 | 1 | 0.74 | 0.94 | 1 | 0.019 | 0.019 | 0.019 | 0.019 | 0.019 | 0.0046 | 0.019 | 0.02 | 0.0088 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.019 | 0.019 | 0.0084 | 0.019 | 0.018 | 0.0074 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.21.105 | Rhomboid protease. | 0.4 | 0.66 | 1 | 0.23 | 1 | 1 | 0.3 | 0.89 | 1 | 0.2 | 0.5 | 1 | 0.042 | 0.8 | 1 | 0.065 | 0.66 | 1 | 0.0095 | 0.0095 | 0.0089 | 0.0084 | 0.0089 | 0.003 | 0.0096 | 0.0079 | 0.0044 | 0.19 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.01 | 0.0078 | 0.0046 | 0.0097 | 0.0092 | 0.0031 | -0.044 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.22 | UDP-glucose 6-dehydrogenase. | 0.76 | 0.89 | 1 | 0.37 | 1 | 1 | 0.41 | 0.92 | 1 | 0.46 | 0.71 | 1 | 0.15 | 0.87 | 1 | 0.15 | 0.71 | 1 | 0.014 | 0.014 | 0.013 | 0.013 | 0.013 | 0.0028 | 0.015 | 0.013 | 0.0033 | 0.21 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.014 | 0.013 | 0.0038 | 0.013 | 0.013 | 0.0041 | -0.11 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.9.1 | Pyruvate. phosphate dikinase. | 0.75 | 0.88 | 1 | 0.34 | 1 | 1 | 0.22 | 0.88 | 1 | 0.75 | 0.9 | 1 | 0.8 | 0.97 | 1 | 0.96 | 0.99 | 1 | 0.033 | 0.033 | 0.033 | 0.034 | 0.034 | 0.0069 | 0.031 | 0.031 | 0.004 | -0.13 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.033 | 0.033 | 0.0076 | 0.034 | 0.032 | 0.0066 | 0.043 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.1.109 | Methylthioribulose 1-phosphate dehydratase. | 0.012 | 0.49 | 1 | 0.64 | 1 | 1 | 0.56 | 0.96 | 1 | 0.072 | 0.43 | 1 | 0.28 | 0.89 | 1 | 0.91 | 0.98 | 1 | 0.00061 | 0.00054 | 0.00025 | 0.001 | 0.00075 | 0.0011 | 0.00097 | 0.00063 | 0.00094 | -0.044 | 32 / 36 (89%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.00018 | 2e-04 | 0.00017 | 0.00032 | 0.00014 | 0.00034 | 0.83 | 9 / 11 (82%) | 10 / 11 (91%) | 0.818 | 0.909 |
| 3.1.6.6 | Choline-sulfatase. | 0.11 | 0.5 | 1 | 0.34 | 1 | 1 | 0.081 | 0.76 | 1 | 0.021 | 0.42 | 1 | 0.41 | 0.9 | 1 | 0.12 | 0.66 | 1 | 0.0022 | 0.0021 | 0.00068 | 0.0025 | 0.0019 | 0.0019 | 0.0018 | 0.00096 | 0.0021 | -0.47 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0011 | 0.00048 | 0.0017 | 0.003 | 0.00057 | 0.0055 | 1.4 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 4.1.1.3 | Transferred entry: 4.1.1.112. | 0.31 | 0.59 | 1 | 0.89 | 1 | 1 | 0.36 | 0.9 | 1 | 0.73 | 0.89 | 1 | 0.22 | 0.88 | 1 | 0.94 | 0.98 | 1 | 0.017 | 0.017 | 0.016 | 0.017 | 0.017 | 0.0033 | 0.017 | 0.018 | 0.0031 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.015 | 0.013 | 0.006 | 0.017 | 0.014 | 0.0043 | 0.18 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.6.1.62 | Adenosylmethionine--8-amino-7-oxononanoate transaminase. | 0.024 | 0.49 | 1 | 0.47 | 1 | 1 | 0.64 | 0.97 | 1 | 0.056 | 0.43 | 1 | 0.37 | 0.9 | 1 | 0.38 | 0.82 | 1 | 0.0034 | 0.0034 | 0.003 | 0.0042 | 0.0037 | 0.0015 | 0.0048 | 0.0056 | 0.0018 | 0.19 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0026 | 0.0023 | 0.0015 | 0.0027 | 0.0025 | 0.0017 | 0.054 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.1.187 | N-acetylglucosaminyldiphosphoundecaprenol N-acetyl-beta-D- | 0.44 | 0.69 | 1 | 0.89 | 1 | 1 | 0.72 | 0.98 | 1 | 0.36 | 0.63 | 1 | 0.57 | 0.93 | 1 | 0.96 | 0.99 | 1 | 0.0031 | 0.0031 | 0.0028 | 0.0026 | 0.0025 | 0.00083 | 0.0027 | 0.0023 | 0.0013 | 0.054 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0032 | 0.003 | 0.0017 | 0.0037 | 0.0029 | 0.0023 | 0.21 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.3.4 | Hydroxymethylglutaryl-CoA lyase. | 0.75 | 0.88 | 1 | 0.63 | 1 | 1 | 0.49 | 0.94 | 1 | 0.69 | 0.87 | 1 | 0.73 | 0.96 | 1 | 0.57 | 0.91 | 1 | 2e-04 | 8.7e-05 | 0 | 8e-05 | 2.8e-05 | 0.00012 | 4.3e-05 | 0 | 5.6e-05 | -0.9 | 16 / 36 (44%) | 4 / 7 (57%) | 3 / 7 (43%) | 0.571 | 0.429 | 8.9e-05 | 0 | 0.00019 | 0.00012 | 0 | 0.00021 | 0.43 | 4 / 11 (36%) | 5 / 11 (45%) | 0.364 | 0.455 |
| 4.1.2.5 | L-threonine aldolase. | 0.38 | 0.65 | 1 | 0.73 | 1 | 1 | 0.85 | 1 | 1 | 0.15 | 0.47 | 1 | 0.41 | 0.9 | 1 | 0.68 | 0.93 | 1 | 0.0043 | 0.0043 | 0.0041 | 0.0036 | 0.0043 | 0.0018 | 0.004 | 0.0035 | 0.0024 | 0.15 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0043 | 0.0035 | 0.0016 | 0.0048 | 0.0041 | 0.002 | 0.16 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.5.1.48 | Cystathionine gamma-synthase. | 0.12 | 0.5 | 1 | 0.63 | 1 | 1 | 0.61 | 0.96 | 1 | 0.061 | 0.43 | 1 | 0.37 | 0.9 | 1 | 0.57 | 0.91 | 1 | 0.013 | 0.013 | 0.01 | 0.0088 | 0.007 | 0.0042 | 0.01 | 0.01 | 0.0065 | 0.18 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.016 | 0.012 | 0.016 | 0.013 | 0.011 | 0.0054 | -0.3 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.2.4 | D-lactate dehydrogenase (cytochrome). | 0.51 | 0.74 | 1 | 0.36 | 1 | 1 | 0.35 | 0.9 | 1 | 0.8 | 0.93 | 1 | 0.49 | 0.92 | 1 | 0.27 | 0.77 | 1 | 0.00046 | 1e-04 | 0 | 0.00016 | 0 | 4e-04 | 0.00027 | 0 | 0.00066 | 0.75 | 8 / 36 (22%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 2.4e-05 | 0 | 4.2e-05 | 4.3e-05 | 0 | 0.00014 | 0.84 | 3 / 11 (27%) | 1 / 11 (9.1%) | 0.273 | 0.0909 |
| 2.8.1.13 | tRNA-uridine 2-sulfurtransferase. | 1 | 1 | 1 | 0.89 | 1 | 1 | 0.61 | 0.96 | 1 | 0.64 | 0.85 | 1 | 0.51 | 0.93 | 1 | 0.77 | 0.95 | 1 | 0.018 | 0.018 | 0.019 | 0.018 | 0.018 | 0.0027 | 0.018 | 0.018 | 0.0035 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.018 | 0.019 | 0.0039 | 0.019 | 0.017 | 0.0042 | 0.078 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.4.99.61 | Precorrin-8X methylmutase. | 0.17 | 0.5 | 1 | 0.18 | 1 | 1 | 0.087 | 0.79 | 1 | 0.71 | 0.89 | 1 | 0.52 | 0.93 | 1 | 0.32 | 0.79 | 1 | 0.0056 | 0.0056 | 0.0055 | 0.006 | 0.0062 | 0.00083 | 0.0054 | 0.0054 | 0.0019 | -0.15 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0052 | 0.0055 | 0.0011 | 0.0057 | 0.0054 | 0.0013 | 0.13 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.266 | dTDP-4-dehydro-6-deoxyglucose reductase. | 0.66 | 0.84 | 1 | 0.89 | 1 | 1 | 0.72 | 0.98 | 1 | 0.89 | 0.96 | 1 | 0.77 | 0.97 | 1 | 0.94 | 0.98 | 1 | 0.00098 | 0.00068 | 0.00043 | 0.00052 | 6e-04 | 0.00054 | 0.00058 | 0.00032 | 0.00067 | 0.16 | 25 / 36 (69%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.00087 | 0.00036 | 0.0012 | 0.00067 | 0.00043 | 8e-04 | -0.38 | 7 / 11 (64%) | 8 / 11 (73%) | 0.636 | 0.727 |
| 4.1.3.38 | Aminodeoxychorismate lyase. | 0.09 | 0.49 | 1 | 0.92 | 1 | 1 | 0.9 | 1 | 1 | 0.033 | 0.43 | 1 | 0.85 | 0.98 | 1 | 0.72 | 0.94 | 1 | 0.002 | 0.002 | 0.0012 | 0.00082 | 0.00077 | 0.00043 | 0.00094 | 0.00047 | 0.00075 | 0.2 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0029 | 0.0013 | 0.005 | 0.0025 | 0.0022 | 0.002 | -0.21 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.151 | Xyloglucan-specific endo-beta-1.4-glucanase. | 0.62 | 0.82 | 1 | 0.53 | 1 | 1 | 0.085 | 0.77 | 1 | 0.67 | 0.87 | 1 | 0.75 | 0.97 | 1 | 0.11 | 0.66 | 1 | 2e-04 | 4.9e-05 | 0 | 4.7e-05 | 0 | 0.00012 | 8.3e-06 | 0 | 1.4e-05 | -2.5 | 9 / 36 (25%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 3e-05 | 0 | 9.9e-05 | 9.7e-05 | 0 | 0.00015 | 1.7 | 1 / 11 (9.1%) | 4 / 11 (36%) | 0.0909 | 0.364 |
| 3.7.1.13 | 2-hydroxy-6-oxo-6-(2-aminophenyl)hexa-2.4-dienoate hydrolase. | 0.3 | 0.58 | 1 | 1 | 1 | 1 | 0.95 | 1 | 1 | 0.17 | 0.49 | 1 | 0.82 | 0.98 | 1 | 0.66 | 0.93 | 1 | 0.00012 | 1e-05 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 3 / 36 (8.3%) | 0 / 7 (0%) | 0 / 7 (0%) | 0 | 0 | 1.2e-05 | 0 | 2.8e-05 | 2.1e-05 | 0 | 6.8e-05 | 0.81 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 1.1.1.128 | Deleted entry. | 0.28 | 0.56 | 1 | 0.26 | 1 | 1 | 0.78 | 1 | 1 | 0.24 | 0.54 | 1 | 0.41 | 0.9 | 1 | 0.94 | 0.98 | 1 | 0.00059 | 4.9e-05 | 0 | 8.4e-05 | 0 | 0.00022 | 0.00015 | 0 | 0.00041 | 0.84 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 8.1e-06 | 0 | 2.7e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.3.3.13 | 2-isopropylmalate synthase. | 0.012 | 0.49 | 1 | 0.66 | 1 | 1 | 0.87 | 1 | 1 | 0.038 | 0.43 | 1 | 0.38 | 0.9 | 1 | 0.25 | 0.76 | 1 | 0.033 | 0.033 | 0.031 | 0.028 | 0.027 | 0.0035 | 0.027 | 0.029 | 0.0051 | -0.052 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.036 | 0.035 | 0.0086 | 0.035 | 0.036 | 0.0061 | -0.041 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.184 | Sulfofructose kinase. | 0.38 | 0.65 | 1 | 0.59 | 1 | 1 | 0.92 | 1 | 1 | 0.14 | 0.47 | 1 | 0.7 | 0.95 | 1 | 0.82 | 0.96 | 1 | 0.00084 | 0.00068 | 5e-04 | 0.00048 | 0.00012 | 0.00067 | 0.00066 | 0.00059 | 0.00087 | 0.46 | 29 / 36 (81%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 7e-04 | 0.00038 | 0.001 | 0.00079 | 0.00068 | 0.00069 | 0.17 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 4.1.1.11 | Aspartate 1-decarboxylase. | 0.66 | 0.84 | 1 | 0.84 | 1 | 1 | 0.45 | 0.93 | 1 | 0.98 | 0.99 | 1 | 0.93 | 0.99 | 1 | 0.45 | 0.86 | 1 | 0.0034 | 0.0034 | 0.0032 | 0.0033 | 0.0027 | 0.0013 | 0.0037 | 0.0036 | 0.0025 | 0.17 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.003 | 0.0032 | 0.00093 | 0.0037 | 0.0035 | 0.0011 | 0.3 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.148 | 4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase. | 0.1 | 0.49 | 1 | 0.8 | 1 | 1 | 0.6 | 0.96 | 1 | 0.15 | 0.47 | 1 | 0.5 | 0.93 | 1 | 0.15 | 0.71 | 1 | 0.016 | 0.016 | 0.016 | 0.015 | 0.014 | 0.0023 | 0.014 | 0.015 | 0.0034 | -0.1 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.017 | 0.017 | 0.0036 | 0.016 | 0.015 | 0.0019 | -0.087 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.3.97 | 3'.5'-nucleoside bisphosphate phosphatase. | 0.24 | 0.55 | 1 | 0.29 | 1 | 1 | 0.85 | 1 | 1 | 0.4 | 0.66 | 1 | 0.96 | 0.99 | 1 | 0.31 | 0.79 | 1 | 0.00052 | 4.3e-05 | 0 | 0.00013 | 0 | 0.00036 | 7.7e-05 | 0 | 2e-04 | -0.76 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 7.3e-06 | 0 | 2.4e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 7.5.2.- | 0.52 | 0.75 | 1 | 0.71 | 1 | 1 | 0.85 | 1 | 1 | 0.32 | 0.59 | 1 | 0.77 | 0.97 | 1 | 0.55 | 0.9 | 1 | 0.00099 | 8e-04 | 0.00049 | 5e-04 | 0.00055 | 0.00049 | 8e-04 | 0.00036 | 0.0011 | 0.68 | 29 / 36 (81%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 0.00084 | 0.00038 | 0.0015 | 0.00094 | 0.00085 | 0.00091 | 0.16 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 | |
| 3.2.1.80 | Fructan beta-fructosidase. | 0.99 | 1 | 1 | 0.18 | 1 | 1 | 0.41 | 0.92 | 1 | 0.8 | 0.93 | 1 | 0.13 | 0.87 | 1 | 0.3 | 0.78 | 1 | 0.007 | 0.007 | 0.0065 | 0.0065 | 0.0063 | 0.0038 | 0.0087 | 0.0066 | 0.0047 | 0.42 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0064 | 0.0065 | 0.0031 | 0.0069 | 0.006 | 0.0035 | 0.11 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.346 | 2.5-didehydrogluconate reductase (2-dehydro-L-gulonate-forming). | 0.3 | 0.58 | 1 | 0.093 | 1 | 1 | 0.77 | 0.99 | 1 | 0.12 | 0.47 | 1 | 0.01 | 0.75 | 1 | 0.26 | 0.76 | 1 | 0.00068 | 0.00049 | 0.00026 | 0.00026 | 0 | 0.00044 | 0.00048 | 0.00024 | 7e-04 | 0.88 | 26 / 36 (72%) | 3 / 7 (43%) | 5 / 7 (71%) | 0.429 | 0.714 | 0.00047 | 0.00019 | 0.00075 | 0.00066 | 0.00045 | 0.00064 | 0.49 | 9 / 11 (82%) | 9 / 11 (82%) | 0.818 | 0.818 |
| 2.7.1.17 | Xylulokinase. | 0.48 | 0.72 | 1 | 0.28 | 1 | 1 | 0.16 | 0.85 | 1 | 0.3 | 0.58 | 1 | 0.045 | 0.8 | 1 | 0.022 | 0.56 | 1 | 0.029 | 0.029 | 0.03 | 0.028 | 0.029 | 0.0066 | 0.031 | 0.034 | 0.0099 | 0.15 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.03 | 0.033 | 0.0081 | 0.028 | 0.03 | 0.0043 | -0.1 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.3.2 | Acid phosphatase. | 0.44 | 0.69 | 1 | 0.35 | 1 | 1 | 0.067 | 0.73 | 1 | 0.55 | 0.78 | 1 | 0.63 | 0.95 | 1 | 0.41 | 0.83 | 1 | 0.0026 | 0.0025 | 0.0018 | 0.0015 | 0.0021 | 0.001 | 0.001 | 0.0011 | 0.00065 | -0.58 | 35 / 36 (97%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.003 | 0.002 | 0.0048 | 0.0037 | 0.0033 | 0.0032 | 0.3 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 3.2.1.39 | Glucan endo-1.3-beta-D-glucosidase. | 0.84 | 0.93 | 1 | 0.39 | 1 | 1 | 0.78 | 1 | 1 | 0.87 | 0.95 | 1 | 0.23 | 0.88 | 1 | 0.35 | 0.81 | 1 | 0.0047 | 0.0047 | 0.0028 | 0.0044 | 0.0034 | 0.0025 | 0.0038 | 0.0024 | 0.0046 | -0.21 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.006 | 0.0033 | 0.0072 | 0.0042 | 0.0029 | 0.0028 | -0.51 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.145 | Deoxynucleoside kinase. | 0.21 | 0.54 | 1 | 0.54 | 1 | 1 | 0.39 | 0.91 | 1 | 0.17 | 0.49 | 1 | 0.41 | 0.9 | 1 | 0.29 | 0.78 | 1 | 0.00011 | 1.2e-05 | 0 | 0 | 0 | 0 | 7.7e-06 | 0 | 2e-05 | Inf | 4 / 36 (11%) | 0 / 7 (0%) | 1 / 7 (14%) | 0 | 0.143 | 2e-05 | 0 | 4.6e-05 | 1.3e-05 | 0 | 4.3e-05 | -0.62 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 2.4.1.15 | Alpha.alpha-trehalose-phosphate synthase (UDP-forming). | 0.66 | 0.84 | 1 | 0.9 | 1 | 1 | 0.21 | 0.87 | 1 | 0.64 | 0.84 | 1 | 0.48 | 0.92 | 1 | 0.73 | 0.94 | 1 | 0.00072 | 0.00052 | 0.00013 | 4e-04 | 0.00012 | 0.00072 | 3e-04 | 0.00024 | 0.00031 | -0.42 | 26 / 36 (72%) | 5 / 7 (71%) | 6 / 7 (86%) | 0.714 | 0.857 | 0.00027 | 8e-05 | 0.00057 | 0.00099 | 0.00014 | 0.002 | 1.9 | 7 / 11 (64%) | 8 / 11 (73%) | 0.636 | 0.727 |
| 2.6.1.53 | Transferred entry: 1.4.1.13. | 0.068 | 0.49 | 1 | 0.083 | 1 | 1 | 0.064 | 0.72 | 1 | 0.028 | 0.43 | 1 | 0.14 | 0.87 | 1 | 0.12 | 0.66 | 1 | 0.00059 | 2e-04 | 0 | 5e-04 | 2.8e-05 | 0.0012 | 0.00043 | 0 | 0.0011 | -0.22 | 12 / 36 (33%) | 5 / 7 (71%) | 2 / 7 (29%) | 0.714 | 0.286 | 1.4e-05 | 0 | 3.3e-05 | 3.3e-05 | 0 | 7e-05 | 1.2 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 3.1.3.8 | 3-phytase. | 0.081 | 0.49 | 1 | 0.02 | 0.92 | 1 | 0.061 | 0.71 | 1 | 0.025 | 0.42 | 1 | 0.078 | 0.86 | 1 | 0.19 | 0.72 | 1 | 0.00084 | 9.4e-05 | 0 | 0.00025 | 0 | 0.00062 | 0.00023 | 0 | 0.00061 | -0.12 | 4 / 36 (11%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 1.4.1.- | 0.45 | 0.69 | 1 | 0.61 | 1 | 1 | 0.63 | 0.96 | 1 | 0.52 | 0.75 | 1 | 0.31 | 0.9 | 1 | 0.7 | 0.94 | 1 | 0.0027 | 0.0027 | 0.0016 | 0.0027 | 0.0024 | 0.0018 | 0.0031 | 0.0022 | 0.0027 | 0.2 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0019 | 0.0013 | 0.0019 | 0.0032 | 0.0012 | 0.0046 | 0.75 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 3.5.2.- | 0.73 | 0.87 | 1 | 0.52 | 1 | 1 | 0.2 | 0.87 | 1 | 0.45 | 0.7 | 1 | 0.39 | 0.9 | 1 | 0.12 | 0.66 | 1 | 0.0046 | 0.0046 | 0.0043 | 0.0045 | 0.0044 | 0.0021 | 0.0052 | 0.0037 | 0.0035 | 0.21 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0047 | 0.0046 | 0.0016 | 0.0041 | 0.0043 | 0.0014 | -0.2 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 2.8.4.1 | Coenzyme-B sulfoethylthiotransferase. | 0.72 | 0.87 | 1 | 0.13 | 1 | 1 | 0.1 | 0.82 | 1 | 0.93 | 0.98 | 1 | 0.7 | 0.95 | 1 | 0.57 | 0.91 | 1 | 0.00063 | 0.00011 | 0 | 0.00019 | 0 | 5e-04 | 7.5e-05 | 0 | 2e-04 | -1.3 | 6 / 36 (17%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 7.1e-05 | 0 | 0.00016 | 0.00011 | 0 | 0.00024 | 0.63 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 7.2.2.6 | P-type K(+) transporter. | 0.22 | 0.54 | 1 | 0.57 | 1 | 1 | 0.34 | 0.89 | 1 | 0.097 | 0.45 | 1 | 0.71 | 0.95 | 1 | 0.4 | 0.83 | 1 | 0.0031 | 0.0031 | 0.0024 | 0.0035 | 0.0031 | 0.0011 | 0.0031 | 0.0023 | 0.0017 | -0.18 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0027 | 0.0014 | 0.0026 | 0.0032 | 0.0021 | 0.003 | 0.25 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.4.2.9 | Phosphoenolpyruvate mutase. | 0.017 | 0.49 | 1 | 0.13 | 1 | 1 | 0.071 | 0.73 | 1 | 0.026 | 0.42 | 1 | 0.19 | 0.87 | 1 | 0.15 | 0.71 | 1 | 0.00051 | 0.00045 | 0.00024 | 0.00073 | 0.00069 | 0.00043 | 0.00049 | 0.00036 | 0.00059 | -0.58 | 32 / 36 (89%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00022 | 0.00016 | 0.00021 | 0.00048 | 0.00018 | 0.00092 | 1.1 | 9 / 11 (82%) | 10 / 11 (91%) | 0.818 | 0.909 |
| 3.6.1.63 | Alpha-D-ribose 1-methylphosphonate 5-triphosphate diphosphatase. | 0.26 | 0.56 | 1 | 0.27 | 1 | 1 | 0.26 | 0.89 | 1 | 0.28 | 0.57 | 1 | 0.36 | 0.9 | 1 | 0.45 | 0.86 | 1 | 0.00028 | 1e-04 | 0 | 0.00015 | 1.6e-05 | 0.00023 | 0.00012 | 0 | 0.00028 | -0.32 | 13 / 36 (36%) | 4 / 7 (57%) | 3 / 7 (43%) | 0.571 | 0.429 | 5.8e-05 | 0 | 0.00012 | 9.5e-05 | 0 | 0.00023 | 0.71 | 3 / 11 (27%) | 3 / 11 (27%) | 0.273 | 0.273 |
| 3.5.1.18 | Succinyl-diaminopimelate desuccinylase. | 0.3 | 0.58 | 1 | 0.95 | 1 | 1 | 0.64 | 0.97 | 1 | 0.18 | 0.5 | 1 | 0.64 | 0.95 | 1 | 0.41 | 0.84 | 1 | 0.014 | 0.014 | 0.011 | 0.012 | 0.012 | 0.005 | 0.012 | 0.01 | 0.0044 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.017 | 0.011 | 0.017 | 0.014 | 0.013 | 0.0054 | -0.28 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.3.1.12 | Glucuronate isomerase. | 0.3 | 0.58 | 1 | 0.71 | 1 | 1 | 0.39 | 0.91 | 1 | 0.74 | 0.9 | 1 | 0.38 | 0.9 | 1 | 0.84 | 0.96 | 1 | 0.0094 | 0.0094 | 0.0093 | 0.0093 | 0.01 | 0.0018 | 0.0099 | 0.01 | 0.0024 | 0.09 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0081 | 0.0074 | 0.0023 | 0.01 | 0.0095 | 0.0036 | 0.3 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.1.117 | 2.3.4.5-tetrahydropyridine-2.6-dicarboxylate N-succinyltransferase. | 0.25 | 0.55 | 1 | 0.69 | 1 | 1 | 0.92 | 1 | 1 | 0.46 | 0.71 | 1 | 0.081 | 0.86 | 1 | 0.12 | 0.66 | 1 | 0.0019 | 0.0018 | 0.00095 | 0.00099 | 6e-04 | 0.00099 | 0.0011 | 6e-04 | 0.0015 | 0.15 | 33 / 36 (92%) | 7 / 7 (100%) | 5 / 7 (71%) | 1 | 0.714 | 0.0025 | 0.0013 | 0.0042 | 0.0019 | 0.00091 | 0.0017 | -0.4 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 1.4.1.24 | 3-dehydroquinate synthase II. | 0.53 | 0.76 | 1 | 0.52 | 1 | 1 | 0.53 | 0.95 | 1 | 0.75 | 0.9 | 1 | 0.81 | 0.98 | 1 | 0.84 | 0.96 | 1 | 0.00026 | 2.9e-05 | 0 | 5.7e-05 | 0 | 0.00015 | 1.3e-05 | 0 | 3.5e-05 | -2.1 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 1.9e-05 | 0 | 6.3e-05 | 3.1e-05 | 0 | 1e-04 | 0.71 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.1.26.8 | Ribonuclease M5. | 0.57 | 0.78 | 1 | 0.75 | 1 | 1 | 0.94 | 1 | 1 | 0.49 | 0.74 | 1 | 0.96 | 0.99 | 1 | 0.93 | 0.98 | 1 | 0.0012 | 0.0012 | 0.0011 | 0.0013 | 0.0011 | 0.00044 | 0.0013 | 0.0011 | 0.00044 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0013 | 0.0012 | 0.00098 | 0.0011 | 0.00095 | 0.00041 | -0.24 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.33 | Amylo-alpha-1.6-glucosidase. | 0.48 | 0.72 | 1 | 0.59 | 1 | 1 | 0.42 | 0.92 | 1 | 0.37 | 0.64 | 1 | 0.46 | 0.92 | 1 | 0.49 | 0.88 | 1 | 0.00031 | 0.00011 | 0 | 9.8e-05 | 0.00011 | 1e-04 | 8.6e-05 | 0 | 0.00016 | -0.19 | 13 / 36 (36%) | 4 / 7 (57%) | 3 / 7 (43%) | 0.571 | 0.429 | 7.5e-05 | 0 | 0.00015 | 0.00017 | 0 | 0.00042 | 1.2 | 3 / 11 (27%) | 3 / 11 (27%) | 0.273 | 0.273 |
| 1.12.1.2 | Hydrogen dehydrogenase. | 0.28 | 0.56 | 1 | 0.22 | 1 | 1 | 0.054 | 0.71 | 1 | 0.19 | 0.5 | 1 | 0.15 | 0.87 | 1 | 0.073 | 0.66 | 1 | 6e-04 | 0.00023 | 0 | 0.00026 | 0.00012 | 0.00043 | 0.00013 | 0 | 0.00025 | -1 | 14 / 36 (39%) | 5 / 7 (71%) | 2 / 7 (29%) | 0.714 | 0.286 | 0.00011 | 0 | 0.00021 | 4e-04 | 0 | 0.00081 | 1.9 | 3 / 11 (27%) | 4 / 11 (36%) | 0.273 | 0.364 |
| 3.6.3.4 | Transferred entry: 7.2.2.9. | 0.07 | 0.49 | 1 | 0.88 | 1 | 1 | 0.75 | 0.99 | 1 | 0.035 | 0.43 | 1 | 0.41 | 0.9 | 1 | 0.48 | 0.87 | 1 | 0.033 | 0.033 | 0.03 | 0.026 | 0.025 | 0.0066 | 0.027 | 0.024 | 0.0089 | 0.054 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.039 | 0.034 | 0.025 | 0.035 | 0.033 | 0.007 | -0.16 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.2.11 | 3-methyl-2-oxobutanoate hydroxymethyltransferase. | 0.51 | 0.74 | 1 | 0.97 | 1 | 1 | 0.21 | 0.87 | 1 | 0.98 | 0.99 | 1 | 0.46 | 0.92 | 1 | 0.64 | 0.92 | 1 | 0.0042 | 0.0042 | 0.004 | 0.0042 | 0.0039 | 0.0016 | 0.0042 | 0.0044 | 0.0015 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0038 | 0.0036 | 0.00098 | 0.0046 | 0.0041 | 0.0015 | 0.28 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.244 | Methanol dehydrogenase. | 0.4 | 0.66 | 1 | 0.54 | 1 | 1 | 0.88 | 1 | 1 | 0.39 | 0.66 | 1 | 0.62 | 0.95 | 1 | 0.75 | 0.95 | 1 | 0.00093 | 0.00093 | 0.00091 | 0.00086 | 0.00072 | 0.00059 | 0.00072 | 7e-04 | 0.00048 | -0.26 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0011 | 0.001 | 0.00061 | 0.00092 | 8e-04 | 0.00062 | -0.26 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.3.10 | Glucose-1-phosphatase. | 0.12 | 0.5 | 1 | 0.12 | 1 | 1 | 0.71 | 0.98 | 1 | 0.16 | 0.49 | 1 | 0.44 | 0.91 | 1 | 0.63 | 0.92 | 1 | 0.00042 | 8.1e-05 | 0 | 0.00025 | 0 | 0.00062 | 0.00013 | 0 | 0.00033 | -0.94 | 7 / 36 (19%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 1.9e-05 | 0 | 4.4e-05 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 3.4.14.12 | Xaa-Xaa-Pro tripeptidyl-peptidase. | 0.71 | 0.87 | 1 | 0.97 | 1 | 1 | 0.71 | 0.98 | 1 | 0.84 | 0.94 | 1 | 0.59 | 0.93 | 1 | 0.69 | 0.94 | 1 | 0.0098 | 0.0098 | 0.0053 | 0.0066 | 0.005 | 0.0039 | 0.0082 | 0.004 | 0.011 | 0.31 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.01 | 0.0061 | 0.012 | 0.013 | 0.0039 | 0.019 | 0.38 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.2.7.8 | Indolepyruvate ferredoxin oxidoreductase. | 0.7 | 0.86 | 1 | 0.84 | 1 | 1 | 0.93 | 1 | 1 | 0.5 | 0.74 | 1 | 0.59 | 0.93 | 1 | 0.46 | 0.86 | 1 | 0.0078 | 0.0078 | 0.007 | 0.0077 | 0.006 | 0.0049 | 0.0073 | 0.0066 | 0.0041 | -0.077 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0082 | 0.0093 | 0.0039 | 0.0078 | 0.0073 | 0.0035 | -0.072 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.99.14 | Spore photoproduct lyase. | 0.41 | 0.67 | 1 | 0.21 | 1 | 1 | 0.15 | 0.84 | 1 | 0.42 | 0.69 | 1 | 0.05 | 0.81 | 1 | 0.058 | 0.66 | 1 | 0.0022 | 0.0022 | 0.0017 | 0.0018 | 0.0019 | 0.0011 | 0.0014 | 0.00053 | 0.0016 | -0.36 | 35 / 36 (97%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.0023 | 0.0031 | 0.0014 | 0.0027 | 0.002 | 0.0019 | 0.23 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.1.198 | 16S rRNA (cytidine(1402)-2'-O)-methyltransferase. | 0.22 | 0.54 | 1 | 0.91 | 1 | 1 | 0.7 | 0.98 | 1 | 0.19 | 0.5 | 1 | 0.25 | 0.88 | 1 | 0.16 | 0.71 | 1 | 0.016 | 0.016 | 0.015 | 0.015 | 0.015 | 0.0022 | 0.015 | 0.016 | 0.0035 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.017 | 0.016 | 0.0045 | 0.016 | 0.015 | 0.0022 | -0.087 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.4.1.21 | S-ribosylhomocysteine lyase. | 0.038 | 0.49 | 1 | 0.48 | 1 | 1 | 0.89 | 1 | 1 | 0.043 | 0.43 | 1 | 0.12 | 0.87 | 1 | 0.33 | 0.8 | 1 | 0.008 | 0.008 | 0.0078 | 0.0063 | 0.0059 | 0.0019 | 0.0069 | 0.0077 | 0.002 | 0.13 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0086 | 0.0078 | 0.0029 | 0.0091 | 0.0092 | 0.0022 | 0.082 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.11.- | 0.7 | 0.86 | 1 | 0.59 | 1 | 1 | 0.82 | 1 | 1 | 0.47 | 0.72 | 1 | 0.42 | 0.91 | 1 | 0.25 | 0.76 | 1 | 0.048 | 0.048 | 0.05 | 0.047 | 0.046 | 0.0093 | 0.046 | 0.046 | 0.011 | -0.031 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.049 | 0.051 | 0.01 | 0.049 | 0.05 | 0.0093 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 6.3.4.5 | Argininosuccinate synthase. | 0.25 | 0.56 | 1 | 0.17 | 1 | 1 | 0.28 | 0.89 | 1 | 0.2 | 0.51 | 1 | 0.73 | 0.96 | 1 | 0.94 | 0.98 | 1 | 0.023 | 0.023 | 0.023 | 0.022 | 0.021 | 0.0039 | 0.02 | 0.019 | 0.0052 | -0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.024 | 0.023 | 0.0048 | 0.024 | 0.025 | 0.0035 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.2.1.17 | Propionate--CoA ligase. | 0.034 | 0.49 | 1 | 0.0093 | 0.72 | 1 | 0.041 | 0.71 | 1 | 0.019 | 0.42 | 1 | 0.04 | 0.8 | 1 | 0.12 | 0.67 | 1 | 0.00034 | 6.7e-05 | 0 | 0.00022 | 1.6e-05 | 0.00048 | 9.7e-05 | 0 | 0.00026 | -1.2 | 7 / 36 (19%) | 4 / 7 (57%) | 1 / 7 (14%) | 0.571 | 0.143 | 1.1e-05 | 0 | 3.6e-05 | 8.1e-06 | 0 | 2.7e-05 | -0.44 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 7.2.2.10 | P-type Ca(2+) transporter. | 0.73 | 0.88 | 1 | 0.19 | 1 | 1 | 0.81 | 1 | 1 | 0.9 | 0.97 | 1 | 0.28 | 0.88 | 1 | 0.87 | 0.97 | 1 | 6.9e-05 | 5.7e-06 | 0 | 1.7e-05 | 0 | 4.5e-05 | 0 | 0 | 0 | -Inf | 3 / 36 (8.3%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 8e-06 | 0 | 1.9e-05 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 7.1.1.7 | Ubiquinol oxidase (electrogenic. proton-motive force generating). | 0.14 | 0.5 | 1 | 0.39 | 1 | 1 | 0.15 | 0.84 | 1 | 0.12 | 0.47 | 1 | 0.74 | 0.96 | 1 | 0.33 | 0.8 | 1 | 7e-04 | 7.8e-05 | 0 | 2e-04 | 0 | 0.00053 | 0.00017 | 0 | 0.00046 | -0.23 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 1.3e-05 | 0 | 4.5e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.5.1.52 | L-mimosine synthase. | 0.088 | 0.49 | 1 | 0.92 | 1 | 1 | 0.65 | 0.97 | 1 | 0.036 | 0.43 | 1 | 0.56 | 0.93 | 1 | 0.79 | 0.95 | 1 | 0.0016 | 0.001 | 0.00025 | 0.00033 | 0 | 0.00055 | 0.00042 | 0 | 0.00093 | 0.35 | 23 / 36 (64%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 0.0019 | 0.00029 | 0.0042 | 0.001 | 0.00053 | 0.0013 | -0.93 | 10 / 11 (91%) | 8 / 11 (73%) | 0.909 | 0.727 |
| 4.1.2.57 | Sulfofructosephosphate aldolase. | 0.26 | 0.56 | 1 | 0.22 | 1 | 1 | 0.69 | 0.98 | 1 | 0.23 | 0.53 | 1 | 0.23 | 0.88 | 1 | 0.75 | 0.95 | 1 | 0.00033 | 2.8e-05 | 0 | 5e-05 | 0 | 0.00013 | 8.7e-05 | 0 | 0.00023 | 0.8 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 5.3.3.3 | Vinylacetyl-CoA Delta-isomerase. | 0.12 | 0.5 | 1 | 0.016 | 0.92 | 1 | 0.01 | 0.61 | 1 | 0.16 | 0.48 | 1 | 0.025 | 0.76 | 1 | 0.019 | 0.55 | 1 | 0.00016 | 4.1e-05 | 0 | 8.7e-05 | 0 | 0.00013 | 1.9e-05 | 0 | 5.1e-05 | -2.2 | 9 / 36 (25%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 1.8e-05 | 0 | 3.9e-05 | 4.8e-05 | 0 | 0.00011 | 1.4 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 5.1.3.8 | N-acylglucosamine 2-epimerase. | 0.21 | 0.53 | 1 | 0.28 | 1 | 1 | 0.92 | 1 | 1 | 0.31 | 0.58 | 1 | 0.056 | 0.82 | 1 | 0.29 | 0.78 | 1 | 0.0025 | 0.0024 | 0.0013 | 0.0015 | 0.0013 | 0.0015 | 0.00096 | 0.00038 | 0.00099 | -0.64 | 35 / 36 (97%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.004 | 0.0018 | 0.0066 | 0.0023 | 0.0012 | 0.0027 | -0.8 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.1.305 | UDP-Glc:alpha-D-GlcNAc-glucosaminyl-diphosphoundecaprenol beta-1.3- | 0.05 | 0.49 | 1 | 0.27 | 1 | 1 | 0.34 | 0.89 | 1 | 0.049 | 0.43 | 1 | 0.34 | 0.9 | 1 | 0.59 | 0.91 | 1 | 0.00043 | 0.00019 | 0 | 0.00039 | 0.00026 | 0.00048 | 0.00025 | 5.4e-05 | 0.00045 | -0.64 | 16 / 36 (44%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 7e-05 | 0 | 0.00015 | 0.00015 | 0 | 0.00046 | 1.1 | 4 / 11 (36%) | 3 / 11 (27%) | 0.364 | 0.273 |
| 1.3.3.1 | Transferred entry: 1.3.98.1. | 0.38 | 0.65 | 1 | 0.086 | 1 | 1 | 0.56 | 0.96 | 1 | 0.1 | 0.46 | 1 | 0.054 | 0.81 | 1 | 0.3 | 0.78 | 1 | 0.0021 | 0.0021 | 0.002 | 0.0015 | 0.00096 | 0.0015 | 0.0024 | 0.003 | 0.0021 | 0.68 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.002 | 0.0015 | 0.0016 | 0.0024 | 0.0022 | 0.0013 | 0.26 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.4.1.17 | Holocytochrome-c synthase. | 0.43 | 0.69 | 1 | 0.77 | 1 | 1 | 0.66 | 0.97 | 1 | 0.82 | 0.93 | 1 | 0.92 | 0.99 | 1 | 0.44 | 0.85 | 1 | 0.00029 | 8e-05 | 0 | 0.00019 | 0 | 0.00044 | 0.00013 | 0 | 0.00026 | -0.55 | 10 / 36 (28%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 4.7e-05 | 0 | 9.7e-05 | 1.1e-05 | 0 | 2.4e-05 | -2.1 | 4 / 11 (36%) | 2 / 11 (18%) | 0.364 | 0.182 |
| 5.5.1.- | 0.24 | 0.55 | 1 | 0.66 | 1 | 1 | 0.97 | 1 | 1 | 0.22 | 0.52 | 1 | 0.97 | 0.99 | 1 | 0.81 | 0.96 | 1 | 0.0027 | 0.002 | 0.00074 | 0.0011 | 5.7e-05 | 0.0015 | 0.00066 | 0.00019 | 0.001 | -0.74 | 26 / 36 (72%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.0034 | 0.00095 | 0.0073 | 0.0019 | 0.0017 | 0.0021 | -0.84 | 9 / 11 (82%) | 9 / 11 (82%) | 0.818 | 0.818 | |
| 1.1.1.392 | 3-alpha-hydroxycholanate dehydrogenase (NADP(+)). | 0.44 | 0.69 | 1 | 0.43 | 1 | 1 | 0.33 | 0.89 | 1 | 0.59 | 0.81 | 1 | 0.35 | 0.9 | 1 | 0.26 | 0.76 | 1 | 8.6e-05 | 1.9e-05 | 0 | 2.8e-05 | 0 | 5e-05 | 4.1e-05 | 0 | 6.1e-05 | 0.55 | 8 / 36 (22%) | 2 / 7 (29%) | 3 / 7 (43%) | 0.286 | 0.429 | 1.2e-05 | 0 | 2.6e-05 | 6.8e-06 | 0 | 2.2e-05 | -0.82 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 6.1.1.24 | Glutamate--tRNA(Gln) ligase. | 0.53 | 0.76 | 1 | 0.27 | 1 | 1 | 0.098 | 0.81 | 1 | 0.49 | 0.73 | 1 | 0.14 | 0.87 | 1 | 0.04 | 0.63 | 1 | 0.00037 | 0.00028 | 0.00018 | 0.00024 | 0.00012 | 0.00038 | 0.00033 | 0.00036 | 0.00017 | 0.46 | 27 / 36 (75%) | 5 / 7 (71%) | 6 / 7 (86%) | 0.714 | 0.857 | 0.00035 | 0.00023 | 0.00053 | 2e-04 | 0.00011 | 0.00039 | -0.81 | 9 / 11 (82%) | 7 / 11 (64%) | 0.818 | 0.636 |
| 1.16.3.1 | Ferroxidase. | 0.3 | 0.58 | 1 | 0.15 | 1 | 1 | 0.1 | 0.82 | 1 | 0.38 | 0.65 | 1 | 0.47 | 0.92 | 1 | 0.46 | 0.86 | 1 | 0.00073 | 0.00069 | 6e-04 | 0.00087 | 6e-04 | 0.00068 | 0.00054 | 0.00053 | 0.00044 | -0.69 | 34 / 36 (94%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 6e-04 | 4e-04 | 0.00045 | 0.00077 | 0.00071 | 0.00058 | 0.36 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 3.2.1.99 | Arabinan endo-1.5-alpha-L-arabinosidase. | 0.84 | 0.93 | 1 | 0.72 | 1 | 1 | 0.13 | 0.82 | 1 | 0.49 | 0.74 | 1 | 0.85 | 0.98 | 1 | 0.2 | 0.73 | 1 | 0.0077 | 0.0077 | 0.0073 | 0.0069 | 0.0078 | 0.0036 | 0.0067 | 0.0061 | 0.0044 | -0.042 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0071 | 0.0066 | 0.003 | 0.0093 | 0.0082 | 0.0042 | 0.39 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.1.255 | N-terminal amino-acid N(alpha)-acetyltransferase NatA. | 0.36 | 0.64 | 1 | 0.41 | 1 | 1 | 0.8 | 1 | 1 | 0.27 | 0.56 | 1 | 0.28 | 0.88 | 1 | 0.51 | 0.89 | 1 | 0.00012 | 1.7e-05 | 0 | 0 | 0 | 0 | 1.2e-05 | 0 | 3.1e-05 | Inf | 5 / 36 (14%) | 0 / 7 (0%) | 1 / 7 (14%) | 0 | 0.143 | 1.5e-05 | 0 | 3.5e-05 | 3.4e-05 | 0 | 7.7e-05 | 1.2 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 1.14.13.39 | Nitric-oxide synthase (NADPH). | 0.14 | 0.5 | 1 | 0.13 | 1 | 1 | 0.044 | 0.71 | 1 | 0.12 | 0.47 | 1 | 0.038 | 0.8 | 1 | 0.02 | 0.55 | 1 | 0.00038 | 0.00025 | 0.00015 | 0.00019 | 0 | 4e-04 | 3e-04 | 0.00024 | 0.00023 | 0.66 | 24 / 36 (67%) | 3 / 7 (43%) | 6 / 7 (86%) | 0.429 | 0.857 | 0.00036 | 0.00036 | 0.00042 | 0.00016 | 0.00015 | 0.00016 | -1.2 | 8 / 11 (73%) | 7 / 11 (64%) | 0.727 | 0.636 |
| 5.4.99.21 | 23S rRNA pseudouridine(2604) synthase. | 0.55 | 0.77 | 1 | 0.18 | 1 | 1 | 0.5 | 0.95 | 1 | 1 | 1 | 1 | 0.59 | 0.93 | 1 | 0.82 | 0.96 | 1 | 3e-04 | 6.7e-05 | 0 | 0.00015 | 0 | 4e-04 | 8.7e-05 | 0 | 0.00023 | -0.79 | 8 / 36 (22%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 3.5e-05 | 0 | 6.1e-05 | 3.2e-05 | 0 | 7.3e-05 | -0.13 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 1.3.1.34 | 2.4-dienoyl-CoA reductase (NADPH). | 0.5 | 0.74 | 1 | 0.24 | 1 | 1 | 0.59 | 0.96 | 1 | 0.4 | 0.67 | 1 | 0.35 | 0.9 | 1 | 0.54 | 0.9 | 1 | 0.0018 | 0.0018 | 0.0017 | 0.0019 | 0.0024 | 0.0011 | 0.0014 | 0.0016 | 0.00086 | -0.44 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0024 | 0.0014 | 0.0018 | 0.0014 | 0.0018 | 0.00064 | -0.78 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.4.1.- | 0.34 | 0.62 | 1 | 0.06 | 1 | 1 | 0.0022 | 0.51 | 1 | 0.47 | 0.72 | 1 | 0.23 | 0.88 | 1 | 0.016 | 0.55 | 1 | 0.002 | 0.002 | 0.0015 | 0.0023 | 0.0018 | 0.0016 | 0.0016 | 0.0011 | 0.0012 | -0.52 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0017 | 0.0012 | 0.0012 | 0.0025 | 0.0023 | 0.0013 | 0.56 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 1.1.1.281 | GDP-4-dehydro-6-deoxy-D-mannose reductase. | 0.75 | 0.88 | 1 | 0.19 | 1 | 1 | 0.11 | 0.82 | 1 | 0.76 | 0.91 | 1 | 0.15 | 0.87 | 1 | 0.083 | 0.66 | 1 | 0.0013 | 0.0011 | 0.00078 | 0.00096 | 0.00042 | 0.00098 | 0.00068 | 0.00021 | 0.00089 | -0.5 | 30 / 36 (83%) | 6 / 7 (86%) | 4 / 7 (57%) | 0.857 | 0.571 | 0.0011 | 0.00096 | 0.001 | 0.0013 | 0.00079 | 0.0011 | 0.24 | 9 / 11 (82%) | 11 / 11 (100%) | 0.818 | 1 |
| 2.8.3.16 | Formyl-CoA transferase. | 0.26 | 0.56 | 1 | 0.83 | 1 | 1 | 0.82 | 1 | 1 | 0.31 | 0.59 | 1 | 0.7 | 0.95 | 1 | 0.92 | 0.98 | 1 | 0.0015 | 0.0015 | 0.0011 | 0.0018 | 0.001 | 0.0013 | 0.002 | 0.0012 | 0.0016 | 0.15 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0012 | 0.0011 | 0.00081 | 0.0014 | 0.00091 | 0.0011 | 0.22 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.2.18 | N-acetylmuramoyl-L-alanyl-D-glutamyl-L-lysyl-(N(6)-triglycine)-D-alanyl- | 0.82 | 0.92 | 1 | 0.4 | 1 | 1 | 0.81 | 1 | 1 | 0.4 | 0.66 | 1 | 0.9 | 0.99 | 1 | 0.37 | 0.82 | 1 | 0.0015 | 0.0012 | 0.00054 | 0.0017 | 0.00012 | 0.0027 | 0.00092 | 0.00038 | 0.0011 | -0.89 | 30 / 36 (83%) | 6 / 7 (86%) | 5 / 7 (71%) | 0.857 | 0.714 | 0.0014 | 6e-04 | 0.0018 | 0.00092 | 0.00087 | 0.0011 | -0.61 | 11 / 11 (100%) | 8 / 11 (73%) | 1 | 0.727 |
| 2.7.1.71 | Shikimate kinase. | 0.13 | 0.5 | 1 | 0.65 | 1 | 1 | 0.66 | 0.97 | 1 | 0.14 | 0.47 | 1 | 0.19 | 0.87 | 1 | 0.19 | 0.72 | 1 | 0.021 | 0.021 | 0.021 | 0.019 | 0.018 | 0.0038 | 0.02 | 0.02 | 0.0063 | 0.074 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.022 | 0.021 | 0.0064 | 0.022 | 0.023 | 0.0035 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.2.1.27 | Methylmalonate-semialdehyde dehydrogenase (CoA acylating). | 0.085 | 0.49 | 1 | 0.045 | 0.97 | 1 | 0.17 | 0.85 | 1 | 0.067 | 0.43 | 1 | 0.012 | 0.76 | 1 | 0.06 | 0.66 | 1 | 0.00099 | 0.00088 | 0.00065 | 0.00071 | 0.00024 | 0.00075 | 0.00028 | 0.00011 | 0.00033 | -1.3 | 32 / 36 (89%) | 6 / 7 (86%) | 4 / 7 (57%) | 0.857 | 0.571 | 0.0012 | 0.0011 | 0.00085 | 0.0011 | 0.00075 | 0.00078 | -0.13 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.1.37 | Uroporphyrinogen decarboxylase. | 0.079 | 0.49 | 1 | 0.11 | 1 | 1 | 0.2 | 0.87 | 1 | 0.29 | 0.58 | 1 | 0.51 | 0.93 | 1 | 0.63 | 0.92 | 1 | 0.00087 | 0.00082 | 5e-04 | 0.0013 | 0.0014 | 8e-04 | 0.00081 | 0.0012 | 0.00049 | -0.68 | 34 / 36 (94%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00057 | 4e-04 | 0.00045 | 0.00079 | 5e-04 | 0.00094 | 0.47 | 11 / 11 (100%) | 9 / 11 (82%) | 1 | 0.818 |
| 3.1.1.85 | Pimeloyl-[acyl-carrier protein] methyl ester esterase. | 0.19 | 0.53 | 1 | 0.48 | 1 | 1 | 0.37 | 0.9 | 1 | 0.096 | 0.45 | 1 | 0.74 | 0.96 | 1 | 0.78 | 0.95 | 1 | 4e-04 | 9.9e-05 | 0 | 0.00011 | 4.8e-05 | 0.00014 | 0.00036 | 0 | 0.00068 | 1.7 | 9 / 36 (25%) | 4 / 7 (57%) | 2 / 7 (29%) | 0.571 | 0.286 | 2.7e-05 | 0 | 7.3e-05 | 2.7e-06 | 0 | 8.9e-06 | -3.3 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 2.2.1.1 | Transketolase. | 0.21 | 0.53 | 1 | 0.57 | 1 | 1 | 0.33 | 0.89 | 1 | 0.15 | 0.47 | 1 | 0.53 | 0.93 | 1 | 0.87 | 0.97 | 1 | 0.031 | 0.031 | 0.031 | 0.028 | 0.027 | 0.0066 | 0.027 | 0.028 | 0.0088 | -0.052 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.033 | 0.036 | 0.0077 | 0.034 | 0.034 | 0.0048 | 0.043 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.1.6 | Beta-ureidopropionase. | 0.15 | 0.5 | 1 | 0.54 | 1 | 1 | 0.52 | 0.95 | 1 | 0.23 | 0.53 | 1 | 0.17 | 0.87 | 1 | 0.19 | 0.72 | 1 | 0.00017 | 3.4e-05 | 0 | 8.9e-05 | 0 | 0.00022 | 7e-05 | 0 | 0.00013 | -0.35 | 7 / 36 (19%) | 2 / 7 (29%) | 3 / 7 (43%) | 0.286 | 0.429 | 6e-06 | 0 | 2e-05 | 2.7e-06 | 0 | 8.9e-06 | -1.2 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.1.4.46 | Glycerophosphodiester phosphodiesterase. | 0.25 | 0.55 | 1 | 0.36 | 1 | 1 | 0.057 | 0.71 | 1 | 0.71 | 0.88 | 1 | 0.88 | 0.99 | 1 | 0.2 | 0.74 | 1 | 0.014 | 0.014 | 0.014 | 0.016 | 0.014 | 0.0073 | 0.014 | 0.012 | 0.0062 | -0.19 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.013 | 0.012 | 0.0037 | 0.015 | 0.015 | 0.0029 | 0.21 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.2.3 | Phosphoribosylaminoimidazolecarboxamide formyltransferase. | 0.12 | 0.5 | 1 | 0.24 | 1 | 1 | 0.39 | 0.91 | 1 | 0.11 | 0.47 | 1 | 0.97 | 0.99 | 1 | 0.82 | 0.96 | 1 | 0.019 | 0.019 | 0.019 | 0.018 | 0.016 | 0.003 | 0.016 | 0.017 | 0.0037 | -0.17 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.021 | 0.019 | 0.0063 | 0.02 | 0.02 | 0.0026 | -0.07 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.57 | Fructuronate reductase. | 0.036 | 0.49 | 1 | 0.51 | 1 | 1 | 0.15 | 0.83 | 1 | 0.1 | 0.46 | 1 | 0.87 | 0.99 | 1 | 0.33 | 0.8 | 1 | 0.0069 | 0.0069 | 0.0069 | 0.0084 | 0.0079 | 0.0034 | 0.0075 | 0.0087 | 0.0028 | -0.16 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0054 | 0.005 | 0.0022 | 0.0071 | 0.0069 | 0.0037 | 0.39 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.6.3.17 | Transferred entry: 7.5.2.8. | 0.52 | 0.75 | 1 | 0.93 | 1 | 1 | 0.92 | 1 | 1 | 0.26 | 0.55 | 1 | 0.76 | 0.97 | 1 | 0.66 | 0.92 | 1 | 0.058 | 0.058 | 0.056 | 0.054 | 0.053 | 0.016 | 0.055 | 0.063 | 0.022 | 0.026 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.062 | 0.055 | 0.034 | 0.058 | 0.057 | 0.013 | -0.096 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.6.1.67 | Dihydroneopterin triphosphate diphosphatase. | 0.18 | 0.52 | 1 | 0.091 | 1 | 1 | 0.24 | 0.88 | 1 | 0.3 | 0.58 | 1 | 0.12 | 0.87 | 1 | 0.3 | 0.78 | 1 | 0.00021 | 2.3e-05 | 0 | 8.6e-05 | 0 | 0.00022 | 0 | 0 | 0 | -Inf | 4 / 36 (11%) | 2 / 7 (29%) | 0 / 7 (0%) | 0.286 | 0 | 1.5e-05 | 0 | 4.9e-05 | 5.4e-06 | 0 | 1.8e-05 | -1.5 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.4.24.70 | Oligopeptidase A. | 0.064 | 0.49 | 1 | 0.36 | 1 | 1 | 0.016 | 0.67 | 1 | 0.0038 | 0.36 | 1 | 0.18 | 0.87 | 1 | 0.0031 | 0.48 | 1 | 0.001 | 0.00082 | 0.00014 | 0.00094 | 0.00057 | 0.00088 | 8e-04 | 0.00012 | 0.0011 | -0.23 | 29 / 36 (81%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00039 | 2.8e-05 | 0.0011 | 0.0012 | 0.00014 | 0.002 | 1.6 | 6 / 11 (55%) | 10 / 11 (91%) | 0.545 | 0.909 |
| 3.5.1.54 | Allophanate hydrolase. | 0.43 | 0.69 | 1 | 0.3 | 1 | 1 | 0.8 | 1 | 1 | 0.78 | 0.92 | 1 | 0.18 | 0.87 | 1 | 0.74 | 0.94 | 1 | 0.0016 | 0.0011 | 0.00019 | 0.00058 | 0.00033 | 0.001 | 0.00025 | 0.00012 | 4e-04 | -1.2 | 25 / 36 (69%) | 6 / 7 (86%) | 4 / 7 (57%) | 0.857 | 0.571 | 0.0023 | 0.00048 | 0.006 | 0.00088 | 0.00022 | 0.0018 | -1.4 | 8 / 11 (73%) | 7 / 11 (64%) | 0.727 | 0.636 |
| 3.1.3.48 | Protein-tyrosine-phosphatase. | 0.65 | 0.84 | 1 | 0.66 | 1 | 1 | 0.51 | 0.95 | 1 | 0.43 | 0.69 | 1 | 0.7 | 0.95 | 1 | 0.97 | 0.99 | 1 | 0.02 | 0.02 | 0.021 | 0.019 | 0.02 | 0.0079 | 0.019 | 0.021 | 0.0071 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.021 | 0.022 | 0.0073 | 0.021 | 0.021 | 0.0046 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.13.5 | Ribonuclease D. | 0.045 | 0.49 | 1 | 0.55 | 1 | 1 | 0.46 | 0.93 | 1 | 0.022 | 0.42 | 1 | 0.8 | 0.97 | 1 | 0.72 | 0.94 | 1 | 0.0026 | 0.0025 | 0.0015 | 0.00079 | 4e-04 | 0.00081 | 0.0013 | 0.00084 | 0.0013 | 0.72 | 35 / 36 (97%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.004 | 0.0018 | 0.007 | 0.0029 | 0.0023 | 0.0026 | -0.46 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.8.8 | CDP-diacylglycerol--serine O-phosphatidyltransferase. | 0.91 | 0.96 | 1 | 0.29 | 1 | 1 | 0.16 | 0.85 | 1 | 0.73 | 0.89 | 1 | 0.41 | 0.9 | 1 | 0.27 | 0.77 | 1 | 0.0029 | 0.0029 | 0.0028 | 0.0029 | 0.0028 | 0.001 | 0.0025 | 0.0021 | 0.0011 | -0.21 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0029 | 0.0027 | 0.001 | 0.0032 | 0.0033 | 0.0011 | 0.14 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.1.290 | N.N'-diacetylbacillosaminyl-diphospho-undecaprenol alpha-1.3-N- | 0.52 | 0.75 | 1 | 0.29 | 1 | 1 | 0.93 | 1 | 1 | 0.54 | 0.77 | 1 | 0.36 | 0.9 | 1 | 0.64 | 0.92 | 1 | 0.00087 | 0.00068 | 0.00026 | 0.00095 | 0.00042 | 0.0011 | 0.00074 | 0.00034 | 0.001 | -0.36 | 28 / 36 (78%) | 6 / 7 (86%) | 5 / 7 (71%) | 0.857 | 0.714 | 0.00075 | 0.00029 | 0.001 | 0.00039 | 0.00017 | 0.00049 | -0.94 | 8 / 11 (73%) | 9 / 11 (82%) | 0.727 | 0.818 |
| 2.4.1.211 | 1.3-beta-galactosyl-N-acetylhexosamine phosphorylase. | 0.49 | 0.73 | 1 | 0.51 | 1 | 1 | 0.32 | 0.89 | 1 | 0.99 | 1 | 1 | 0.86 | 0.99 | 1 | 0.85 | 0.96 | 1 | 0.014 | 0.014 | 0.014 | 0.015 | 0.015 | 0.0037 | 0.014 | 0.016 | 0.0046 | -0.1 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.014 | 0.013 | 0.0038 | 0.015 | 0.014 | 0.0037 | 0.1 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.1.47 | Tartronate-semialdehyde synthase. | 0.15 | 0.5 | 1 | 0.21 | 1 | 1 | 0.07 | 0.73 | 1 | 0.16 | 0.49 | 1 | 0.27 | 0.88 | 1 | 0.098 | 0.66 | 1 | 0.00043 | 0.00012 | 0 | 0.00029 | 0 | 0.00071 | 0.00027 | 0 | 0.00072 | -0.1 | 10 / 36 (28%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 9.9e-06 | 0 | 2.5e-05 | 2.9e-05 | 0 | 4.5e-05 | 1.6 | 2 / 11 (18%) | 4 / 11 (36%) | 0.182 | 0.364 |
| 1.1.1.369 | D-chiro-inositol 1-dehydrogenase. | 0.025 | 0.49 | 1 | 0.41 | 1 | 1 | 0.073 | 0.73 | 1 | 0.031 | 0.43 | 1 | 0.29 | 0.89 | 1 | 0.061 | 0.66 | 1 | 0.001 | 0.001 | 0.00095 | 0.0012 | 0.00096 | 0.00093 | 0.0014 | 0.00095 | 0.0012 | 0.22 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00047 | 0.00026 | 0.00048 | 0.0012 | 0.0011 | 0.00059 | 1.4 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.3.4.14 | Biotin carboxylase. | 0.022 | 0.49 | 1 | 0.63 | 1 | 1 | 0.16 | 0.85 | 1 | 0.014 | 0.42 | 1 | 0.2 | 0.87 | 1 | 0.034 | 0.63 | 1 | 0.017 | 0.017 | 0.016 | 0.013 | 0.01 | 0.0053 | 0.014 | 0.012 | 0.006 | 0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.022 | 0.017 | 0.015 | 0.017 | 0.017 | 0.0031 | -0.37 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.6.1.50 | Glutamine--scyllo-inositol transaminase. | 0.79 | 0.9 | 1 | 0.16 | 1 | 1 | 0.82 | 1 | 1 | 0.56 | 0.79 | 1 | 0.19 | 0.87 | 1 | 0.54 | 0.9 | 1 | 0.00075 | 0.00071 | 0.00048 | 0.00053 | 0.00041 | 0.00049 | 0.00089 | 5e-04 | 0.0012 | 0.75 | 34 / 36 (94%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00059 | 0.00031 | 0.00058 | 0.00082 | 0.00071 | 5e-04 | 0.47 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.19.1 | Acylaminoacyl-peptidase. | 0.97 | 0.99 | 1 | 0.9 | 1 | 1 | 0.44 | 0.93 | 1 | 0.99 | 1 | 1 | 0.63 | 0.95 | 1 | 0.87 | 0.97 | 1 | 0.00082 | 0.00057 | 0.00017 | 4e-04 | 0.00022 | 0.00058 | 0.00025 | 0.00013 | 0.00032 | -0.68 | 25 / 36 (69%) | 4 / 7 (57%) | 6 / 7 (86%) | 0.571 | 0.857 | 0.00047 | 0.00021 | 0.00076 | 0.00098 | 0.00015 | 0.0019 | 1.1 | 6 / 11 (55%) | 9 / 11 (82%) | 0.545 | 0.818 |
| 5.4.99.25 | tRNA pseudouridine(55) synthase. | 0.15 | 0.5 | 1 | 0.55 | 1 | 1 | 0.9 | 1 | 1 | 0.18 | 0.5 | 1 | 0.85 | 0.98 | 1 | 0.52 | 0.89 | 1 | 0.015 | 0.015 | 0.015 | 0.014 | 0.013 | 0.0017 | 0.013 | 0.013 | 0.0027 | -0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.016 | 0.015 | 0.0051 | 0.015 | 0.015 | 0.0024 | -0.093 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.371 | Scyllo-inositol 2-dehydrogenase (NADP(+)). | 0.38 | 0.65 | 1 | 0.81 | 1 | 1 | 0.85 | 1 | 1 | 0.96 | 0.99 | 1 | 0.14 | 0.87 | 1 | 0.31 | 0.79 | 1 | 0.00036 | 0.00011 | 0 | 0.00029 | 0 | 0.00076 | 0.00015 | 0 | 3e-04 | -0.95 | 11 / 36 (31%) | 1 / 7 (14%) | 3 / 7 (43%) | 0.143 | 0.429 | 1.7e-05 | 0 | 3e-05 | 6.5e-05 | 0 | 0.00017 | 1.9 | 4 / 11 (36%) | 3 / 11 (27%) | 0.364 | 0.273 |
| 1.6.1.1 | NAD(P)(+) transhydrogenase (Si-specific). | 0.12 | 0.5 | 1 | 0.92 | 1 | 1 | 0.21 | 0.87 | 1 | 0.11 | 0.47 | 1 | 0.84 | 0.98 | 1 | 0.32 | 0.8 | 1 | 0.00042 | 0.00018 | 0 | 0.00029 | 4.8e-05 | 0.00056 | 0.00032 | 0.00011 | 7e-04 | 0.14 | 15 / 36 (42%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 3.5e-05 | 0 | 8.4e-05 | 0.00015 | 0 | 3e-04 | 2.1 | 2 / 11 (18%) | 5 / 11 (45%) | 0.182 | 0.455 |
| 5.1.3.1 | Ribulose-phosphate 3-epimerase. | 0.0069 | 0.49 | 1 | 0.98 | 1 | 1 | 0.66 | 0.97 | 1 | 0.042 | 0.43 | 1 | 0.51 | 0.93 | 1 | 0.27 | 0.77 | 1 | 0.014 | 0.014 | 0.014 | 0.012 | 0.012 | 0.0016 | 0.012 | 0.013 | 0.0038 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.016 | 0.016 | 0.0033 | 0.015 | 0.015 | 0.0021 | -0.093 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.1.3.- | 0.1 | 0.49 | 1 | 0.78 | 1 | 1 | 0.5 | 0.95 | 1 | 0.08 | 0.44 | 1 | 0.78 | 0.97 | 1 | 0.33 | 0.8 | 1 | 0.0019 | 0.0019 | 0.0018 | 0.0023 | 0.0019 | 0.0012 | 0.0026 | 0.0024 | 0.0022 | 0.18 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0015 | 0.0012 | 0.0013 | 0.0018 | 0.0019 | 0.00081 | 0.26 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 2.7.1.25 | Adenylyl-sulfate kinase. | 0.21 | 0.53 | 1 | 0.32 | 1 | 1 | 0.67 | 0.98 | 1 | 0.16 | 0.48 | 1 | 0.043 | 0.8 | 1 | 0.19 | 0.72 | 1 | 0.0069 | 0.0069 | 0.0063 | 0.0055 | 0.0058 | 0.0028 | 0.0063 | 0.0056 | 0.0036 | 0.2 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0073 | 0.0067 | 0.0034 | 0.0076 | 0.0066 | 0.0029 | 0.058 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.11.1.7 | Peroxidase. | 0.15 | 0.5 | 1 | 0.087 | 1 | 1 | 0.07 | 0.73 | 1 | 0.26 | 0.55 | 1 | 0.38 | 0.9 | 1 | 0.25 | 0.76 | 1 | 0.00042 | 9.3e-05 | 0 | 0.00028 | 0 | 0.00071 | 0.00015 | 0 | 0.00038 | -0.9 | 8 / 36 (22%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 9.9e-06 | 0 | 2.5e-05 | 2.2e-05 | 0 | 4.6e-05 | 1.2 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 1.1.1.41 | Isocitrate dehydrogenase (NAD(+)). | 0.074 | 0.49 | 1 | 0.51 | 1 | 1 | 0.21 | 0.87 | 1 | 0.11 | 0.47 | 1 | 0.49 | 0.92 | 1 | 0.23 | 0.75 | 1 | 0.00016 | 7.8e-05 | 1.6e-05 | 0.00012 | 0.00012 | 0.00011 | 1e-04 | 0 | 0.00014 | -0.26 | 18 / 36 (50%) | 5 / 7 (71%) | 3 / 7 (43%) | 0.714 | 0.429 | 3e-05 | 0 | 4.5e-05 | 8.4e-05 | 3.3e-05 | 0.00011 | 1.5 | 4 / 11 (36%) | 6 / 11 (55%) | 0.364 | 0.545 |
| 2.7.7.9 | UTP--glucose-1-phosphate uridylyltransferase. | 0.15 | 0.5 | 1 | 0.18 | 1 | 1 | 0.63 | 0.96 | 1 | 0.074 | 0.43 | 1 | 0.29 | 0.89 | 1 | 0.91 | 0.98 | 1 | 0.0075 | 0.0075 | 0.0066 | 0.0064 | 0.0063 | 0.0016 | 0.0051 | 0.0052 | 0.0011 | -0.33 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0093 | 0.0074 | 0.0068 | 0.0081 | 0.0075 | 0.0036 | -0.2 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.14.11.- | 0.019 | 0.49 | 1 | 0.025 | 0.94 | 1 | 0.011 | 0.61 | 1 | 0.007 | 0.37 | 1 | 0.052 | 0.81 | 1 | 0.022 | 0.56 | 1 | 0.00044 | 0.00011 | 0 | 3e-04 | 3.2e-05 | 0.00056 | 0.00019 | 0 | 0.00051 | -0.66 | 9 / 36 (25%) | 4 / 7 (57%) | 1 / 7 (14%) | 0.571 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 4.1e-05 | 0 | 8e-05 | 4 | 1 / 11 (9.1%) | 3 / 11 (27%) | 0.0909 | 0.273 | |
| 3.7.1.22 | 3D-(3.5/4)-trihydroxycyclohexane-1.2-dione acylhydrolase (decyclizing). | 0.51 | 0.74 | 1 | 0.54 | 1 | 1 | 0.57 | 0.96 | 1 | 0.73 | 0.9 | 1 | 0.32 | 0.9 | 1 | 0.72 | 0.94 | 1 | 0.0018 | 0.0018 | 0.0017 | 0.0019 | 0.0014 | 0.0015 | 0.0021 | 0.002 | 0.0014 | 0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0015 | 0.0013 | 0.0011 | 0.002 | 0.0022 | 0.00086 | 0.42 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.40 | Pyruvate kinase. | 0.1 | 0.49 | 1 | 0.72 | 1 | 1 | 0.95 | 1 | 1 | 0.14 | 0.47 | 1 | 0.19 | 0.87 | 1 | 0.29 | 0.78 | 1 | 0.027 | 0.027 | 0.028 | 0.025 | 0.026 | 0.0029 | 0.026 | 0.027 | 0.0066 | 0.057 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.028 | 0.028 | 0.004 | 0.029 | 0.03 | 0.0032 | 0.051 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.1.51 | Alpha-L-fucosidase. | 0.55 | 0.77 | 1 | 0.24 | 1 | 1 | 0.2 | 0.87 | 1 | 0.29 | 0.57 | 1 | 0.31 | 0.9 | 1 | 0.62 | 0.92 | 1 | 0.006 | 0.006 | 0.0038 | 0.0047 | 0.0054 | 0.0033 | 0.0031 | 0.0018 | 0.0028 | -0.6 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0061 | 0.0038 | 0.0052 | 0.0084 | 0.0042 | 0.012 | 0.46 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.13.1 | Exoribonuclease II. | 0.32 | 0.6 | 1 | 0.51 | 1 | 1 | 0.45 | 0.93 | 1 | 0.29 | 0.57 | 1 | 0.59 | 0.93 | 1 | 0.62 | 0.92 | 1 | 0.026 | 0.026 | 0.026 | 0.025 | 0.024 | 0.0051 | 0.024 | 0.021 | 0.0065 | -0.059 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.027 | 0.028 | 0.0051 | 0.028 | 0.026 | 0.0054 | 0.052 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.3.1.7 | Ethanolamine ammonia-lyase. | 0.71 | 0.86 | 1 | 0.72 | 1 | 1 | 0.88 | 1 | 1 | 0.58 | 0.8 | 1 | 0.55 | 0.93 | 1 | 0.76 | 0.95 | 1 | 0.00069 | 0.00065 | 0.00046 | 0.00085 | 0.00034 | 0.001 | 0.00082 | 0.00075 | 0.00064 | -0.052 | 34 / 36 (94%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00054 | 0.00036 | 0.00048 | 0.00053 | 0.00045 | 0.00032 | -0.027 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.3.4.- | 0.57 | 0.79 | 1 | 0.4 | 1 | 1 | 0.65 | 0.97 | 1 | 0.49 | 0.74 | 1 | 0.12 | 0.87 | 1 | 0.22 | 0.74 | 1 | 0.014 | 0.014 | 0.015 | 0.013 | 0.012 | 0.0023 | 0.014 | 0.015 | 0.0039 | 0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.014 | 0.015 | 0.004 | 0.014 | 0.015 | 0.0037 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 3.2.1.172 | Unsaturated rhamnogalacturonyl hydrolase. | 0.27 | 0.56 | 1 | 0.18 | 1 | 1 | 0.054 | 0.71 | 1 | 0.26 | 0.55 | 1 | 0.17 | 0.87 | 1 | 0.12 | 0.66 | 1 | 0.0085 | 0.0085 | 0.0063 | 0.0093 | 0.0091 | 0.0036 | 0.0066 | 0.004 | 0.0048 | -0.49 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0066 | 0.0048 | 0.0046 | 0.011 | 0.0074 | 0.011 | 0.74 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.8.2.4 | Dimethyl sulfide:cytochrome c2 reductase. | 0.81 | 0.91 | 1 | 0.8 | 1 | 1 | 0.3 | 0.89 | 1 | 0.52 | 0.76 | 1 | 0.43 | 0.91 | 1 | 0.11 | 0.66 | 1 | 0.00017 | 6.2e-05 | 0 | 9.5e-05 | 0 | 0.00021 | 6.4e-05 | 3.1e-05 | 8.8e-05 | -0.57 | 13 / 36 (36%) | 2 / 7 (29%) | 4 / 7 (57%) | 0.286 | 0.571 | 7.4e-05 | 0 | 1e-04 | 2.7e-05 | 0 | 6e-05 | -1.5 | 5 / 11 (45%) | 2 / 11 (18%) | 0.455 | 0.182 |
| 1.16.-.- | 0.32 | 0.6 | 1 | 0.85 | 1 | 1 | 0.39 | 0.91 | 1 | 0.3 | 0.58 | 1 | 0.63 | 0.95 | 1 | 0.4 | 0.83 | 1 | 0.00081 | 0.00072 | 0.00064 | 0.00078 | 0.00069 | 0.00064 | 0.00086 | 0.00062 | 8e-04 | 0.14 | 32 / 36 (89%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00055 | 0.00056 | 0.00047 | 0.00075 | 0.00085 | 0.00055 | 0.45 | 9 / 11 (82%) | 10 / 11 (91%) | 0.818 | 0.909 | |
| 1.4.99.1 | Transferred entry: 1.4.99.6. | 0.14 | 0.5 | 1 | 0.079 | 1 | 1 | 0.39 | 0.91 | 1 | 0.19 | 0.5 | 1 | 0.44 | 0.91 | 1 | 0.87 | 0.97 | 1 | 0.00028 | 3.1e-05 | 0 | 1e-04 | 0 | 0.00027 | 4.8e-05 | 0 | 0.00013 | -1.1 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 5.1e-06 | 0 | 1.7e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 6.3.2.3 | Glutathione synthase. | 0.99 | 1 | 1 | 0.72 | 1 | 1 | 0.66 | 0.97 | 1 | 0.96 | 0.99 | 1 | 0.62 | 0.95 | 1 | 0.84 | 0.96 | 1 | 0.0013 | 0.0011 | 0.00032 | 0.00064 | 0.00069 | 0.00047 | 0.00048 | 0.00036 | 0.00053 | -0.42 | 31 / 36 (86%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00069 | 0.00049 | 0.00074 | 0.0023 | 0.00022 | 0.0071 | 1.7 | 10 / 11 (91%) | 9 / 11 (82%) | 0.909 | 0.818 |
| 3.5.1.10 | Formyltetrahydrofolate deformylase. | 0.2 | 0.53 | 1 | 0.64 | 1 | 1 | 0.25 | 0.89 | 1 | 0.17 | 0.49 | 1 | 0.86 | 0.99 | 1 | 0.56 | 0.9 | 1 | 0.00059 | 0.00048 | 0.00021 | 0.00056 | 0.00048 | 0.00057 | 0.00043 | 0.00032 | 0.00037 | -0.38 | 29 / 36 (81%) | 6 / 7 (86%) | 6 / 7 (86%) | 0.857 | 0.857 | 0.00026 | 0.00013 | 0.00039 | 0.00067 | 0.00014 | 0.0013 | 1.4 | 8 / 11 (73%) | 9 / 11 (82%) | 0.727 | 0.818 |
| 2.7.1.92 | 5-dehydro-2-deoxygluconokinase. | 0.59 | 0.79 | 1 | 0.66 | 1 | 1 | 0.2 | 0.87 | 1 | 0.86 | 0.95 | 1 | 0.76 | 0.97 | 1 | 0.46 | 0.86 | 1 | 0.022 | 0.022 | 0.021 | 0.022 | 0.021 | 0.0044 | 0.021 | 0.022 | 0.0075 | -0.067 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.021 | 0.019 | 0.0052 | 0.023 | 0.022 | 0.0038 | 0.13 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.21.92 | Endopeptidase Clp. | 0.061 | 0.49 | 1 | 0.74 | 1 | 1 | 0.83 | 1 | 1 | 0.061 | 0.43 | 1 | 0.57 | 0.93 | 1 | 0.31 | 0.79 | 1 | 0.015 | 0.015 | 0.015 | 0.013 | 0.014 | 0.0015 | 0.013 | 0.012 | 0.0028 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.016 | 0.015 | 0.0042 | 0.015 | 0.015 | 0.002 | -0.093 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.1.1.17 | Glutamate--tRNA ligase. | 0.17 | 0.51 | 1 | 0.73 | 1 | 1 | 0.54 | 0.95 | 1 | 0.12 | 0.47 | 1 | 0.65 | 0.95 | 1 | 0.85 | 0.96 | 1 | 0.028 | 0.028 | 0.027 | 0.025 | 0.023 | 0.0039 | 0.024 | 0.021 | 0.0059 | -0.059 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.029 | 0.028 | 0.0095 | 0.031 | 0.03 | 0.0067 | 0.096 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.2.53 | 2-keto-3-deoxy-L-rhamnonate aldolase. | 0.16 | 0.5 | 1 | 0.31 | 1 | 1 | 0.15 | 0.84 | 1 | 0.17 | 0.49 | 1 | 0.78 | 0.97 | 1 | 0.67 | 0.93 | 1 | 0.00034 | 2.8e-05 | 0 | 8.4e-05 | 0 | 0.00022 | 5.8e-05 | 0 | 0.00015 | -0.53 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.1.1.304 | Diacetyl reductase ((S)-acetoin forming). | 0.27 | 0.56 | 1 | 0.16 | 1 | 1 | 0.27 | 0.89 | 1 | 0.23 | 0.53 | 1 | 0.46 | 0.92 | 1 | 0.3 | 0.78 | 1 | 0.00056 | 0.00048 | 0.00036 | 0.00043 | 0.00033 | 0.00044 | 0.00018 | 0.00021 | 0.00015 | -1.3 | 31 / 36 (86%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.00064 | 4e-04 | 0.00059 | 0.00055 | 0.00044 | 0.00044 | -0.22 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 1.1.1.38 | Malate dehydrogenase (oxaloacetate-decarboxylating). | 0.096 | 0.49 | 1 | 0.008 | 0.7 | 1 | 0.00098 | 0.51 | 1 | 0.51 | 0.74 | 1 | 0.023 | 0.76 | 1 | 0.0021 | 0.48 | 1 | 0.0076 | 0.0076 | 0.0078 | 0.0086 | 0.0089 | 0.0024 | 0.0064 | 0.0054 | 0.0019 | -0.43 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0069 | 0.0072 | 0.0018 | 0.0084 | 0.0085 | 0.0023 | 0.28 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.3.1.128 | Transferred entry: 2.3.1.267. | 0.033 | 0.49 | 1 | 0.65 | 1 | 1 | 0.34 | 0.89 | 1 | 0.029 | 0.43 | 1 | 0.95 | 0.99 | 1 | 0.71 | 0.94 | 1 | 0.0052 | 0.0052 | 0.0047 | 0.0031 | 0.0032 | 0.0016 | 0.0039 | 0.0025 | 0.003 | 0.33 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.007 | 0.0055 | 0.0067 | 0.0054 | 0.0051 | 0.0024 | -0.37 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.4.99.- | 0.15 | 0.5 | 1 | 0.63 | 1 | 1 | 0.6 | 0.96 | 1 | 0.16 | 0.48 | 1 | 0.36 | 0.9 | 1 | 0.32 | 0.8 | 1 | 0.061 | 0.061 | 0.06 | 0.058 | 0.059 | 0.0077 | 0.056 | 0.055 | 0.012 | -0.051 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.064 | 0.064 | 0.0075 | 0.065 | 0.06 | 0.0095 | 0.022 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 3.2.1.183 | UDP-N-acetylglucosamine 2-epimerase (hydrolyzing). | 0.38 | 0.65 | 1 | 0.53 | 1 | 1 | 0.67 | 0.98 | 1 | 0.2 | 0.51 | 1 | 0.036 | 0.8 | 1 | 0.14 | 0.7 | 1 | 0.00088 | 0.00081 | 0.00057 | 0.00061 | 0.00045 | 7e-04 | 0.00075 | 0.00011 | 0.0012 | 0.3 | 33 / 36 (92%) | 7 / 7 (100%) | 4 / 7 (57%) | 1 | 0.571 | 0.00084 | 0.00087 | 0.00049 | 0.00093 | 0.00057 | 0.0011 | 0.15 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 7.5.2.10 | ABC-type D-xylose transporter. | 0.069 | 0.49 | 1 | 0.036 | 0.97 | 1 | 0.062 | 0.71 | 1 | 0.089 | 0.45 | 1 | 0.16 | 0.87 | 1 | 0.17 | 0.71 | 1 | 0.00055 | 0.00011 | 0 | 0.00039 | 0 | 0.00097 | 0.00012 | 0 | 0.00031 | -1.7 | 7 / 36 (19%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 1.1e-05 | 0 | 3.6e-05 | 1.5e-05 | 0 | 3.3e-05 | 0.45 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 3.4.11.5 | Prolyl aminopeptidase. | 0.086 | 0.49 | 1 | 0.78 | 1 | 1 | 0.73 | 0.99 | 1 | 0.062 | 0.43 | 1 | 0.54 | 0.93 | 1 | 0.66 | 0.93 | 1 | 0.0054 | 0.0054 | 0.0043 | 0.0033 | 0.0026 | 0.0016 | 0.0038 | 0.0034 | 0.0026 | 0.2 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0071 | 0.0049 | 0.0082 | 0.006 | 0.0058 | 0.0031 | -0.24 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.17.1.5 | Nicotinate dehydrogenase. | 0.49 | 0.73 | 1 | 0.92 | 1 | 1 | 0.84 | 1 | 1 | 0.67 | 0.86 | 1 | 0.79 | 0.97 | 1 | 0.72 | 0.94 | 1 | 0.0032 | 0.0032 | 0.0029 | 0.0036 | 0.0038 | 0.0016 | 0.0036 | 0.0029 | 0.002 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0032 | 0.0027 | 0.0018 | 0.0028 | 0.0027 | 0.0012 | -0.19 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.1.17 | Ornithine decarboxylase. | 0.92 | 0.96 | 1 | 0.44 | 1 | 1 | 0.12 | 0.82 | 1 | 0.7 | 0.88 | 1 | 0.18 | 0.87 | 1 | 0.079 | 0.66 | 1 | 0.0013 | 0.0013 | 0.001 | 0.0013 | 0.00048 | 0.0019 | 0.0012 | 0.00024 | 0.0021 | -0.12 | 35 / 36 (97%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.0011 | 0.0014 | 0.00071 | 0.0016 | 0.0017 | 0.00083 | 0.54 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.5.1.4 | RNA 3'-terminal-phosphate cyclase (ATP). | 0.44 | 0.69 | 1 | 0.27 | 1 | 1 | 0.48 | 0.94 | 1 | 0.78 | 0.92 | 1 | 0.47 | 0.92 | 1 | 0.3 | 0.78 | 1 | 0.00024 | 4.6e-05 | 0 | 0.00011 | 0 | 0.00026 | 8.7e-05 | 0 | 0.00023 | -0.34 | 7 / 36 (19%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.4e-05 | 0 | 4.1e-05 | 0 | 0 | 0 | -Inf | 4 / 11 (36%) | 0 / 11 (0%) | 0.364 | 0 |
| 3.6.3.6 | Transferred entry: 7.1.2.1. | 0.8 | 0.91 | 1 | 0.52 | 1 | 1 | 0.81 | 1 | 1 | 0.94 | 0.98 | 1 | 0.43 | 0.91 | 1 | 0.9 | 0.98 | 1 | 0.00024 | 5.9e-05 | 0 | 2.1e-05 | 0 | 4.5e-05 | 5.9e-05 | 0 | 0.00011 | 1.5 | 9 / 36 (25%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 8.2e-05 | 0 | 0.00026 | 6.2e-05 | 0 | 0.00011 | -0.4 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 2.6.1.17 | Succinyldiaminopimelate transaminase. | 0.33 | 0.6 | 1 | 0.61 | 1 | 1 | 0.71 | 0.98 | 1 | 0.27 | 0.56 | 1 | 0.55 | 0.93 | 1 | 0.66 | 0.93 | 1 | 0.004 | 0.004 | 0.0029 | 0.0027 | 0.0029 | 0.0012 | 0.0034 | 0.0028 | 0.0022 | 0.33 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0049 | 0.0026 | 0.0068 | 0.0044 | 0.0033 | 0.003 | -0.16 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.3.7 | 3'(2').5'-bisphosphate nucleotidase. | 0.074 | 0.49 | 1 | 0.57 | 1 | 1 | 0.06 | 0.71 | 1 | 0.008 | 0.38 | 1 | 0.26 | 0.88 | 1 | 0.023 | 0.56 | 1 | 0.00096 | 0.00069 | 0.00023 | 0.00074 | 0.00071 | 5e-04 | 0.00081 | 0.00064 | 0.00098 | 0.13 | 26 / 36 (72%) | 7 / 7 (100%) | 5 / 7 (71%) | 1 | 0.714 | 0.00036 | 0 | 0.00082 | 0.00091 | 1e-04 | 0.0015 | 1.3 | 5 / 11 (45%) | 9 / 11 (82%) | 0.455 | 0.818 |
| 1.2.1.10 | Acetaldehyde dehydrogenase (acetylating). | 0.44 | 0.69 | 1 | 0.8 | 1 | 1 | 0.97 | 1 | 1 | 0.5 | 0.74 | 1 | 0.54 | 0.93 | 1 | 0.63 | 0.92 | 1 | 0.0034 | 0.0034 | 0.0023 | 0.0026 | 0.0026 | 0.0015 | 0.0026 | 0.0023 | 0.002 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0044 | 0.0024 | 0.006 | 0.0034 | 0.002 | 0.0025 | -0.37 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.2.1.19 | Aminobutyraldehyde dehydrogenase. | 0.031 | 0.49 | 1 | 0.21 | 1 | 1 | 0.32 | 0.89 | 1 | 0.025 | 0.42 | 1 | 0.56 | 0.93 | 1 | 0.68 | 0.93 | 1 | 0.00019 | 3.6e-05 | 0 | 0.00012 | 0 | 0.00026 | 5.4e-05 | 0 | 1e-04 | -1.2 | 7 / 36 (19%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 2.6e-06 | 0 | 8.5e-06 | 2.7e-06 | 0 | 8.9e-06 | 0.054 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 1.8.1.2 | Assimilatory sulfite reductase (NADPH). | 0.63 | 0.83 | 1 | 0.41 | 1 | 1 | 0.79 | 1 | 1 | 0.8 | 0.93 | 1 | 0.37 | 0.9 | 1 | 0.64 | 0.92 | 1 | 7e-04 | 0.00058 | 0.00025 | 0.00082 | 4e-04 | 0.0014 | 0.00073 | 0.00021 | 0.0015 | -0.17 | 30 / 36 (83%) | 6 / 7 (86%) | 5 / 7 (71%) | 0.857 | 0.714 | 0.00056 | 0.00022 | 0.00081 | 0.00036 | 0.00026 | 0.00033 | -0.64 | 10 / 11 (91%) | 9 / 11 (82%) | 0.909 | 0.818 |
| 2.7.1.200 | Protein-N(pi)-phosphohistidine--galactitol phosphotransferase. | 0.1 | 0.5 | 1 | 0.38 | 1 | 1 | 0.61 | 0.96 | 1 | 0.16 | 0.48 | 1 | 0.59 | 0.93 | 1 | 0.42 | 0.85 | 1 | 0.00067 | 9.3e-05 | 0 | 0.00034 | 0 | 0.00089 | 0.00013 | 0 | 3e-04 | -1.4 | 5 / 36 (14%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 2.6e-06 | 0 | 8.5e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 3.6.1.19 | Transferred entry: 3.6.1.9. | 0.87 | 0.95 | 1 | 0.63 | 1 | 1 | 0.2 | 0.87 | 1 | 0.51 | 0.75 | 1 | 0.69 | 0.95 | 1 | 0.41 | 0.84 | 1 | 0.00087 | 0.00087 | 0.00066 | 0.00072 | 0.00055 | 0.00067 | 0.00085 | 5e-04 | 0.00087 | 0.24 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00072 | 0.00057 | 0.00057 | 0.0011 | 0.00097 | 0.00084 | 0.61 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.14.11.47 | [50S ribosomal protein L16]-arginine 3-hydroxylase. | 0.026 | 0.49 | 1 | 0.035 | 0.97 | 1 | 0.015 | 0.67 | 1 | 0.02 | 0.42 | 1 | 0.14 | 0.87 | 1 | 0.053 | 0.65 | 1 | 0.00024 | 3.9e-05 | 0 | 0.00013 | 0 | 0.00026 | 4.8e-05 | 0 | 0.00013 | -1.4 | 6 / 36 (17%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 0 | 0 | 0 | 1.8e-05 | 0 | 4e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 2.7.1.22 | Ribosylnicotinamide kinase. | 0.81 | 0.91 | 1 | 0.71 | 1 | 1 | 0.77 | 0.99 | 1 | 0.81 | 0.93 | 1 | 0.62 | 0.95 | 1 | 0.82 | 0.96 | 1 | 0.00043 | 3e-04 | 0.00011 | 0.00044 | 8.1e-05 | 0.00095 | 0.00037 | 0.00013 | 0.00069 | -0.25 | 25 / 36 (69%) | 6 / 7 (86%) | 4 / 7 (57%) | 0.857 | 0.571 | 0.00032 | 2e-04 | 0.00052 | 0.00015 | 0.00011 | 0.00017 | -1.1 | 7 / 11 (64%) | 8 / 11 (73%) | 0.636 | 0.727 |
| 2.2.1.- | 0.36 | 0.64 | 1 | 0.085 | 1 | 1 | 0.0082 | 0.61 | 1 | 0.45 | 0.71 | 1 | 0.15 | 0.87 | 1 | 0.018 | 0.55 | 1 | 0.0015 | 0.0011 | 0.00035 | 0.0012 | 0.00067 | 0.0013 | 0.00051 | 0.00022 | 0.00076 | -1.2 | 27 / 36 (75%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 0.00069 | 0.00038 | 0.0011 | 0.0018 | 3e-04 | 0.0028 | 1.4 | 7 / 11 (64%) | 11 / 11 (100%) | 0.636 | 1 | |
| 3.2.1.177 | Alpha-D-xyloside xylohydrolase. | 0.79 | 0.9 | 1 | 0.55 | 1 | 1 | 0.95 | 1 | 1 | 0.43 | 0.69 | 1 | 0.091 | 0.86 | 1 | 0.26 | 0.76 | 1 | 0.06 | 0.06 | 0.063 | 0.058 | 0.053 | 0.02 | 0.061 | 0.057 | 0.019 | 0.073 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.06 | 0.067 | 0.014 | 0.062 | 0.066 | 0.01 | 0.047 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.16.1.- | 0.02 | 0.49 | 1 | 0.0027 | 0.63 | 1 | 0.044 | 0.71 | 1 | 0.0083 | 0.38 | 1 | 0.011 | 0.75 | 1 | 0.13 | 0.68 | 1 | 0.00046 | 7.7e-05 | 0 | 0.00027 | 3.2e-05 | 0.00056 | 9.7e-05 | 0 | 0.00026 | -1.5 | 6 / 36 (17%) | 4 / 7 (57%) | 1 / 7 (14%) | 0.571 | 0.143 | 1.8e-05 | 0 | 5.9e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 | |
| 1.5.1.20 | Methylenetetrahydrofolate reductase (NAD(P)H). | 0.71 | 0.86 | 1 | 0.26 | 1 | 1 | 0.091 | 0.79 | 1 | 0.4 | 0.66 | 1 | 0.94 | 0.99 | 1 | 0.4 | 0.83 | 1 | 0.015 | 0.015 | 0.015 | 0.014 | 0.014 | 0.0021 | 0.013 | 0.013 | 0.0024 | -0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.015 | 0.014 | 0.0034 | 0.016 | 0.016 | 0.0019 | 0.093 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.1.2.20 | 2-dehydro-3-deoxyglucarate aldolase. | 0.079 | 0.49 | 1 | 0.78 | 1 | 1 | 0.89 | 1 | 1 | 0.091 | 0.45 | 1 | 0.85 | 0.98 | 1 | 0.86 | 0.96 | 1 | 0.00017 | 5.5e-05 | 0 | 9.7e-05 | 3.2e-05 | 0.00017 | 0.00012 | 0 | 0.00017 | 0.31 | 12 / 36 (33%) | 4 / 7 (57%) | 3 / 7 (43%) | 0.571 | 0.429 | 1.4e-05 | 0 | 3.7e-05 | 2.6e-05 | 0 | 4.9e-05 | 0.89 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 3.5.1.28 | N-acetylmuramoyl-L-alanine amidase. | 0.77 | 0.89 | 1 | 0.46 | 1 | 1 | 0.57 | 0.96 | 1 | 0.44 | 0.7 | 1 | 0.21 | 0.87 | 1 | 0.27 | 0.76 | 1 | 0.047 | 0.047 | 0.047 | 0.045 | 0.047 | 0.012 | 0.05 | 0.057 | 0.023 | 0.15 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.046 | 0.045 | 0.012 | 0.046 | 0.047 | 0.0067 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.1.56 | Lipopolysaccharide N-acetylglucosaminyltransferase. | 0.052 | 0.49 | 1 | 0.78 | 1 | 1 | 0.36 | 0.9 | 1 | 0.03 | 0.43 | 1 | 0.32 | 0.9 | 1 | 0.16 | 0.71 | 1 | 0.0019 | 0.0013 | 0.00046 | 0.00037 | 0 | 0.00056 | 0.00047 | 0.00012 | 0.00079 | 0.35 | 25 / 36 (69%) | 3 / 7 (43%) | 4 / 7 (57%) | 0.429 | 0.571 | 0.0025 | 0.00089 | 0.005 | 0.0013 | 0.00045 | 0.0015 | -0.94 | 9 / 11 (82%) | 9 / 11 (82%) | 0.818 | 0.818 |
| 2.7.1.206 | Protein-N(pi)-phosphohistidine--L-sorbose phosphotransferase. | 0.23 | 0.54 | 1 | 0.35 | 1 | 1 | 0.75 | 0.99 | 1 | 0.28 | 0.57 | 1 | 0.24 | 0.88 | 1 | 0.93 | 0.98 | 1 | 0.00054 | 7.5e-05 | 0 | 0.00015 | 0 | 4e-04 | 0.00018 | 0 | 0.00043 | 0.26 | 5 / 36 (14%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 0 | 0 | 0 | 3.4e-05 | 0 | 8.7e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 3.1.22.- | 0.92 | 0.96 | 1 | 0.57 | 1 | 1 | 0.76 | 0.99 | 1 | 0.73 | 0.89 | 1 | 0.94 | 0.99 | 1 | 0.43 | 0.85 | 1 | 1e-04 | 1.4e-05 | 0 | 2e-05 | 0 | 5.4e-05 | 8.8e-06 | 0 | 2.3e-05 | -1.2 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2e-05 | 0 | 4.5e-05 | 9e-06 | 0 | 3e-05 | -1.2 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 | |
| 1.14.12.21 | Transferred entry: 1.14.13.208. | 0.83 | 0.92 | 1 | 0.3 | 1 | 1 | 0.12 | 0.82 | 1 | 0.58 | 0.8 | 1 | 0.52 | 0.93 | 1 | 0.32 | 0.8 | 1 | 0.00028 | 0.00018 | 0.00011 | 0.00019 | 6.4e-05 | 0.00027 | 7.7e-05 | 2.7e-05 | 0.00013 | -1.3 | 24 / 36 (67%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.00017 | 0.00013 | 0.00019 | 0.00027 | 0.00021 | 0.00025 | 0.67 | 8 / 11 (73%) | 8 / 11 (73%) | 0.727 | 0.727 |
| 2.5.1.46 | Deoxyhypusine synthase. | 0.96 | 0.98 | 1 | 0.14 | 1 | 1 | 0.12 | 0.82 | 1 | 0.9 | 0.97 | 1 | 0.3 | 0.9 | 1 | 0.36 | 0.82 | 1 | 0.00013 | 1.1e-05 | 0 | 8.1e-06 | 0 | 2.1e-05 | 0 | 0 | 0 | -Inf | 3 / 36 (8.3%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 1.1e-05 | 0 | 3.6e-05 | 2.1e-05 | 0 | 6.8e-05 | 0.93 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 6.1.2.1 | D-alanine--(R)-lactate ligase. | 0.72 | 0.87 | 1 | 0.98 | 1 | 1 | 0.61 | 0.96 | 1 | 0.43 | 0.69 | 1 | 0.71 | 0.95 | 1 | 0.38 | 0.82 | 1 | 0.00013 | 3.2e-05 | 0 | 5e-05 | 0 | 0.00013 | 2.5e-05 | 0 | 4.7e-05 | -1 | 9 / 36 (25%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 3.8e-05 | 0 | 6.9e-05 | 1.7e-05 | 0 | 3.9e-05 | -1.2 | 4 / 11 (36%) | 2 / 11 (18%) | 0.364 | 0.182 |
| 1.1.1.64 | Testosterone 17-beta-dehydrogenase (NADP(+)). | 0.075 | 0.49 | 1 | 0.93 | 1 | 1 | 0.47 | 0.93 | 1 | 0.058 | 0.43 | 1 | 0.49 | 0.92 | 1 | 0.15 | 0.71 | 1 | 0.0018 | 0.0012 | 0.00038 | 4e-04 | 0 | 0.00055 | 0.00046 | 6.8e-05 | 0.00091 | 0.2 | 24 / 36 (67%) | 3 / 7 (43%) | 4 / 7 (57%) | 0.429 | 0.571 | 0.0021 | 0.00096 | 0.004 | 0.0013 | 0.00044 | 0.0018 | -0.69 | 9 / 11 (82%) | 8 / 11 (73%) | 0.818 | 0.727 |
| 2.7.8.37 | Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase. | 0.52 | 0.75 | 1 | 1 | 1 | 1 | 0.99 | 1 | 1 | 0.77 | 0.92 | 1 | 0.71 | 0.95 | 1 | 0.64 | 0.92 | 1 | 0.00046 | 0.00024 | 4.3e-05 | 0.00038 | 5.7e-05 | 0.00078 | 0.00029 | 0.00011 | 0.00046 | -0.39 | 19 / 36 (53%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.00016 | 2.8e-05 | 0.00029 | 0.00021 | 0 | 0.00052 | 0.39 | 6 / 11 (55%) | 5 / 11 (45%) | 0.545 | 0.455 |
| 2.4.1.166 | Raffinose--raffinose alpha-galactosyltransferase. | 0.61 | 0.81 | 1 | 0.54 | 1 | 1 | 0.68 | 0.98 | 1 | 0.68 | 0.87 | 1 | 0.61 | 0.95 | 1 | 0.64 | 0.92 | 1 | 0.00012 | 2e-05 | 0 | 1.4e-05 | 0 | 3e-05 | 3.9e-06 | 0 | 1e-05 | -1.8 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 4.8e-05 | 0 | 0.00012 | 6.8e-06 | 0 | 2.2e-05 | -2.8 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 2.5.1.19 | 3-phosphoshikimate 1-carboxyvinyltransferase. | 0.26 | 0.56 | 1 | 0.79 | 1 | 1 | 0.92 | 1 | 1 | 0.17 | 0.49 | 1 | 0.25 | 0.88 | 1 | 0.32 | 0.8 | 1 | 0.019 | 0.019 | 0.018 | 0.017 | 0.017 | 0.0021 | 0.018 | 0.018 | 0.0045 | 0.082 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.02 | 0.018 | 0.0048 | 0.02 | 0.02 | 0.0026 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.4.13 | dCTP deaminase. | 0.12 | 0.5 | 1 | 0.7 | 1 | 1 | 0.45 | 0.93 | 1 | 0.14 | 0.47 | 1 | 0.86 | 0.99 | 1 | 0.31 | 0.79 | 1 | 0.00021 | 0.00013 | 4.9e-05 | 8.2e-05 | 3.2e-05 | 1e-04 | 4.8e-05 | 3.1e-05 | 5.4e-05 | -0.77 | 22 / 36 (61%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.00021 | 0.00017 | 2e-04 | 0.00013 | 0 | 0.00018 | -0.69 | 9 / 11 (82%) | 5 / 11 (45%) | 0.818 | 0.455 |
| 1.1.1.290 | 4-phosphoerythronate dehydrogenase. | 0.62 | 0.82 | 1 | 0.81 | 1 | 1 | 0.78 | 1 | 1 | 0.43 | 0.69 | 1 | 0.41 | 0.9 | 1 | 0.91 | 0.98 | 1 | 0.018 | 0.018 | 0.015 | 0.015 | 0.015 | 0.0043 | 0.016 | 0.015 | 0.0049 | 0.093 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.019 | 0.015 | 0.015 | 0.019 | 0.018 | 0.0078 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.6.1.11 | Acetylornithine transaminase. | 0.17 | 0.5 | 1 | 0.95 | 1 | 1 | 0.99 | 1 | 1 | 0.14 | 0.47 | 1 | 0.43 | 0.91 | 1 | 0.48 | 0.87 | 1 | 0.024 | 0.024 | 0.024 | 0.021 | 0.021 | 0.0037 | 0.022 | 0.026 | 0.007 | 0.067 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.026 | 0.025 | 0.0071 | 0.026 | 0.026 | 0.0049 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.6.2.1 | DNA topoisomerase. | 0.21 | 0.54 | 1 | 0.74 | 1 | 1 | 0.91 | 1 | 1 | 0.16 | 0.48 | 1 | 0.47 | 0.92 | 1 | 0.26 | 0.76 | 1 | 0.042 | 0.042 | 0.041 | 0.039 | 0.039 | 0.0075 | 0.039 | 0.038 | 0.01 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.045 | 0.042 | 0.011 | 0.043 | 0.044 | 0.0049 | -0.066 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.6.3.- | 0.21 | 0.54 | 1 | 0.9 | 1 | 1 | 0.68 | 0.98 | 1 | 0.17 | 0.49 | 1 | 0.4 | 0.9 | 1 | 0.78 | 0.95 | 1 | 0.012 | 0.012 | 0.012 | 0.01 | 0.0091 | 0.0039 | 0.01 | 0.011 | 0.0046 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.013 | 0.012 | 0.0044 | 0.014 | 0.014 | 0.005 | 0.11 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 2.7.1.162 | N-acetylhexosamine 1-kinase. | 0.94 | 0.97 | 1 | 0.54 | 1 | 1 | 0.089 | 0.79 | 1 | 0.71 | 0.89 | 1 | 0.83 | 0.98 | 1 | 0.8 | 0.96 | 1 | 0.0012 | 0.0011 | 0.00054 | 0.00095 | 0.0012 | 0.00069 | 0.00075 | 0.00036 | 0.00079 | -0.34 | 34 / 36 (94%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.00096 | 0.00052 | 0.0012 | 0.0016 | 0.0016 | 0.0014 | 0.74 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 6.3.2.29 | Cyanophycin synthase (L-aspartate-adding). | 0.15 | 0.5 | 1 | 0.58 | 1 | 1 | 0.73 | 0.99 | 1 | 0.033 | 0.43 | 1 | 0.1 | 0.86 | 1 | 0.24 | 0.76 | 1 | 0.0027 | 0.0022 | 0.0011 | 0.00096 | 2e-04 | 0.0014 | 0.0012 | 6e-04 | 0.0015 | 0.32 | 30 / 36 (83%) | 4 / 7 (57%) | 5 / 7 (71%) | 0.571 | 0.714 | 0.0034 | 0.00087 | 0.007 | 0.0025 | 0.0015 | 0.0024 | -0.44 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 2.7.7.49 | RNA-directed DNA polymerase. | 0.27 | 0.56 | 1 | 0.29 | 1 | 1 | 0.12 | 0.82 | 1 | 0.67 | 0.86 | 1 | 0.44 | 0.91 | 1 | 0.21 | 0.74 | 1 | 0.049 | 0.049 | 0.048 | 0.052 | 0.051 | 0.015 | 0.047 | 0.044 | 0.019 | -0.15 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.045 | 0.046 | 0.012 | 0.051 | 0.048 | 0.014 | 0.18 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.3.1.6 | Fumarate reductase (NADH). | 0.52 | 0.75 | 1 | 0.98 | 1 | 1 | 0.61 | 0.96 | 1 | 0.29 | 0.57 | 1 | 0.76 | 0.97 | 1 | 0.58 | 0.91 | 1 | 0.00021 | 7.7e-05 | 0 | 5.9e-05 | 0 | 0.00013 | 9.7e-05 | 0 | 0.00026 | 0.72 | 13 / 36 (36%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 7.9e-05 | 2.8e-05 | 0.00013 | 7.4e-05 | 0 | 0.00014 | -0.094 | 6 / 11 (55%) | 4 / 11 (36%) | 0.545 | 0.364 |
| 3.5.1.46 | 6-aminohexanoate-oligomer exohydrolase. | 0.084 | 0.49 | 1 | 0.91 | 1 | 1 | 0.79 | 1 | 1 | 0.44 | 0.69 | 1 | 0.29 | 0.89 | 1 | 0.27 | 0.77 | 1 | 0.0063 | 0.0063 | 0.0057 | 0.0078 | 0.0072 | 0.0042 | 0.0077 | 0.0074 | 0.0033 | -0.019 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0055 | 0.0057 | 0.0017 | 0.0053 | 0.0045 | 0.0011 | -0.053 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.41 | Glucose-1-phosphate phosphodismutase. | 0.68 | 0.85 | 1 | 0.83 | 1 | 1 | 0.75 | 0.99 | 1 | 0.95 | 0.99 | 1 | 0.59 | 0.93 | 1 | 0.96 | 0.99 | 1 | 0.00013 | 4e-05 | 0 | 5.5e-05 | 0 | 0.00013 | 5.3e-05 | 0 | 9.6e-05 | -0.053 | 11 / 36 (31%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 2.2e-05 | 0 | 3.8e-05 | 4e-05 | 0 | 7.6e-05 | 0.86 | 3 / 11 (27%) | 4 / 11 (36%) | 0.273 | 0.364 |
| 3.6.3.14 | Transferred entry: 7.1.2.2. | 0.8 | 0.9 | 1 | 0.99 | 1 | 1 | 0.39 | 0.91 | 1 | 0.36 | 0.63 | 1 | 0.26 | 0.88 | 1 | 0.8 | 0.96 | 1 | 0.043 | 0.043 | 0.043 | 0.041 | 0.043 | 0.01 | 0.041 | 0.04 | 0.011 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.042 | 0.039 | 0.011 | 0.046 | 0.048 | 0.0069 | 0.13 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.21.89 | Signal peptidase I. | 0.25 | 0.56 | 1 | 0.99 | 1 | 1 | 0.91 | 1 | 1 | 0.23 | 0.53 | 1 | 0.13 | 0.87 | 1 | 0.089 | 0.66 | 1 | 0.028 | 0.028 | 0.028 | 0.027 | 0.026 | 0.0033 | 0.027 | 0.026 | 0.0057 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.029 | 0.028 | 0.0032 | 0.028 | 0.029 | 0.0028 | -0.051 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.27.- | 0.95 | 0.98 | 1 | 0.19 | 1 | 1 | 0.61 | 0.96 | 1 | 0.99 | 1 | 1 | 0.21 | 0.88 | 1 | 0.61 | 0.92 | 1 | 0.00038 | 0.00029 | 0.00024 | 0.00024 | 0.00024 | 2e-04 | 4e-04 | 0.00025 | 0.00042 | 0.74 | 28 / 36 (78%) | 5 / 7 (71%) | 6 / 7 (86%) | 0.714 | 0.857 | 0.00025 | 0.00012 | 0.00028 | 0.00031 | 0.00021 | 3e-04 | 0.31 | 8 / 11 (73%) | 9 / 11 (82%) | 0.727 | 0.818 | |
| 1.16.1.9 | Ferric-chelate reductase (NADPH). | 0.19 | 0.53 | 1 | 0.32 | 1 | 1 | 0.95 | 1 | 1 | 0.21 | 0.51 | 1 | 0.53 | 0.93 | 1 | 0.74 | 0.94 | 1 | 0.00037 | 4.2e-05 | 0 | 0.00011 | 0 | 0.00027 | 9.7e-05 | 0 | 0.00026 | -0.18 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 7.3e-06 | 0 | 2.4e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 6.1.1.11 | Serine--tRNA ligase. | 0.023 | 0.49 | 1 | 0.98 | 1 | 1 | 0.83 | 1 | 1 | 0.056 | 0.43 | 1 | 0.28 | 0.88 | 1 | 0.42 | 0.85 | 1 | 0.023 | 0.023 | 0.024 | 0.019 | 0.018 | 0.0045 | 0.019 | 0.022 | 0.0047 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.025 | 0.025 | 0.0063 | 0.025 | 0.025 | 0.0033 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.6.1.65 | (d)CTP diphosphatase. | 0.25 | 0.56 | 1 | 0.17 | 1 | 1 | 0.62 | 0.96 | 1 | 0.27 | 0.56 | 1 | 0.17 | 0.87 | 1 | 0.59 | 0.91 | 1 | 0.00012 | 3.8e-05 | 0 | 3.6e-05 | 0 | 5.7e-05 | 1e-04 | 5.4e-05 | 0.00015 | 1.5 | 12 / 36 (33%) | 3 / 7 (43%) | 4 / 7 (57%) | 0.429 | 0.571 | 5.4e-06 | 0 | 1.2e-05 | 3.4e-05 | 0 | 7.3e-05 | 2.7 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 1.14.13.81 | Magnesium-protoporphyrin IX monomethyl ester (oxidative) cyclase. | 0.037 | 0.49 | 1 | 0.25 | 1 | 1 | 0.18 | 0.86 | 1 | 0.032 | 0.43 | 1 | 0.4 | 0.9 | 1 | 0.25 | 0.76 | 1 | 0.00067 | 0.00067 | 0.00048 | 0.00099 | 0.00079 | 0.00067 | 0.00073 | 0.00071 | 0.00054 | -0.44 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.00049 | 0.00048 | 0.00051 | 6e-04 | 0.00041 | 0.00059 | 0.29 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.90 | Aryl-alcohol dehydrogenase. | 0.011 | 0.49 | 1 | 0.3 | 1 | 1 | 0.047 | 0.71 | 1 | 0.0054 | 0.36 | 1 | 0.27 | 0.88 | 1 | 0.035 | 0.63 | 1 | 9.2e-05 | 3.3e-05 | 0 | 2.3e-06 | 0 | 6.1e-06 | 3.8e-05 | 0 | 8.8e-05 | 4 | 13 / 36 (36%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 6.1e-05 | 5.7e-05 | 5.8e-05 | 2.3e-05 | 0 | 4e-05 | -1.4 | 7 / 11 (64%) | 3 / 11 (27%) | 0.636 | 0.273 |
| 2.3.3.14 | Homocitrate synthase. | 0.24 | 0.55 | 1 | 0.29 | 1 | 1 | 0.12 | 0.82 | 1 | 0.21 | 0.52 | 1 | 0.23 | 0.88 | 1 | 0.12 | 0.67 | 1 | 0.00023 | 1.9e-05 | 0 | 2.1e-05 | 0 | 4.2e-05 | 0 | 0 | 0 | -Inf | 3 / 36 (8.3%) | 2 / 7 (29%) | 0 / 7 (0%) | 0.286 | 0 | 0 | 0 | 0 | 5e-05 | 0 | 0.00016 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 3.5.2.12 | 6-aminohexanoate-cyclic-dimer hydrolase. | 0.17 | 0.51 | 1 | 0.019 | 0.92 | 1 | 0.021 | 0.71 | 1 | 0.21 | 0.51 | 1 | 0.022 | 0.76 | 1 | 0.04 | 0.63 | 1 | 0.00021 | 1e-04 | 1.7e-05 | 2.9e-05 | 0 | 5e-05 | 0.00019 | 0.00014 | 2e-04 | 2.7 | 18 / 36 (50%) | 2 / 7 (29%) | 5 / 7 (71%) | 0.286 | 0.714 | 0.00014 | 0 | 2e-04 | 6.2e-05 | 3.3e-05 | 7.6e-05 | -1.2 | 5 / 11 (45%) | 6 / 11 (55%) | 0.455 | 0.545 |
| 3.5.1.89 | N-acetylglucosaminylphosphatidylinositol deacetylase. | 0.63 | 0.82 | 1 | 0.13 | 1 | 1 | 0.28 | 0.89 | 1 | 0.71 | 0.88 | 1 | 0.19 | 0.87 | 1 | 0.21 | 0.74 | 1 | 0.00029 | 0.00012 | 0 | 2e-04 | 0 | 0.00031 | 2.4e-05 | 0 | 4.3e-05 | -3.1 | 15 / 36 (42%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 0.00017 | 0 | 0.00036 | 8.1e-05 | 3e-05 | 0.00011 | -1.1 | 4 / 11 (36%) | 6 / 11 (55%) | 0.364 | 0.545 |
| 1.1.1.88 | Hydroxymethylglutaryl-CoA reductase. | 0.72 | 0.87 | 1 | 0.56 | 1 | 1 | 0.18 | 0.86 | 1 | 0.97 | 0.99 | 1 | 0.39 | 0.9 | 1 | 0.11 | 0.66 | 1 | 0.00042 | 0.00032 | 0.00019 | 0.00026 | 0.00022 | 0.00023 | 0.00034 | 0.00025 | 0.00034 | 0.39 | 28 / 36 (78%) | 6 / 7 (86%) | 7 / 7 (100%) | 0.857 | 1 | 0.00043 | 0.00029 | 0.00055 | 0.00025 | 7.1e-05 | 0.00043 | -0.78 | 8 / 11 (73%) | 7 / 11 (64%) | 0.727 | 0.636 |
| 2.7.1.166 | 3-deoxy-D-manno-octulosonic acid kinase. | 0.73 | 0.87 | 1 | 0.62 | 1 | 1 | 0.19 | 0.87 | 1 | 0.69 | 0.87 | 1 | 0.65 | 0.95 | 1 | 0.33 | 0.8 | 1 | 0.00037 | 0.00013 | 0 | 9e-05 | 0 | 0.00014 | 5.5e-05 | 0 | 0.00011 | -0.71 | 13 / 36 (36%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 7.5e-05 | 0 | 0.00016 | 0.00027 | 0 | 0.00055 | 1.8 | 3 / 11 (27%) | 5 / 11 (45%) | 0.273 | 0.455 |
| 2.6.-.- | 0.83 | 0.92 | 1 | 0.77 | 1 | 1 | 0.66 | 0.97 | 1 | 0.93 | 0.98 | 1 | 0.31 | 0.9 | 1 | 0.36 | 0.81 | 1 | 0.00084 | 0.00082 | 0.00063 | 0.00067 | 6e-04 | 0.00041 | 0.00077 | 6e-04 | 0.00081 | 0.2 | 35 / 36 (97%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00086 | 0.00053 | 9e-04 | 0.00091 | 7e-04 | 0.00071 | 0.082 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 3.2.1.170 | Mannosylglycerate hydrolase. | 0.67 | 0.84 | 1 | 0.89 | 1 | 1 | 0.4 | 0.92 | 1 | 0.92 | 0.97 | 1 | 0.56 | 0.93 | 1 | 0.87 | 0.97 | 1 | 0.0097 | 0.0097 | 0.0097 | 0.0099 | 0.013 | 0.0041 | 0.0098 | 0.0087 | 0.0043 | -0.015 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0092 | 0.01 | 0.0033 | 0.01 | 0.01 | 0.0027 | 0.12 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.2.7.4 | Anaerobic carbon-monoxide dehydrogenase. | 0.13 | 0.5 | 1 | 0.39 | 1 | 1 | 0.87 | 1 | 1 | 0.54 | 0.77 | 1 | 0.94 | 0.99 | 1 | 0.26 | 0.76 | 1 | 0.0067 | 0.0067 | 0.0064 | 0.0085 | 0.0092 | 0.0035 | 0.0077 | 0.0079 | 0.0036 | -0.14 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0063 | 0.0064 | 0.0019 | 0.0054 | 0.0058 | 0.0017 | -0.22 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.1.17 | Glucuronosyltransferase. | 0.2 | 0.53 | 1 | 0.35 | 1 | 1 | 0.22 | 0.88 | 1 | 0.32 | 0.59 | 1 | 0.97 | 0.99 | 1 | 0.65 | 0.92 | 1 | 0.00036 | 5e-05 | 0 | 0.00015 | 0 | 4e-04 | 8.7e-05 | 0 | 0.00023 | -0.79 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 9.5e-06 | 0 | 2.3e-05 | 1.9 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 2.2.1.9 | 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylic-acid | 0.32 | 0.6 | 1 | 0.4 | 1 | 1 | 0.036 | 0.71 | 1 | 0.2 | 0.5 | 1 | 0.43 | 0.91 | 1 | 0.032 | 0.62 | 1 | 0.0027 | 0.0027 | 0.0019 | 0.0026 | 0.0019 | 0.0012 | 0.0022 | 0.0022 | 0.0013 | -0.24 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0021 | 0.0016 | 0.0021 | 0.0036 | 0.0026 | 0.0036 | 0.78 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.1.1.18 | Glutamine--tRNA ligase. | 0.55 | 0.77 | 1 | 0.96 | 1 | 1 | 0.96 | 1 | 1 | 0.5 | 0.74 | 1 | 0.39 | 0.9 | 1 | 0.45 | 0.86 | 1 | 0.024 | 0.024 | 0.024 | 0.023 | 0.023 | 0.0026 | 0.023 | 0.024 | 0.0044 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.025 | 0.026 | 0.0066 | 0.024 | 0.024 | 0.0055 | -0.059 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.2.7.- | 0.95 | 0.98 | 1 | 0.56 | 1 | 1 | 0.1 | 0.82 | 1 | 0.79 | 0.92 | 1 | 0.99 | 1 | 1 | 0.3 | 0.78 | 1 | 0.025 | 0.025 | 0.025 | 0.024 | 0.024 | 0.0045 | 0.023 | 0.021 | 0.0077 | -0.061 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.024 | 0.024 | 0.0092 | 0.028 | 0.026 | 0.0088 | 0.22 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 2.7.1.33 | Pantothenate kinase. | 0.073 | 0.49 | 1 | 0.43 | 1 | 1 | 0.47 | 0.93 | 1 | 0.06 | 0.43 | 1 | 0.12 | 0.87 | 1 | 0.16 | 0.71 | 1 | 0.019 | 0.019 | 0.017 | 0.016 | 0.016 | 0.0026 | 0.017 | 0.017 | 0.0044 | 0.087 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.02 | 0.018 | 0.008 | 0.02 | 0.019 | 0.0033 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.2.1.41 | Glutamate-5-semialdehyde dehydrogenase. | 0.04 | 0.49 | 1 | 0.69 | 1 | 1 | 0.72 | 0.98 | 1 | 0.093 | 0.45 | 1 | 0.19 | 0.87 | 1 | 0.22 | 0.74 | 1 | 0.017 | 0.017 | 0.017 | 0.015 | 0.014 | 0.0024 | 0.016 | 0.014 | 0.0036 | 0.093 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.018 | 0.017 | 0.0046 | 0.018 | 0.018 | 0.002 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 6.3.2.2 | Glutamate--cysteine ligase. | 0.27 | 0.56 | 1 | 0.82 | 1 | 1 | 0.95 | 1 | 1 | 0.23 | 0.53 | 1 | 0.66 | 0.95 | 1 | 0.83 | 0.96 | 1 | 0.0047 | 0.0047 | 0.0035 | 0.0027 | 0.0025 | 0.00076 | 0.0031 | 0.0035 | 0.0014 | 0.2 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0056 | 0.0038 | 0.0072 | 0.0062 | 0.0047 | 0.0066 | 0.15 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.6.3.31 | Transferred entry: 7.6.2.11. | 0.14 | 0.5 | 1 | 0.98 | 1 | 1 | 0.79 | 1 | 1 | 0.071 | 0.43 | 1 | 0.82 | 0.98 | 1 | 0.74 | 0.94 | 1 | 0.016 | 0.016 | 0.011 | 0.0097 | 0.0079 | 0.0068 | 0.01 | 0.0084 | 0.0078 | 0.044 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.023 | 0.012 | 0.034 | 0.018 | 0.012 | 0.013 | -0.35 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.4.1.9 | Leucine dehydrogenase. | 0.92 | 0.96 | 1 | 0.88 | 1 | 1 | 0.81 | 1 | 1 | 0.94 | 0.98 | 1 | 0.62 | 0.95 | 1 | 0.59 | 0.91 | 1 | 0.00085 | 0.00083 | 0.00059 | 0.00081 | 6e-04 | 0.00065 | 0.00083 | 6e-04 | 0.00061 | 0.035 | 35 / 36 (97%) | 7 / 7 (100%) | 6 / 7 (86%) | 1 | 0.857 | 0.00084 | 0.00052 | 9e-04 | 0.00082 | 7e-04 | 0.00058 | -0.035 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.5.1.88 | Peptide deformylase. | 0.049 | 0.49 | 1 | 0.97 | 1 | 1 | 0.43 | 0.93 | 1 | 0.039 | 0.43 | 1 | 0.4 | 0.9 | 1 | 0.12 | 0.66 | 1 | 0.017 | 0.017 | 0.016 | 0.015 | 0.014 | 0.0029 | 0.015 | 0.016 | 0.0044 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.02 | 0.019 | 0.0084 | 0.018 | 0.017 | 0.0036 | -0.15 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.48 | Uridine kinase. | 0.56 | 0.78 | 1 | 0.57 | 1 | 1 | 0.12 | 0.82 | 1 | 0.97 | 0.99 | 1 | 0.92 | 0.99 | 1 | 0.31 | 0.79 | 1 | 0.016 | 0.016 | 0.016 | 0.015 | 0.016 | 0.0028 | 0.015 | 0.016 | 0.0046 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.015 | 0.015 | 0.004 | 0.017 | 0.017 | 0.004 | 0.18 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.7.1.56 | 1-phosphofructokinase. | 0.44 | 0.69 | 1 | 0.79 | 1 | 1 | 0.27 | 0.89 | 1 | 0.4 | 0.66 | 1 | 0.89 | 0.99 | 1 | 0.54 | 0.9 | 1 | 0.00054 | 0.00046 | 0.00034 | 4e-04 | 0.00036 | 0.00038 | 0.00054 | 0.00034 | 0.00061 | 0.43 | 31 / 36 (86%) | 6 / 7 (86%) | 5 / 7 (71%) | 0.857 | 0.714 | 0.00062 | 0.00043 | 0.00075 | 3e-04 | 3e-04 | 2e-04 | -1 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 1.1.1.385 | Dihydroanticapsin 7-dehydrogenase. | 0.51 | 0.74 | 1 | 0.37 | 1 | 1 | 0.44 | 0.93 | 1 | 0.82 | 0.93 | 1 | 0.58 | 0.93 | 1 | 0.56 | 0.9 | 1 | 0.00021 | 9.7e-05 | 0 | 0.00013 | 0 | 0.00019 | 0.00021 | 0 | 0.00029 | 0.69 | 17 / 36 (47%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 4.9e-05 | 2.8e-05 | 6e-05 | 5e-05 | 0 | 6.1e-05 | 0.029 | 6 / 11 (55%) | 5 / 11 (45%) | 0.545 | 0.455 |
| 2.7.7.4 | Sulfate adenylyltransferase. | 0.011 | 0.49 | 1 | 0.67 | 1 | 1 | 0.21 | 0.87 | 1 | 0.033 | 0.43 | 1 | 0.086 | 0.86 | 1 | 0.012 | 0.55 | 1 | 0.021 | 0.021 | 0.02 | 0.018 | 0.017 | 0.0036 | 0.018 | 0.019 | 0.0039 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.024 | 0.026 | 0.0067 | 0.021 | 0.021 | 0.0032 | -0.19 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 5.1.1.7 | Diaminopimelate epimerase. | 0.39 | 0.65 | 1 | 0.69 | 1 | 1 | 0.84 | 1 | 1 | 0.33 | 0.6 | 1 | 0.68 | 0.95 | 1 | 0.46 | 0.86 | 1 | 0.021 | 0.021 | 0.02 | 0.019 | 0.019 | 0.003 | 0.019 | 0.02 | 0.0053 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.022 | 0.02 | 0.0052 | 0.021 | 0.02 | 0.0055 | -0.067 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 1.1.1.282 | Quinate/shikimate dehydrogenase. | 0.5 | 0.74 | 1 | 0.13 | 1 | 1 | 0.63 | 0.96 | 1 | 0.93 | 0.98 | 1 | 0.11 | 0.87 | 1 | 0.69 | 0.94 | 1 | 0.0019 | 0.0019 | 0.0018 | 0.0019 | 0.0016 | 0.0013 | 0.0025 | 0.0027 | 0.0017 | 0.4 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0015 | 0.0014 | 0.00084 | 0.0021 | 0.0021 | 0.00042 | 0.49 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.4.2.11 | Transferred entry: 6.3.4.21. | 0.91 | 0.96 | 1 | 0.35 | 1 | 1 | 0.028 | 0.71 | 1 | 0.61 | 0.82 | 1 | 0.29 | 0.89 | 1 | 0.019 | 0.55 | 1 | 2e-04 | 5.5e-05 | 0 | 9.3e-05 | 0 | 0.00022 | 0.00012 | 0 | 0.00022 | 0.37 | 10 / 36 (28%) | 2 / 7 (29%) | 3 / 7 (43%) | 0.286 | 0.429 | 4.3e-05 | 0 | 6e-05 | 0 | 0 | 0 | -Inf | 5 / 11 (45%) | 0 / 11 (0%) | 0.455 | 0 |
| 1.13.11.27 | 4-hydroxyphenylpyruvate dioxygenase. | 0.21 | 0.53 | 1 | 0.48 | 1 | 1 | 0.9 | 1 | 1 | 0.31 | 0.58 | 1 | 0.27 | 0.88 | 1 | 0.65 | 0.92 | 1 | 0.00021 | 2.4e-05 | 0 | 5e-05 | 0 | 0.00013 | 5.5e-05 | 0 | 1e-04 | 0.14 | 4 / 36 (11%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 0 | 0 | 0 | 9.9e-06 | 0 | 3.3e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 3.6.3.8 | Transferred entry: 7.2.2.10. | 0.11 | 0.5 | 1 | 0.63 | 1 | 1 | 0.33 | 0.89 | 1 | 0.078 | 0.44 | 1 | 0.28 | 0.88 | 1 | 0.15 | 0.71 | 1 | 0.038 | 0.038 | 0.035 | 0.031 | 0.028 | 0.0088 | 0.034 | 0.028 | 0.015 | 0.13 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.045 | 0.043 | 0.029 | 0.038 | 0.033 | 0.0083 | -0.24 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.1.12.1 | 5' to 3' exodeoxyribonuclease (nucleoside 3'-phosphate-forming). | 0.37 | 0.64 | 1 | 0.75 | 1 | 1 | 0.64 | 0.97 | 1 | 0.69 | 0.87 | 1 | 0.49 | 0.92 | 1 | 0.92 | 0.98 | 1 | 0.0025 | 0.0025 | 0.0024 | 0.0026 | 0.0021 | 0.00081 | 0.0028 | 0.0026 | 0.0011 | 0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0022 | 0.0024 | 0.00084 | 0.0027 | 0.0026 | 0.00098 | 0.3 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.5.1.47 | Cysteine synthase. | 0.0044 | 0.49 | 1 | 0.7 | 1 | 1 | 0.59 | 0.96 | 1 | 0.051 | 0.43 | 1 | 0.1 | 0.86 | 1 | 0.075 | 0.66 | 1 | 0.02 | 0.02 | 0.02 | 0.018 | 0.016 | 0.003 | 0.018 | 0.019 | 0.0027 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.022 | 0.022 | 0.0031 | 0.021 | 0.021 | 0.0026 | -0.067 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.4.21.107 | Peptidase Do. | 0.94 | 0.97 | 1 | 0.89 | 1 | 1 | 0.62 | 0.96 | 1 | 0.62 | 0.83 | 1 | 0.71 | 0.95 | 1 | 0.33 | 0.8 | 1 | 0.019 | 0.019 | 0.018 | 0.019 | 0.022 | 0.0065 | 0.02 | 0.017 | 0.0095 | 0.074 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.02 | 0.02 | 0.0095 | 0.017 | 0.018 | 0.0044 | -0.23 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 4.2.99.21 | Isochorismate lyase. | 0.29 | 0.57 | 1 | 0.23 | 1 | 1 | 0.5 | 0.95 | 1 | 0.65 | 0.85 | 1 | 0.79 | 0.97 | 1 | 0.83 | 0.96 | 1 | 0.00031 | 4.3e-05 | 0 | 0.00015 | 0 | 4e-04 | 3.9e-05 | 0 | 1e-04 | -1.9 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 1.4e-05 | 0 | 3.7e-05 | 6.8e-06 | 0 | 2.3e-05 | -1 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 2.5.1.22 | Spermine synthase. | 0.11 | 0.5 | 1 | 0.068 | 1 | 1 | 0.14 | 0.83 | 1 | 0.14 | 0.47 | 1 | 0.38 | 0.9 | 1 | 0.55 | 0.9 | 1 | 0.00031 | 4.3e-05 | 0 | 0.00015 | 0 | 4e-04 | 5.8e-05 | 0 | 0.00015 | -1.4 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 2.7e-06 | 0 | 8.9e-06 | 0.054 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 1.6.99.3 | NADH dehydrogenase. | 0.67 | 0.84 | 1 | 0.62 | 1 | 1 | 0.58 | 0.96 | 1 | 0.24 | 0.54 | 1 | 0.35 | 0.9 | 1 | 0.91 | 0.98 | 1 | 0.0036 | 0.0036 | 0.0031 | 0.003 | 0.0026 | 0.0018 | 0.0034 | 0.0033 | 0.0022 | 0.18 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0033 | 0.0032 | 0.0014 | 0.0042 | 0.0031 | 0.0025 | 0.35 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 2.5.1.6 | Methionine adenosyltransferase. | 0.54 | 0.76 | 1 | 0.82 | 1 | 1 | 0.99 | 1 | 1 | 0.32 | 0.59 | 1 | 0.26 | 0.88 | 1 | 0.17 | 0.71 | 1 | 0.028 | 0.028 | 0.028 | 0.027 | 0.022 | 0.01 | 0.027 | 0.024 | 0.009 | 0 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.029 | 0.029 | 0.0052 | 0.028 | 0.029 | 0.0037 | -0.051 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| 3.2.2.- | 0.072 | 0.49 | 1 | 0.73 | 1 | 1 | 0.4 | 0.92 | 1 | 0.019 | 0.42 | 1 | 0.32 | 0.9 | 1 | 0.18 | 0.72 | 1 | 0.0078 | 0.0078 | 0.007 | 0.0061 | 0.0053 | 0.002 | 0.0066 | 0.0059 | 0.003 | 0.11 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0094 | 0.0083 | 0.0059 | 0.008 | 0.008 | 0.0021 | -0.23 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 7.2.3.- | 0.032 | 0.49 | 1 | 0.075 | 1 | 1 | 0.22 | 0.88 | 1 | 0.13 | 0.47 | 1 | 0.25 | 0.88 | 1 | 0.63 | 0.92 | 1 | 0.0062 | 0.0062 | 0.0055 | 0.0083 | 0.0086 | 0.0023 | 0.0064 | 0.0052 | 0.0028 | -0.38 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0056 | 0.0049 | 0.0024 | 0.0055 | 0.0041 | 0.0031 | -0.026 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 | |
| 4.1.99.22 | GTP 3'.8-cyclase. | 0.36 | 0.63 | 1 | 0.0054 | 0.7 | 1 | 0.28 | 0.89 | 1 | 0.22 | 0.52 | 1 | 0.066 | 0.86 | 1 | 0.95 | 0.99 | 1 | 0.0044 | 0.0044 | 0.0042 | 0.0045 | 0.0044 | 0.0018 | 0.0032 | 0.0028 | 0.00072 | -0.49 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 0.0051 | 0.0047 | 0.0018 | 0.0044 | 0.0042 | 0.0017 | -0.21 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
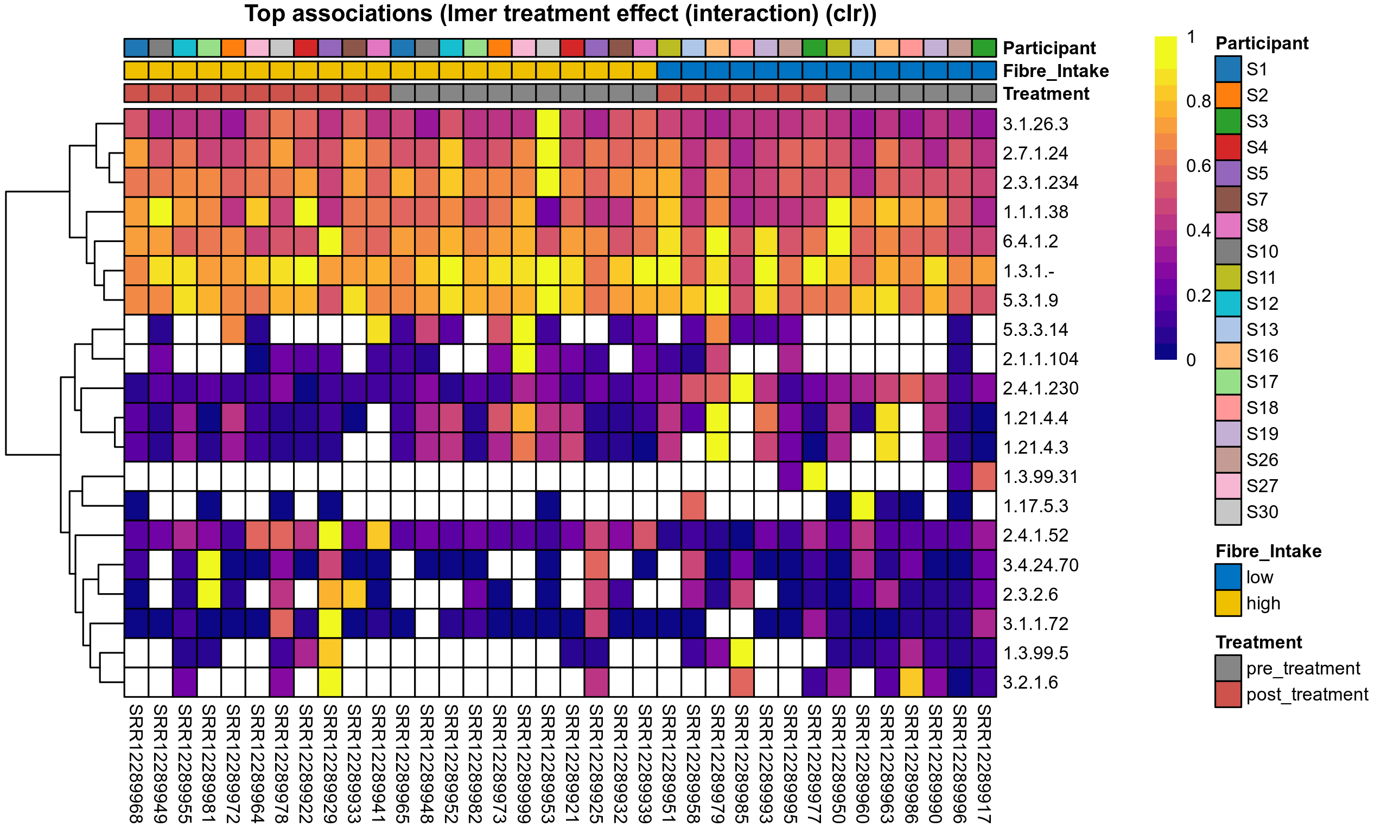
Click here to open full-sized image in new window.
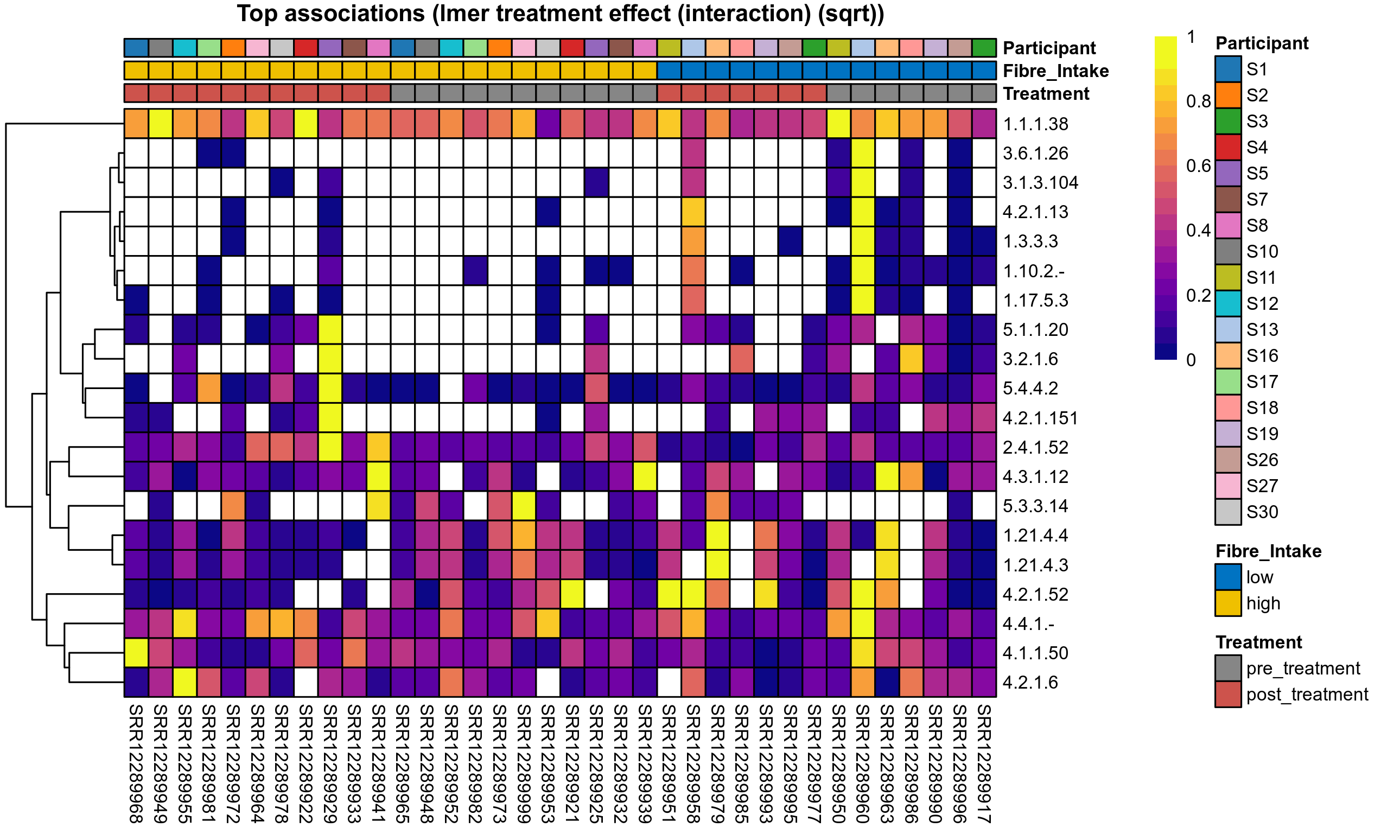
Click here to open full-sized image in new window.
P lmer study group effect (sqrt): Differentially abundant functions were identified by linear mixed effect regression without confounders. Random effects: Participant (intercept). Fixed effects: Fibre_Intake, Treatment, Fibre_Intake vs Treatment. Effect tested: Fibre_Intake.
P lmer study group effect (clr): Differentially abundant functions were identified by linear mixed effect regression without confounders. Random effects: Participant (intercept). Fixed effects: Fibre_Intake, Treatment, Fibre_Intake vs Treatment. Effect tested: Fibre_Intake.
P lmer time effect (sqrt): Differentially abundant functions were identified by linear mixed effect regression without confounders. Random effects: Participant (intercept). Fixed effects: Fibre_Intake, Treatment, Fibre_Intake vs Treatment. Effect tested: Treatment.
P lmer time effect (clr): Differentially abundant functions were identified by linear mixed effect regression without confounders. Random effects: Participant (intercept). Fixed effects: Fibre_Intake, Treatment, Fibre_Intake vs Treatment. Effect tested: Treatment.
P lmer treatment effect (interaction) (sqrt): Differentially abundant functions were identified by linear mixed effect regression without confounders. Random effects: Participant (intercept). Fixed effects: Fibre_Intake, Treatment, Fibre_Intake vs Treatment. Effect tested: Fibre_Intake vs Treatment.
P lmer treatment effect (interaction) (clr): Differentially abundant functions were identified by linear mixed effect regression without confounders. Random effects: Participant (intercept). Fixed effects: Fibre_Intake, Treatment, Fibre_Intake vs Treatment. Effect tested: Fibre_Intake vs Treatment.
The following plots present the distribution of the top most differentially abundant microbial functions across all applied statistical analysis. Plots are ordered alphabetically.