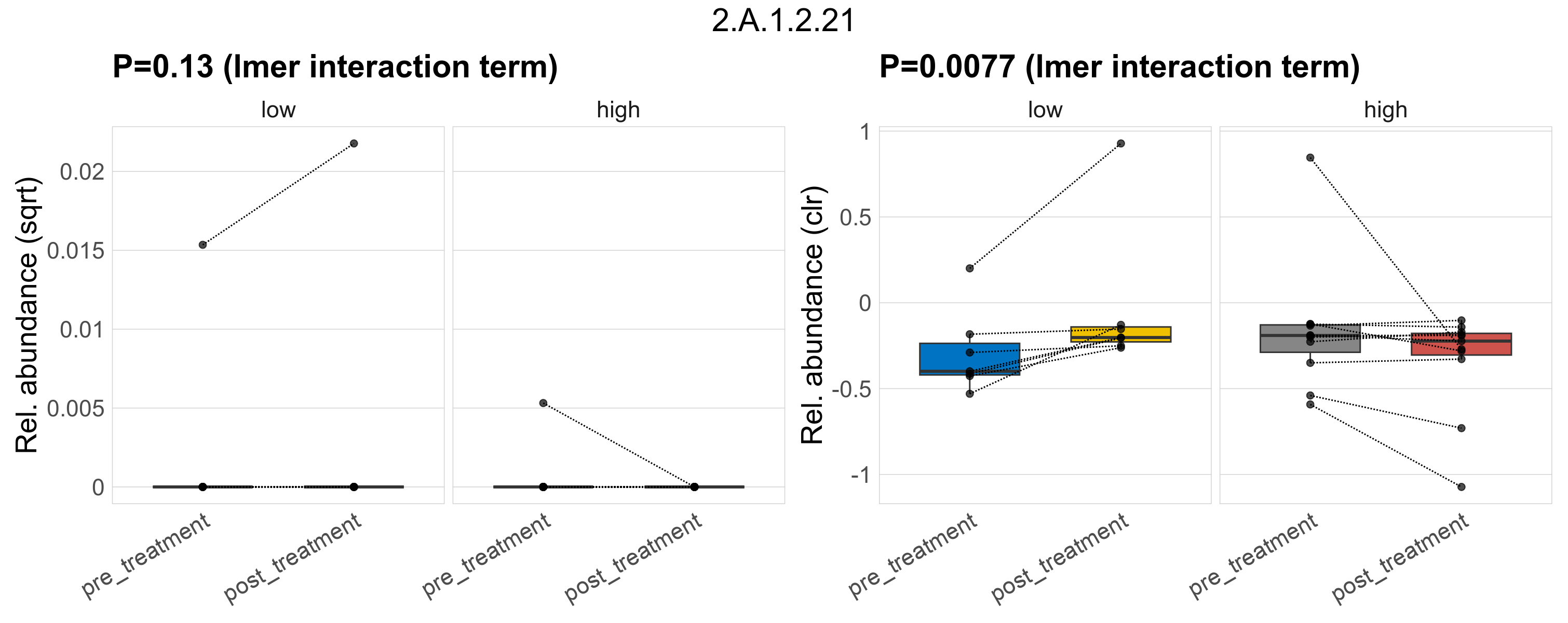
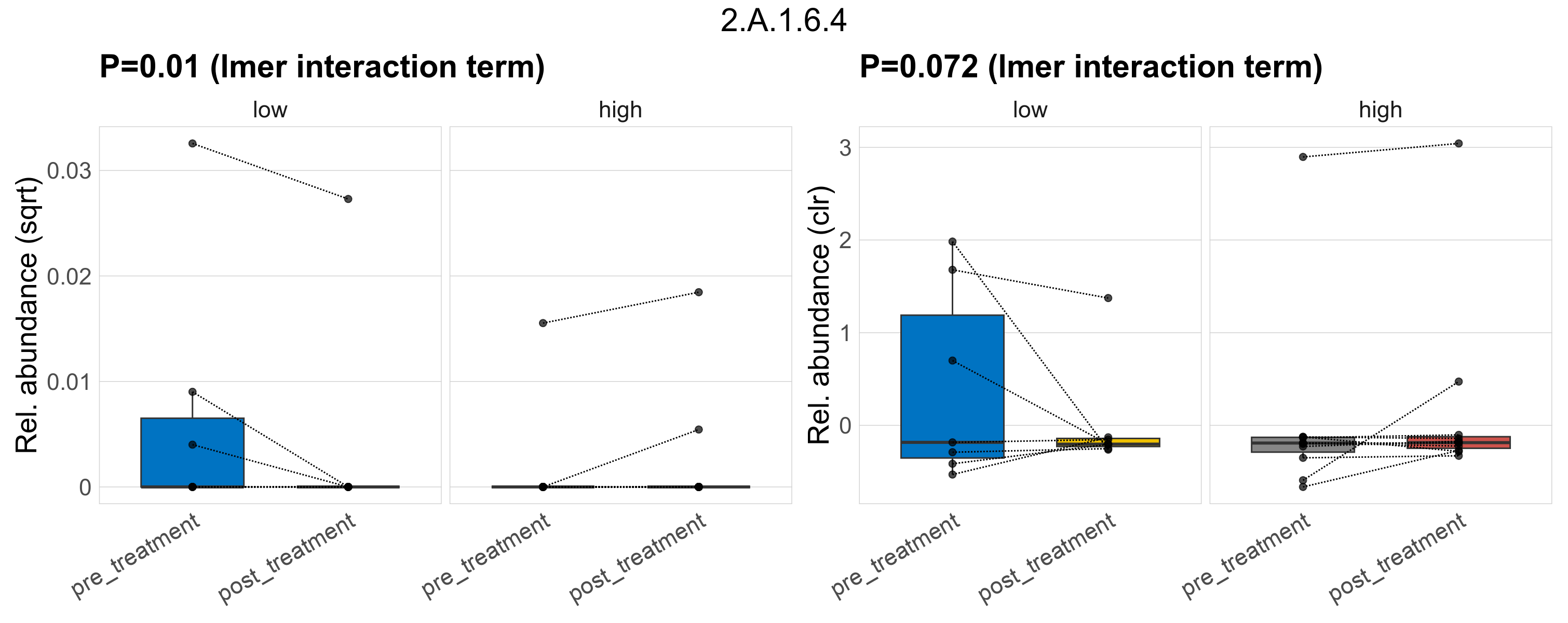
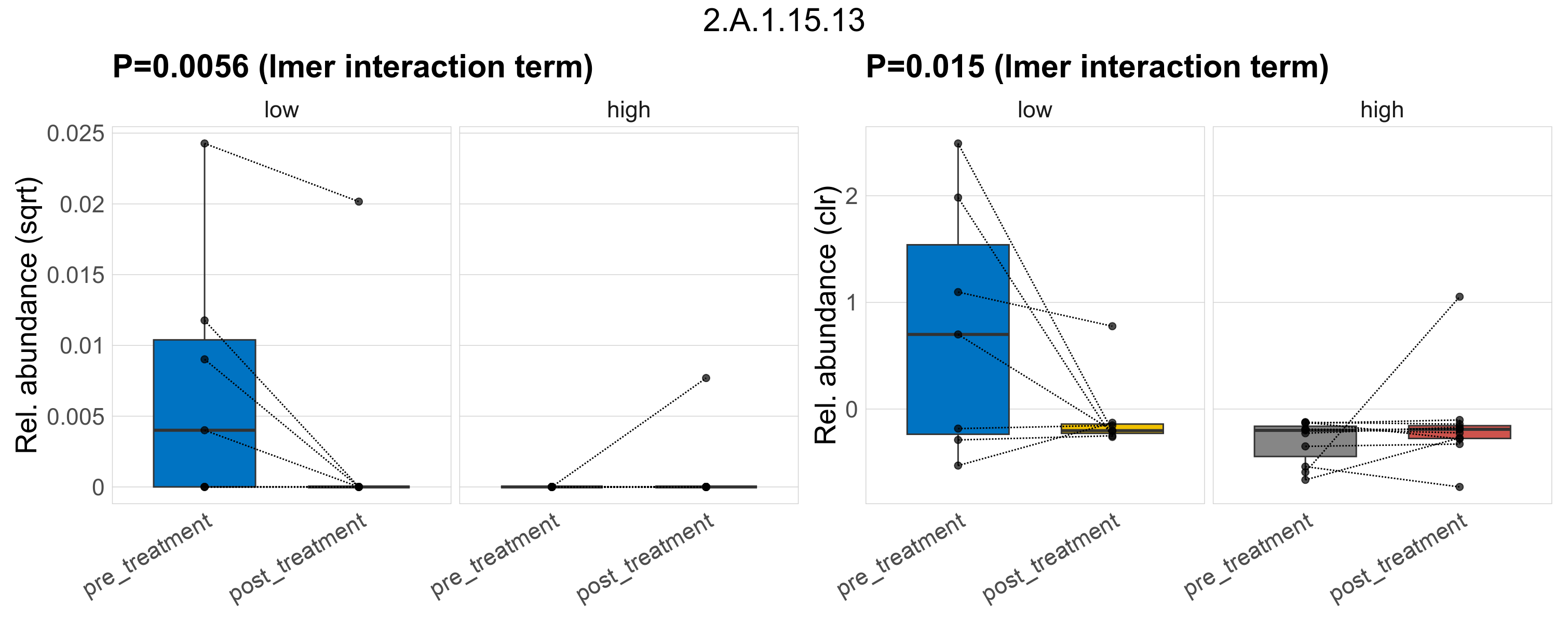
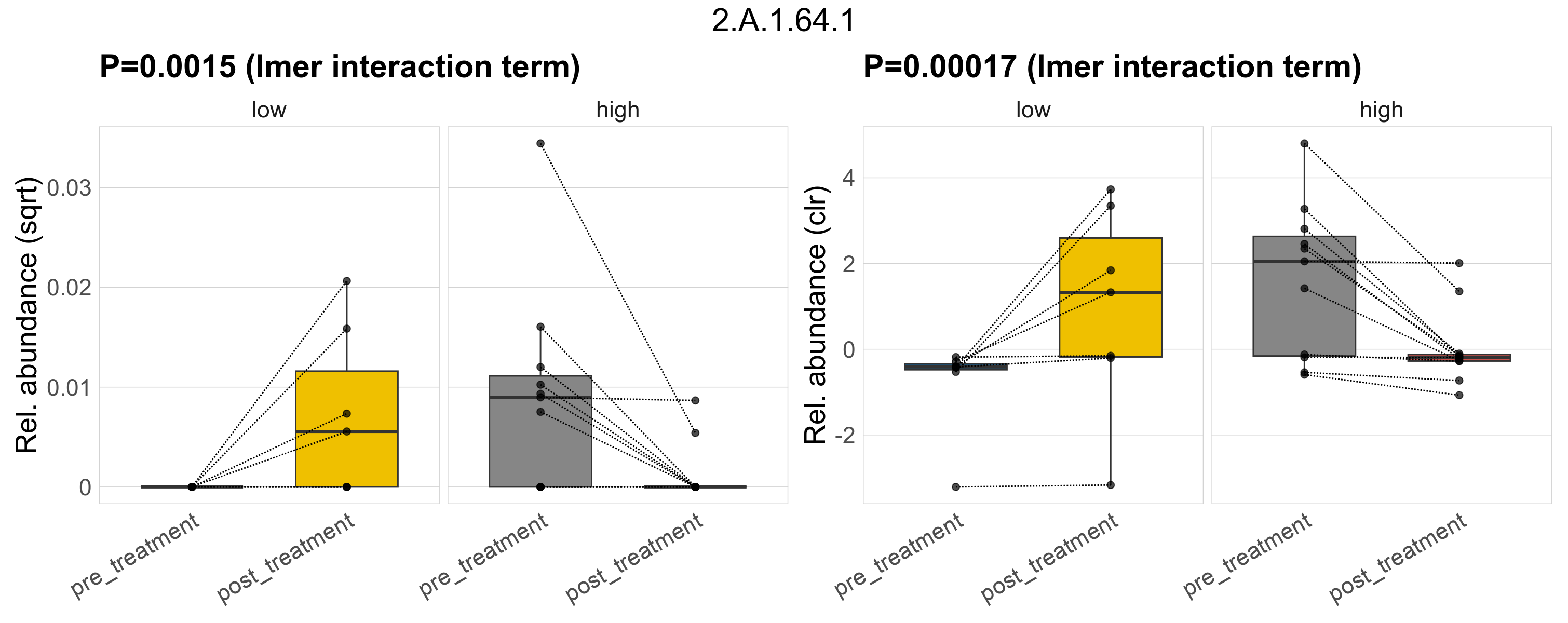
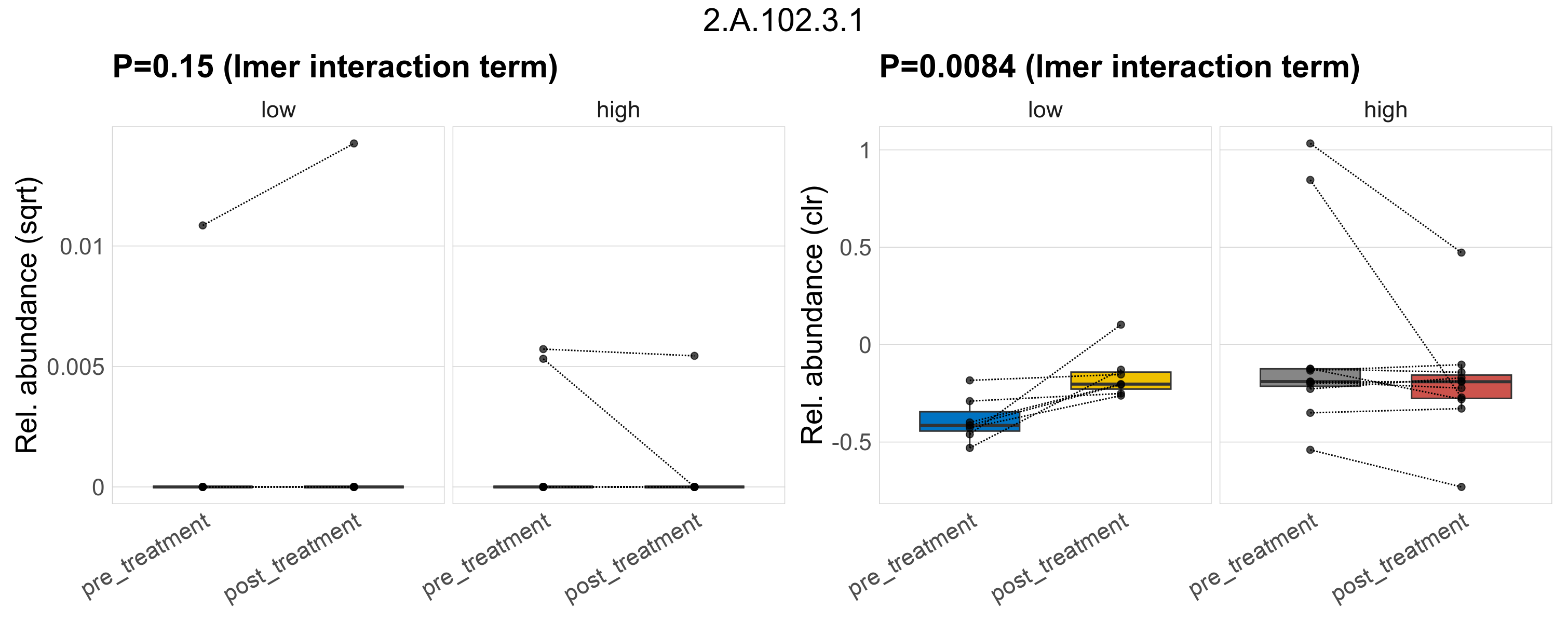
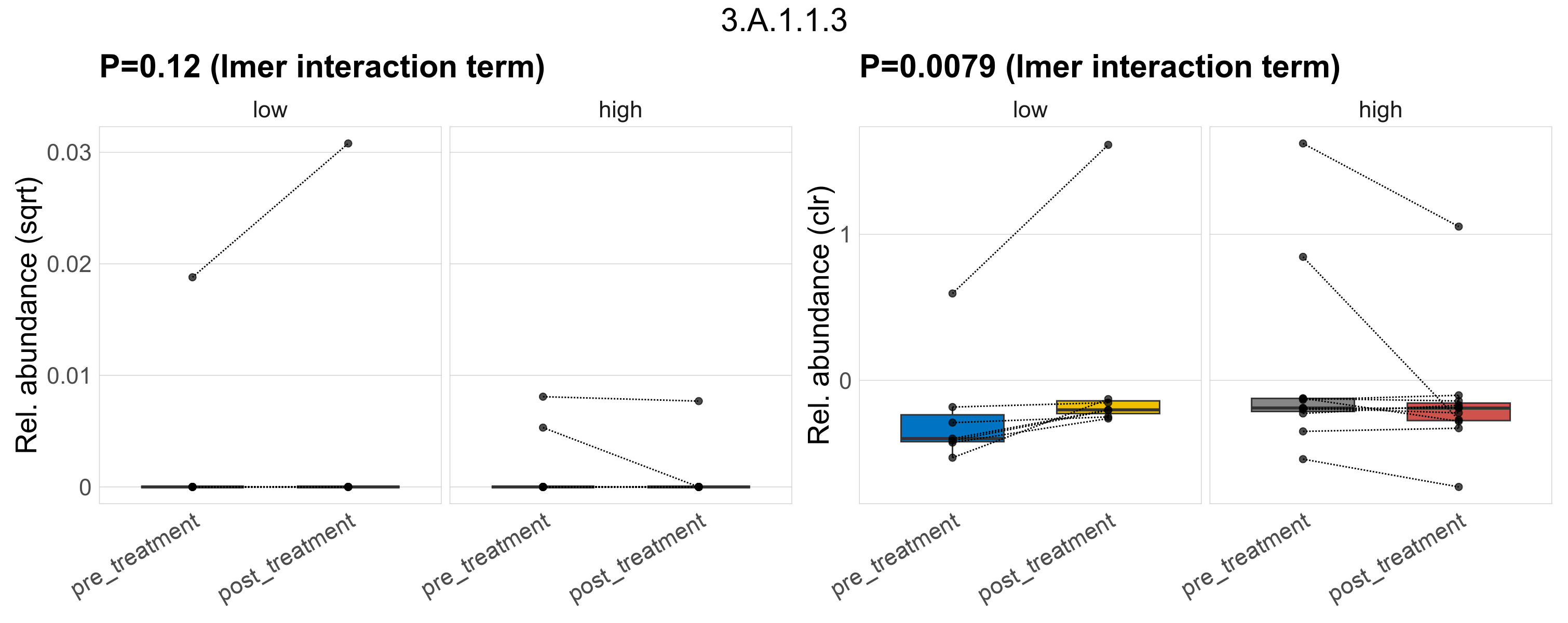
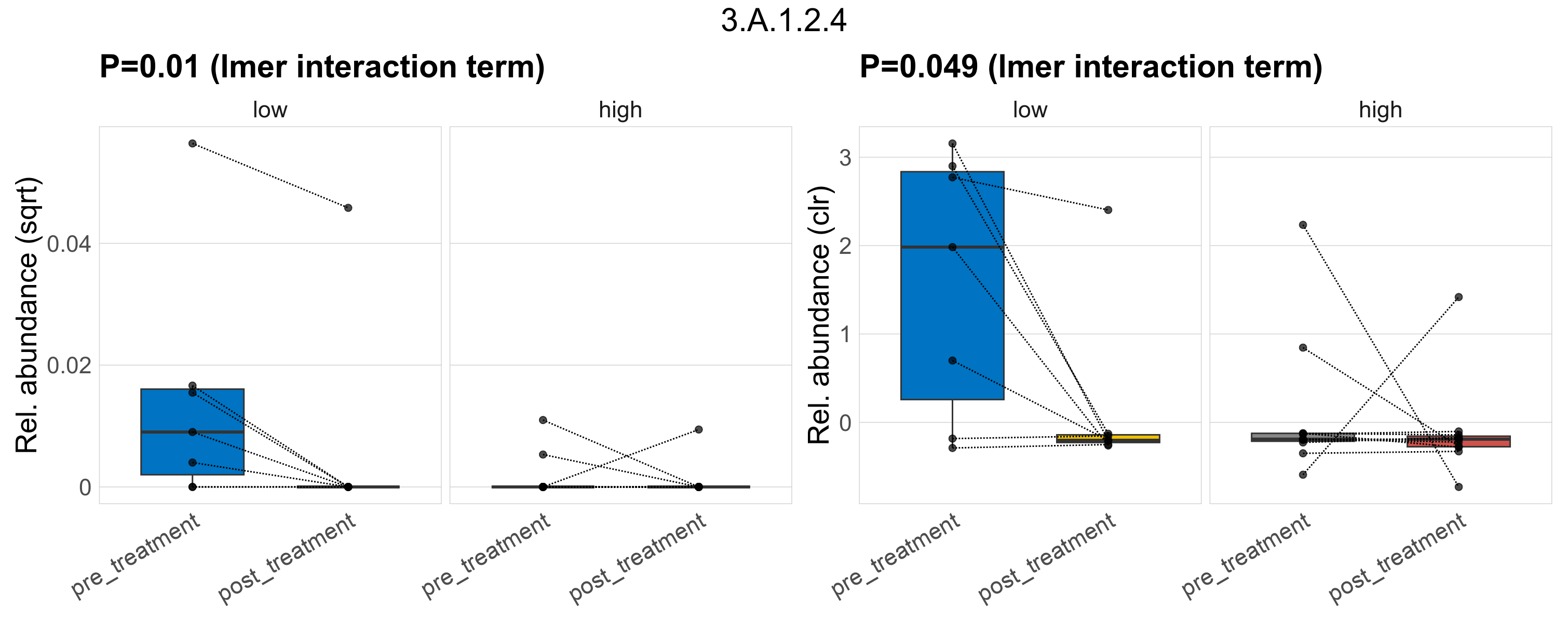
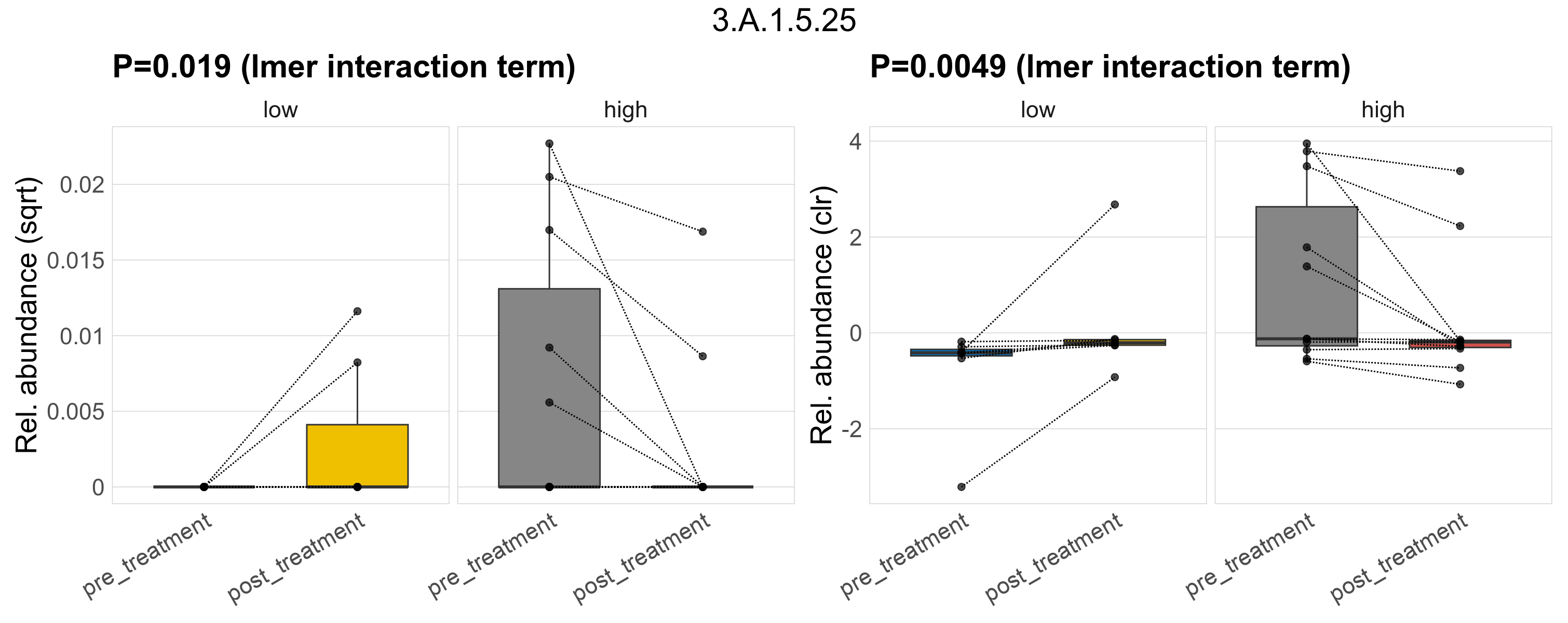
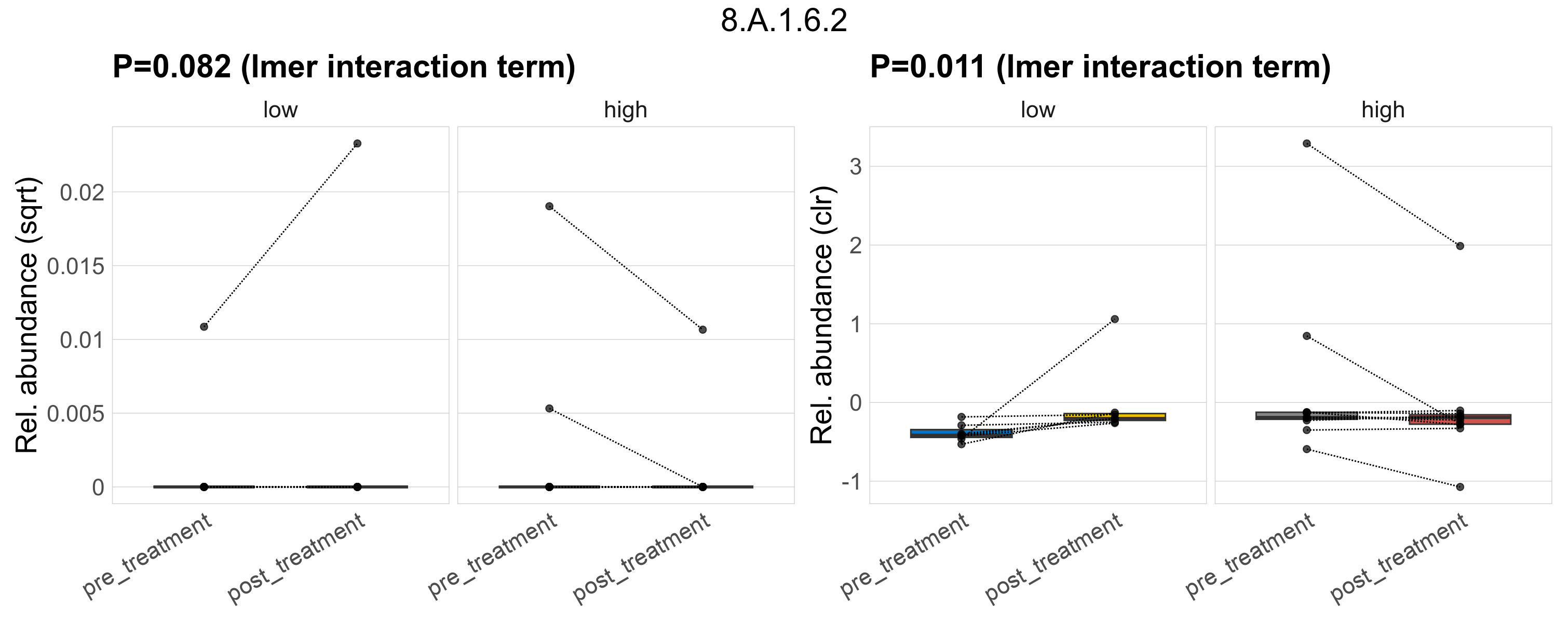
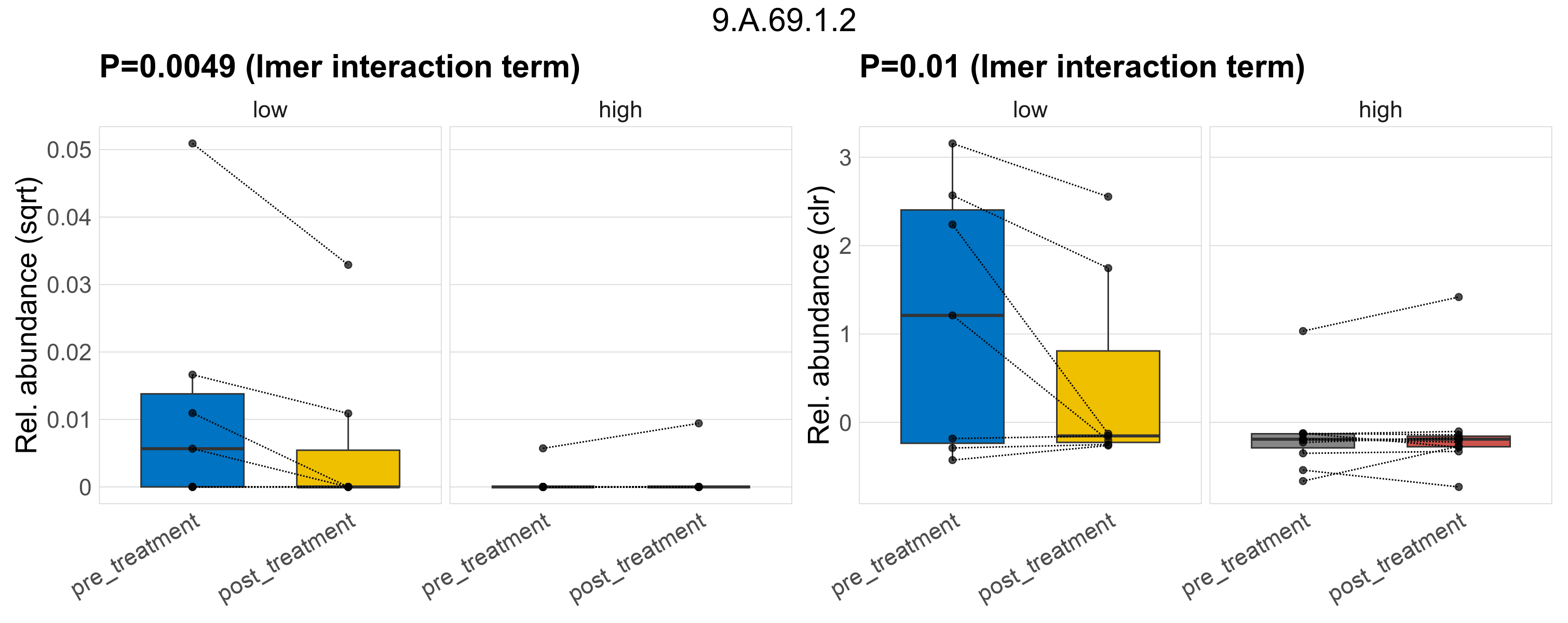
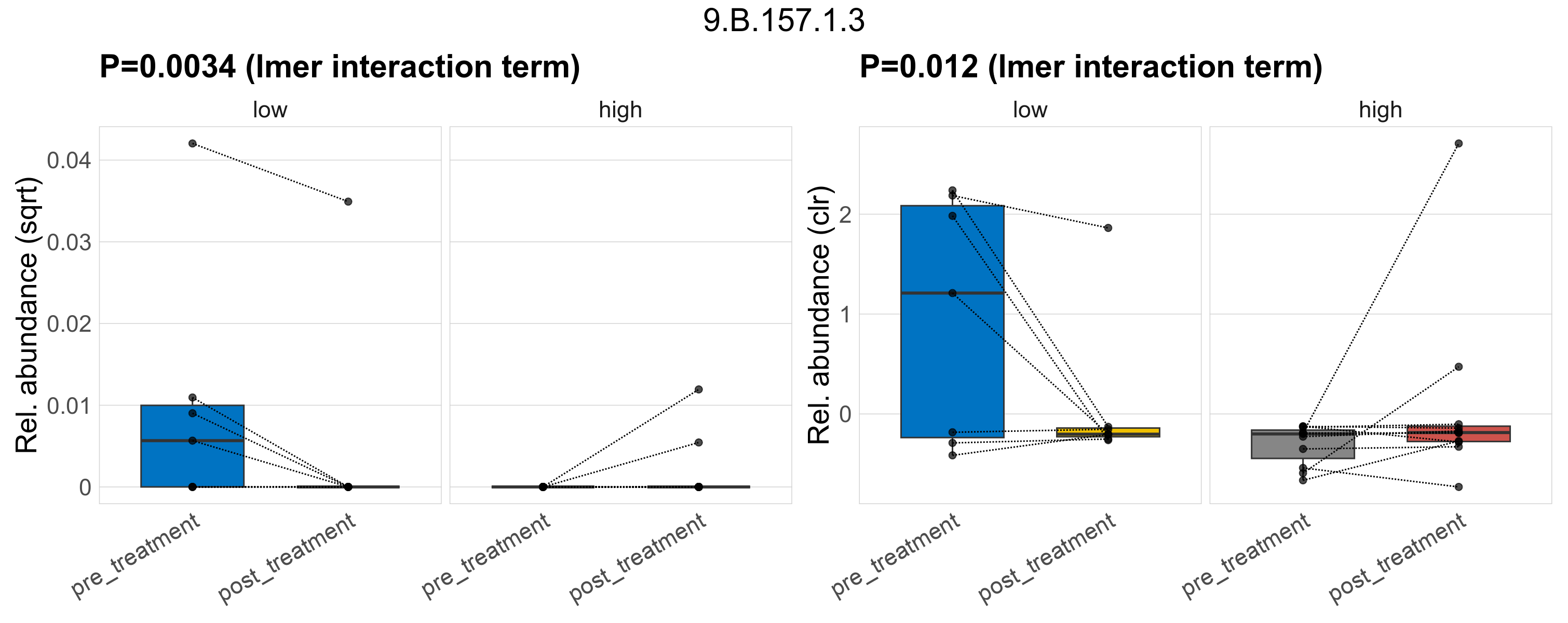
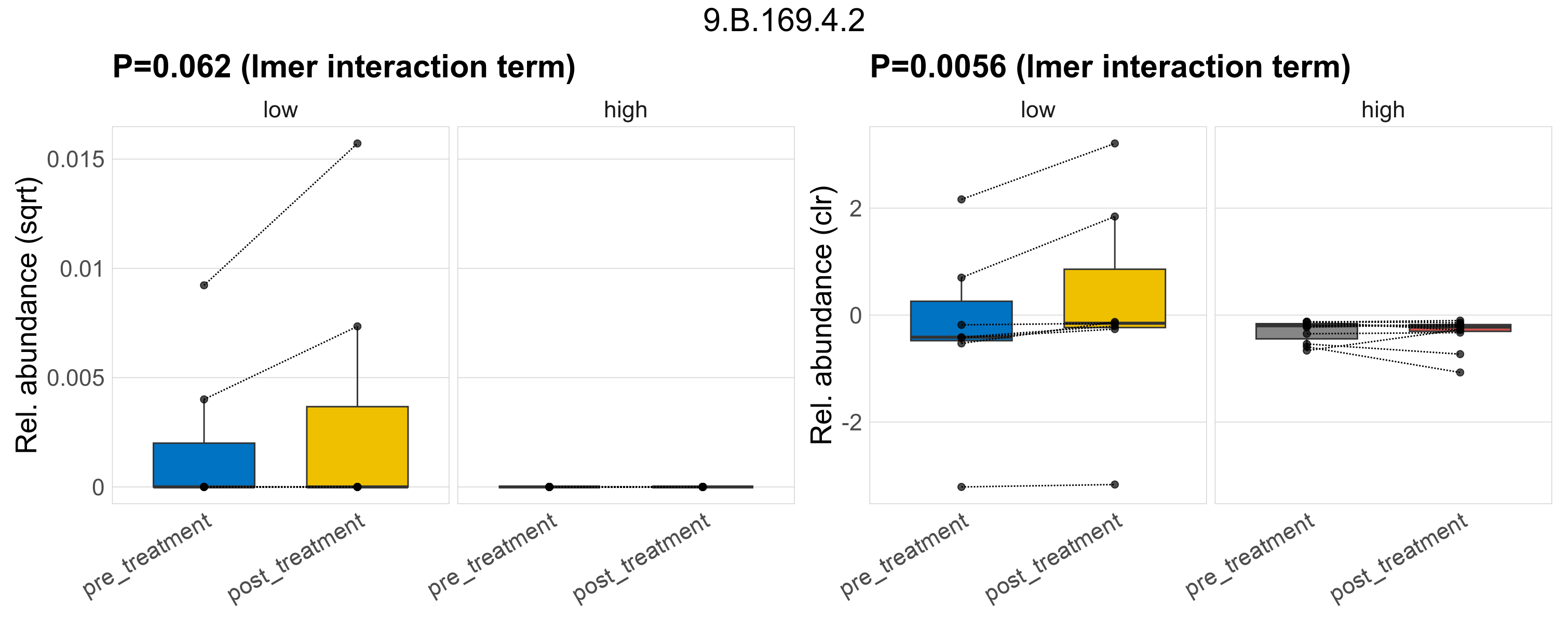
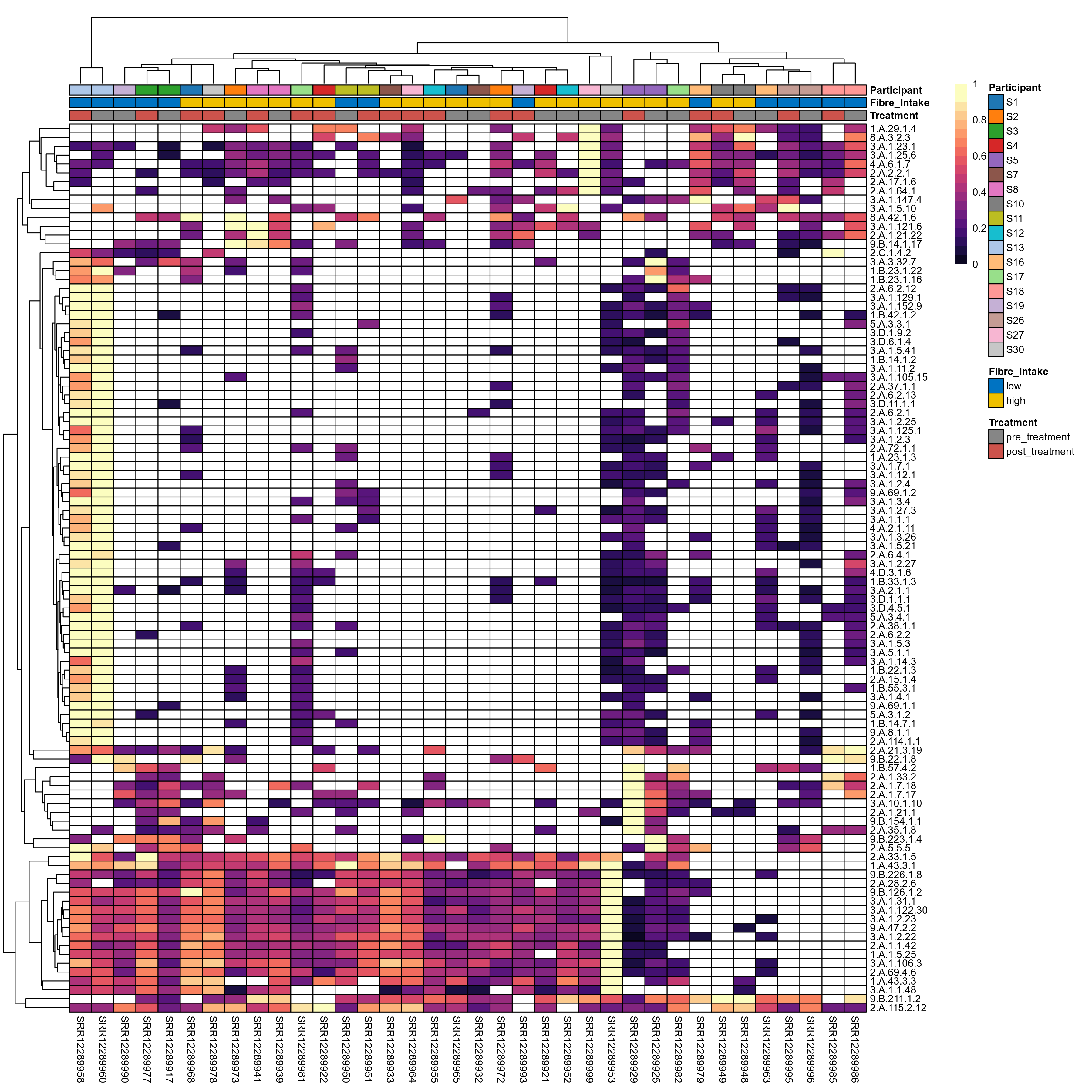
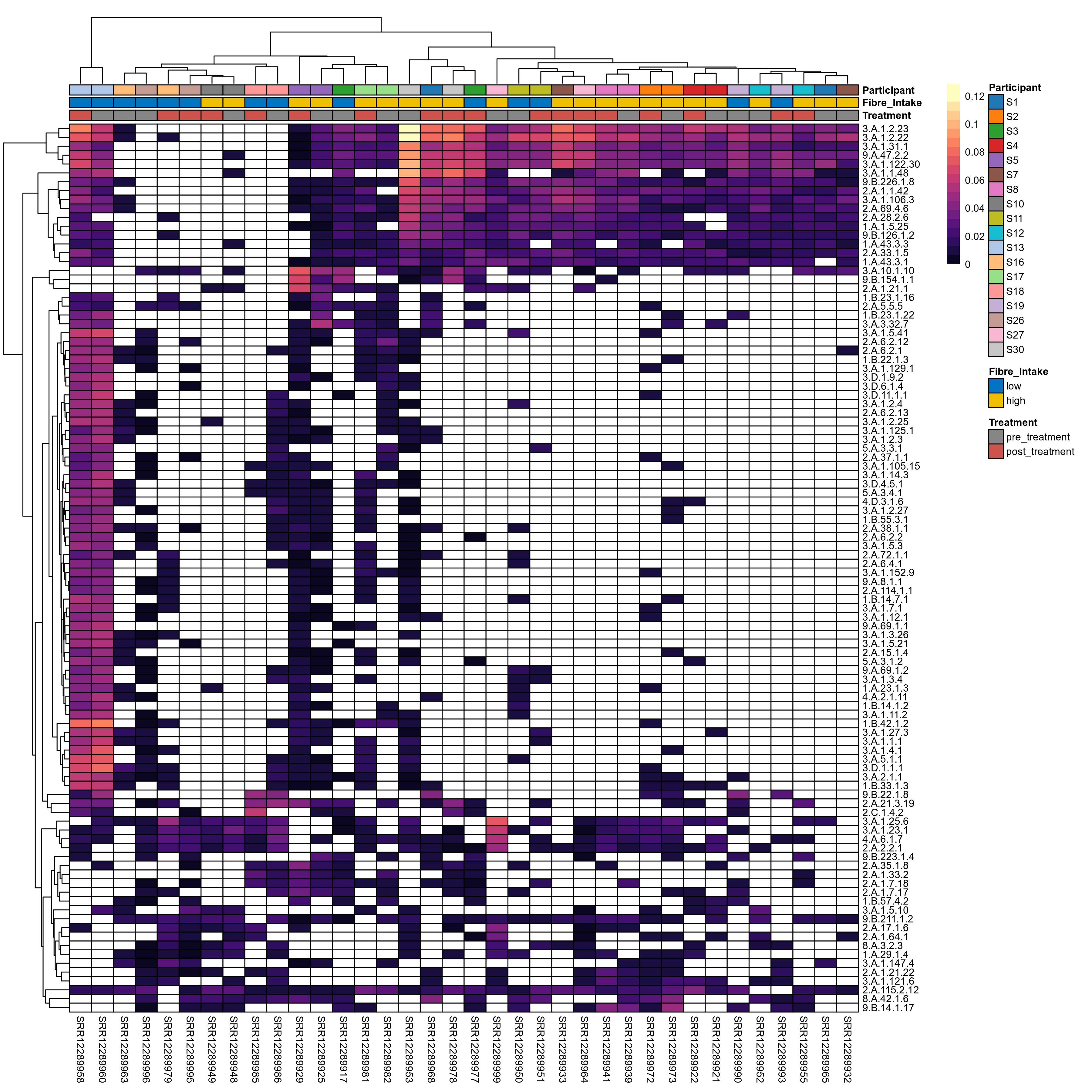
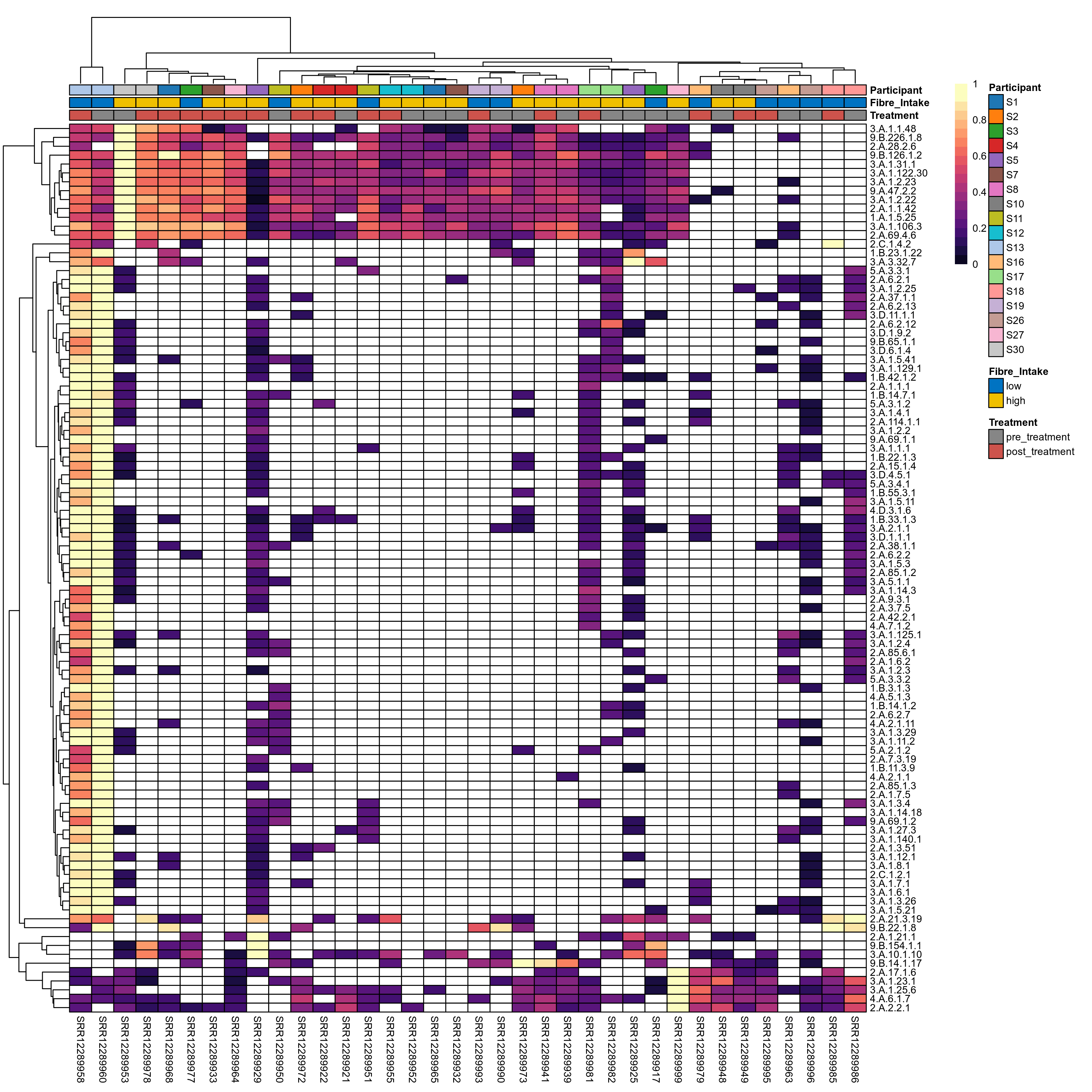
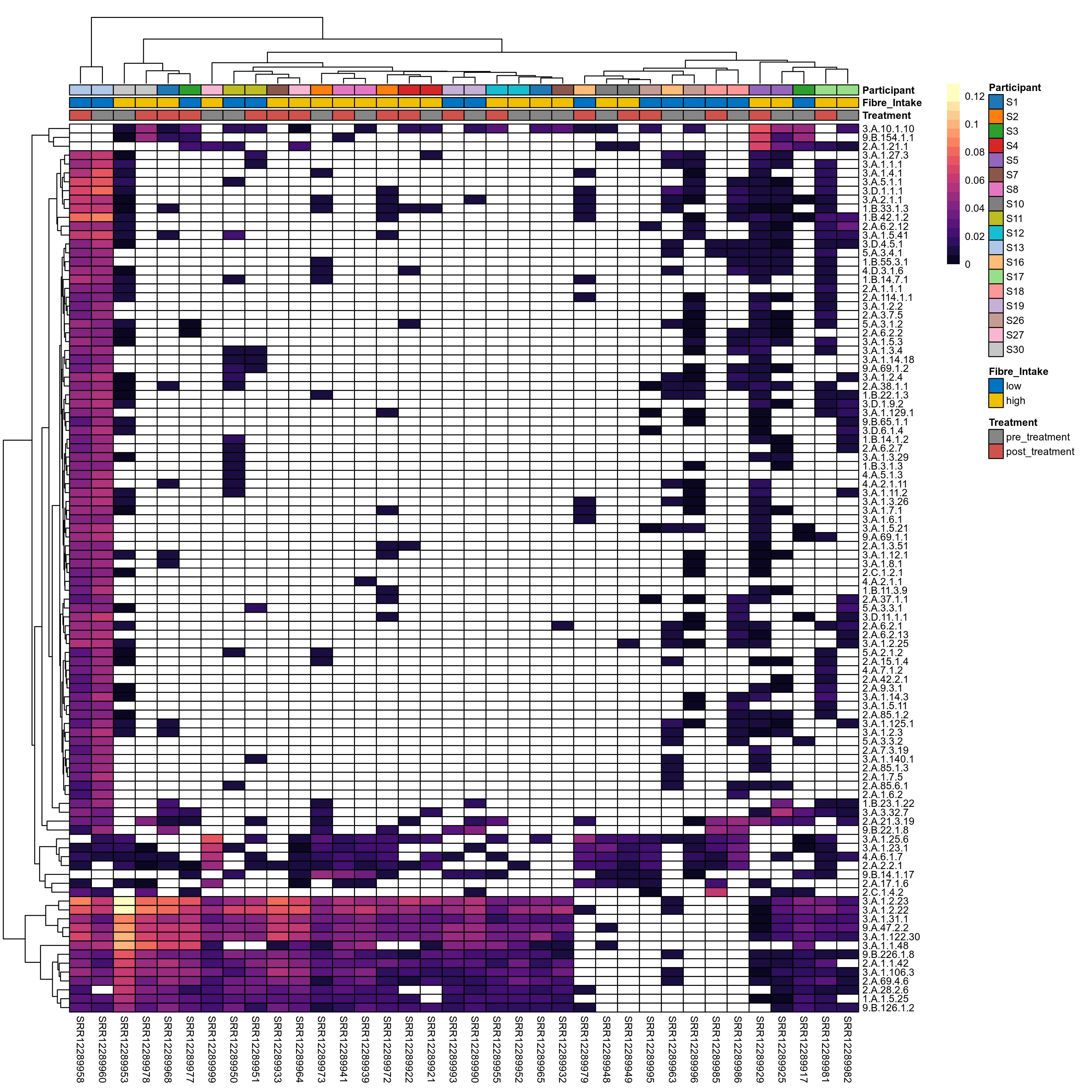
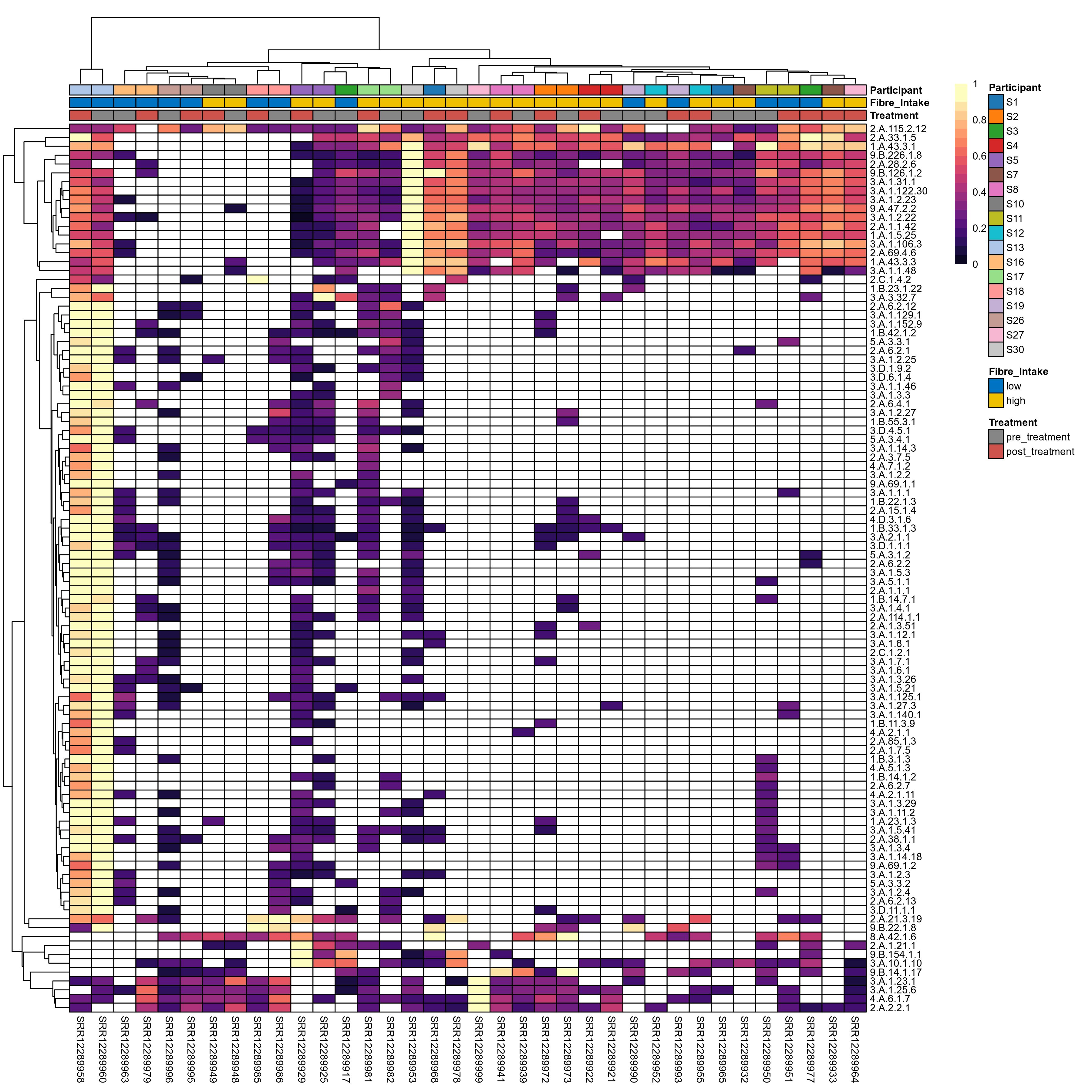
Quantitative visualisation of the top most abundant microbial functions identified in the analysed samples.
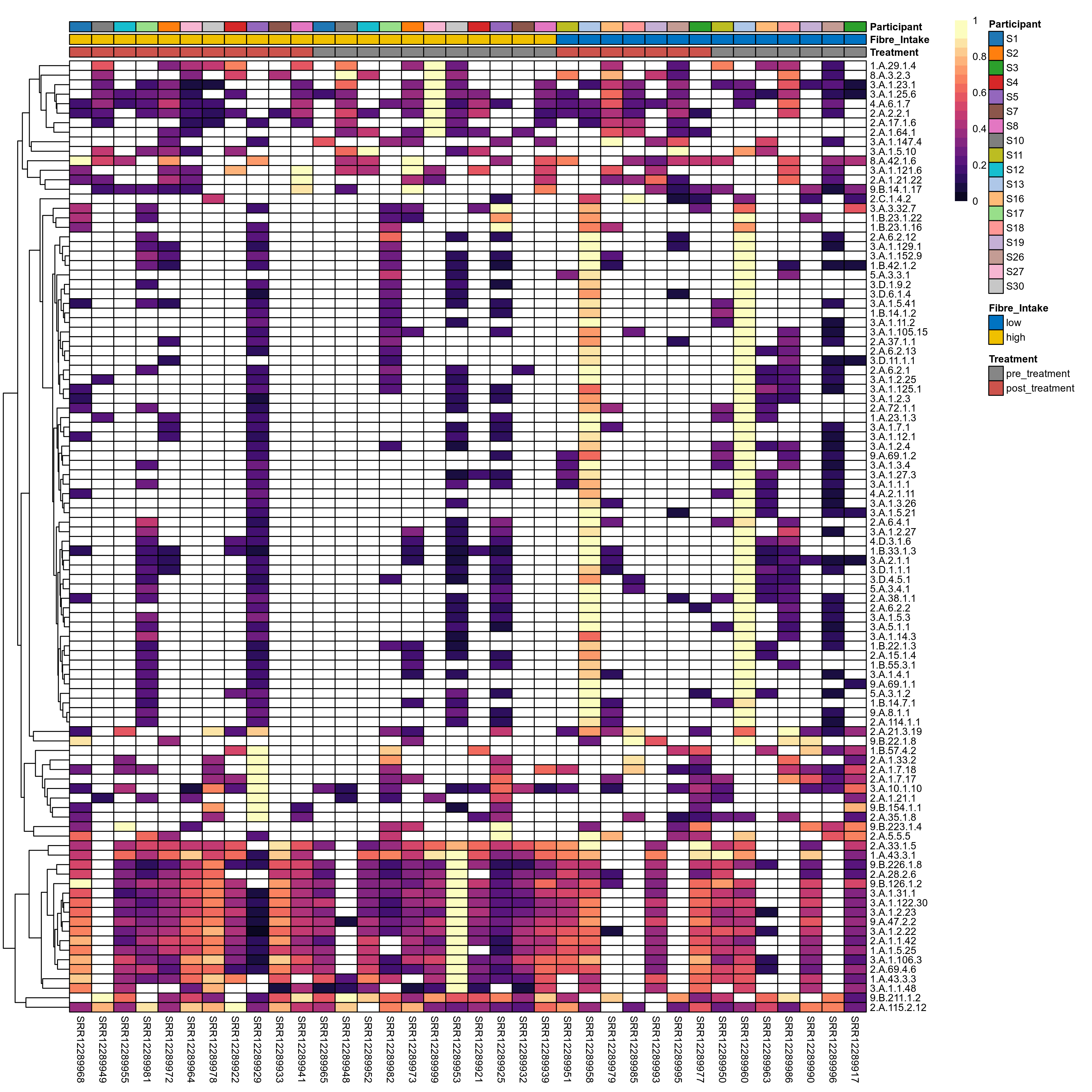
Click here to open full-sized image in new window.
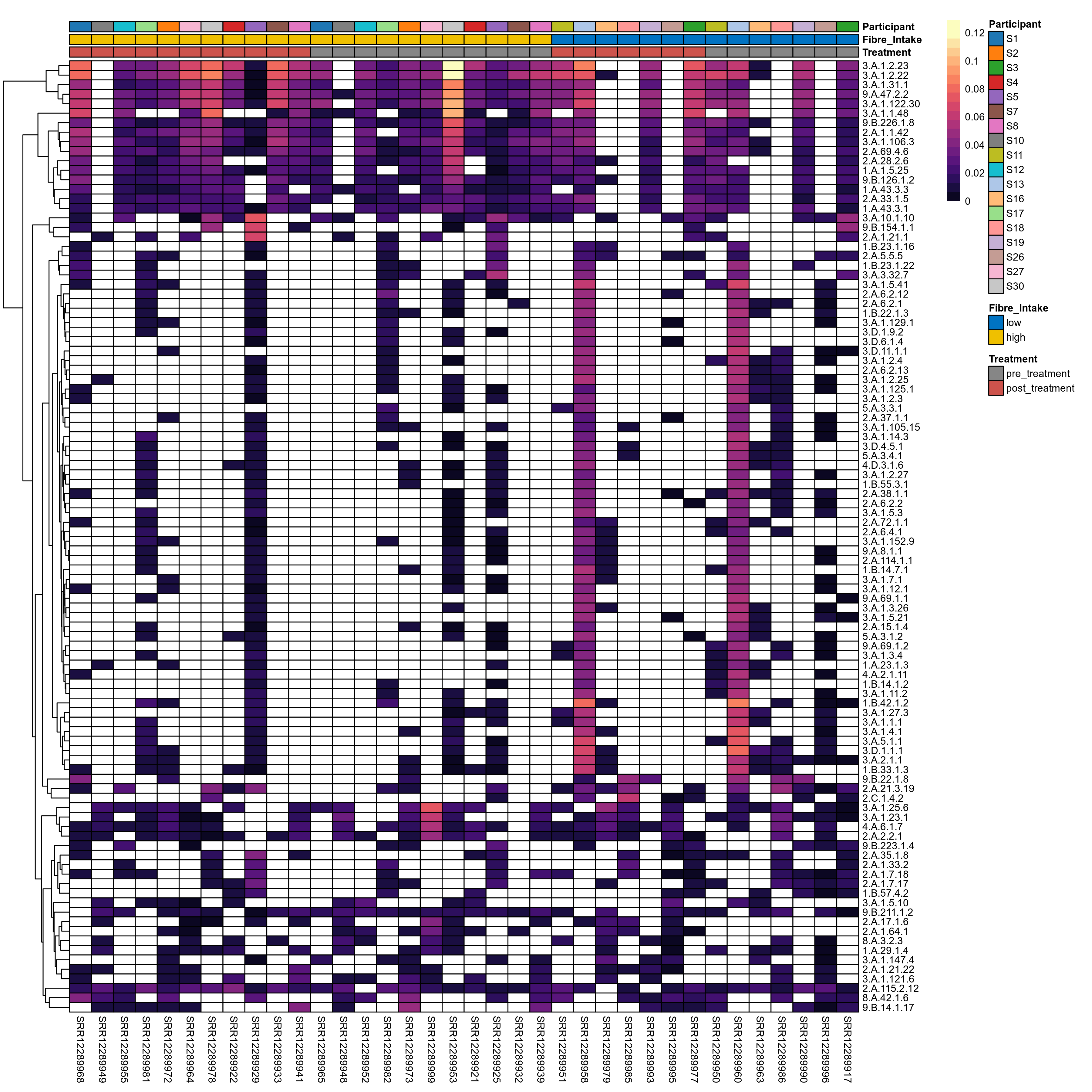
Click here to open full-sized image in new window.
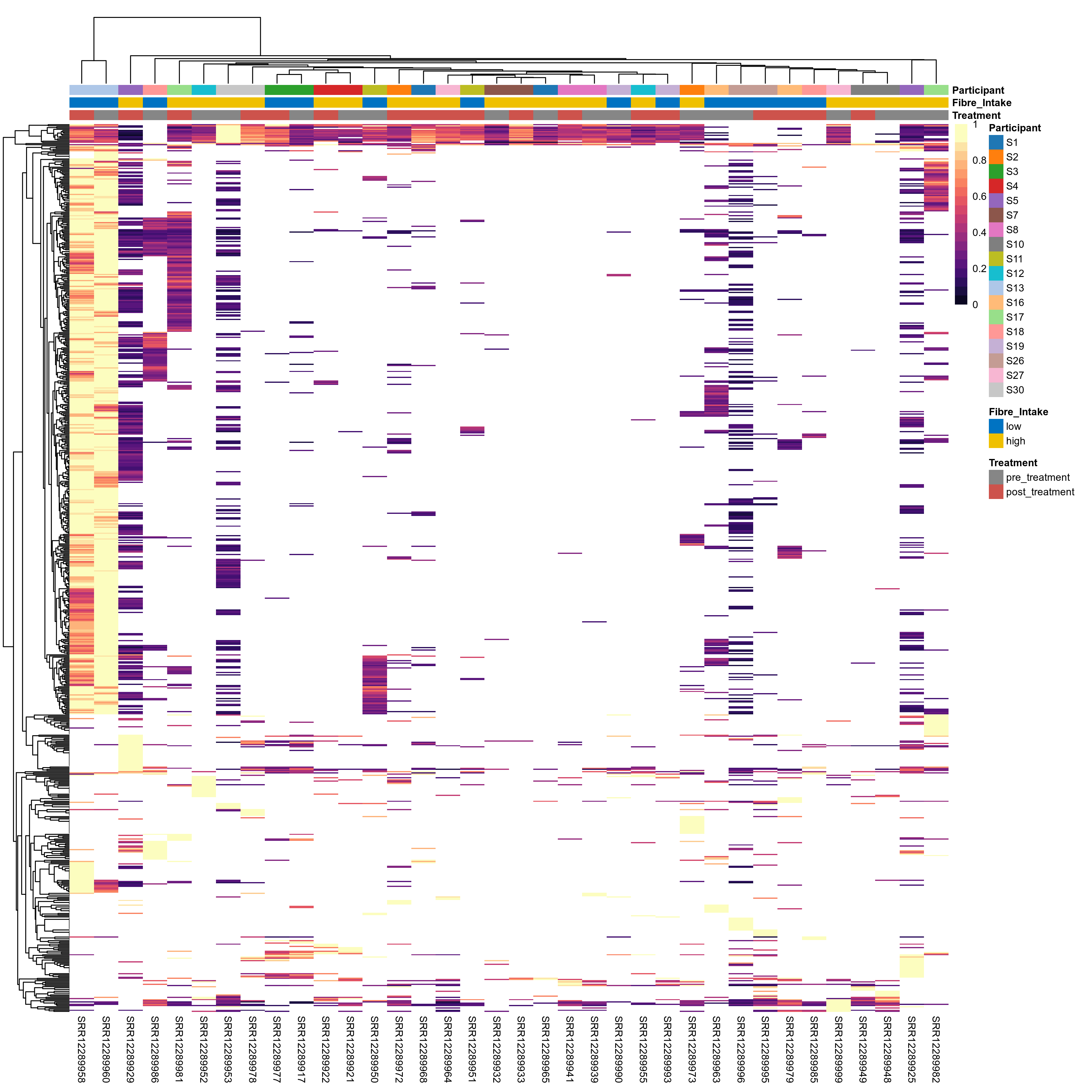
Click here to open full-sized image in new window.
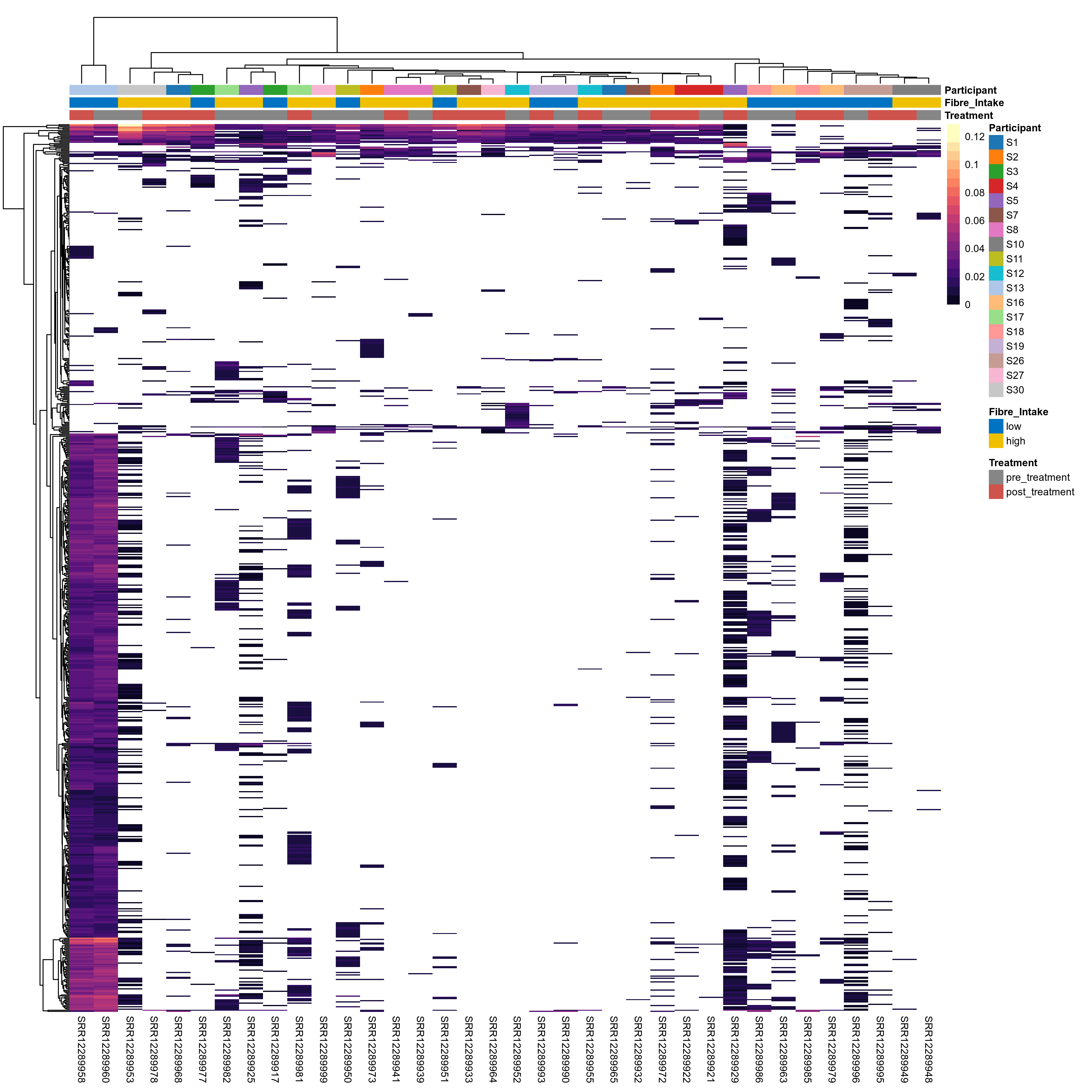
Click here to open full-sized image in new window.
Click here to open full-sized image in new window.
Click here to open full-sized image in new window.
Click here to open full-sized image in new window.
Click here to open full-sized image in new window.
Click here to open full-sized image in new window.
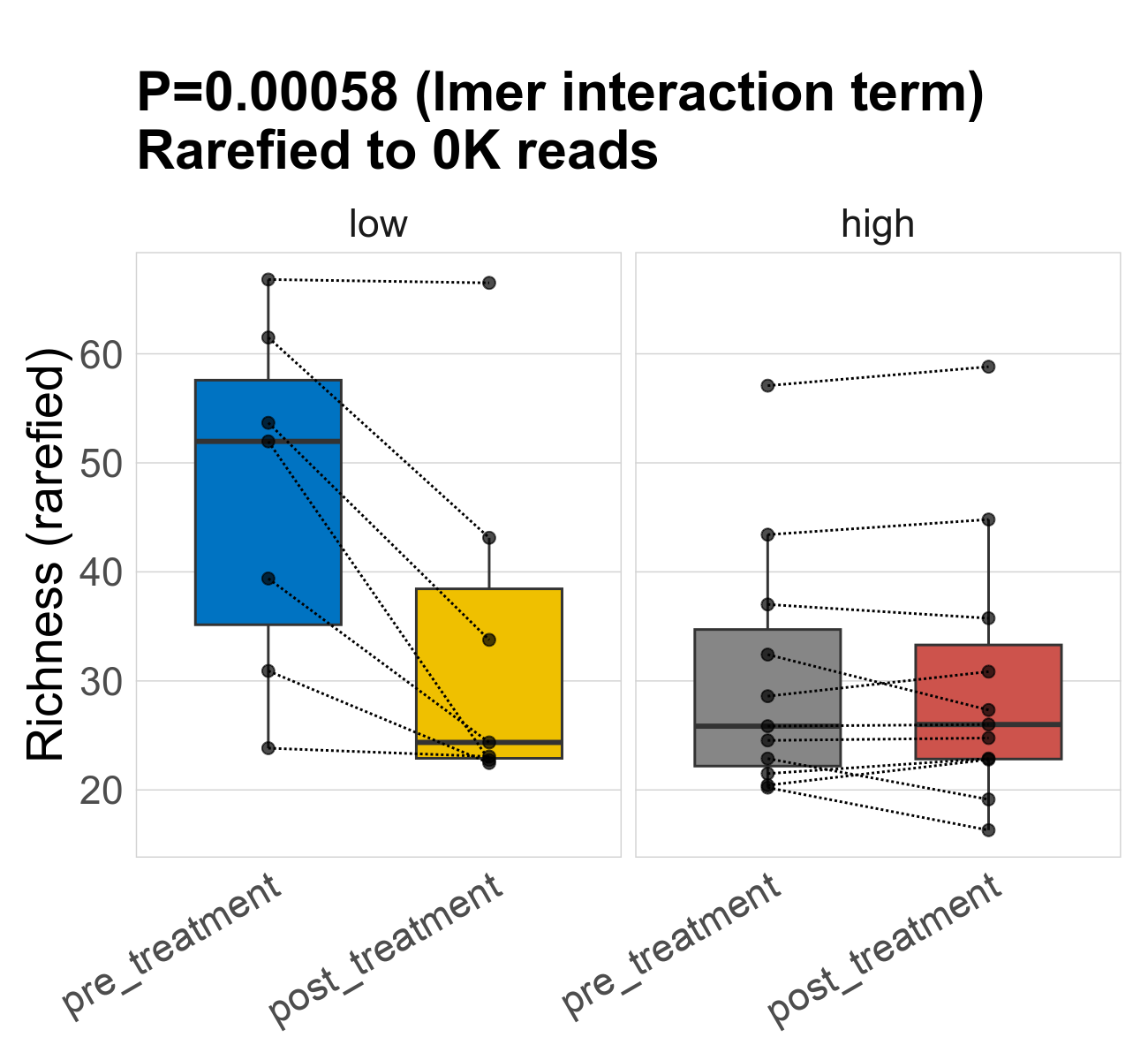
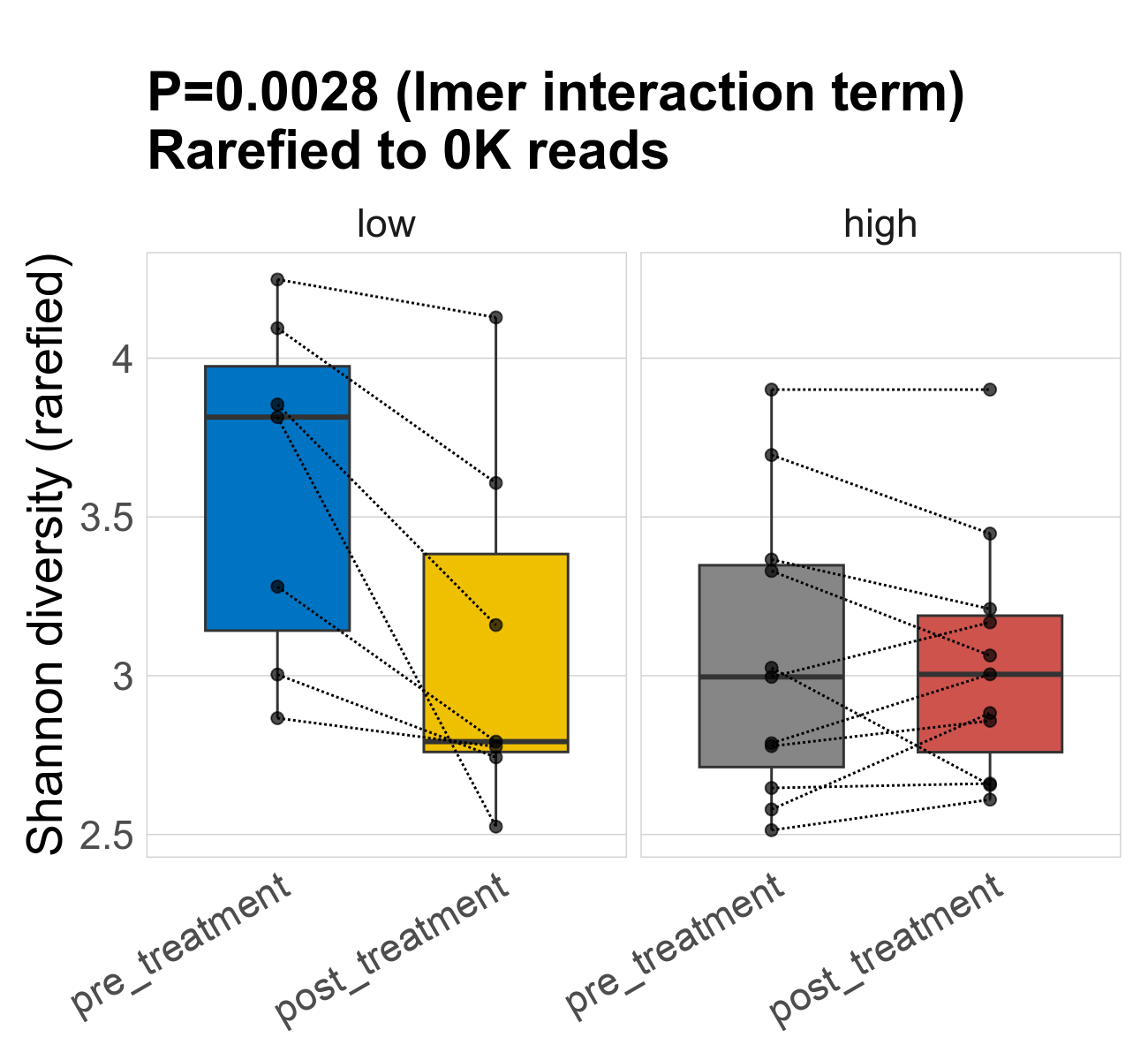
This page provides an overview of the functional alpha diversity of the analysed sample. Alpha diversity was measured using the Shannon index and functional richnes. Richness measures the total number of gene functions present in each sample. Shannon index combines richness and evenness.
| Index | rarefiedTo | P lmer treatment effect (interaction) | Mean Pos | Mean Abundance | Median Abundance | Mean Fibre_Intakelow:Treatmentpre_treatment | Median Fibre_Intakelow:Treatmentpre_treatment | SD Fibre_Intakelow:Treatmentpre_treatment | Mean Fibre_Intakelow:Treatmentpost_treatment | Median Fibre_Intakelow:Treatmentpost_treatment | SD Fibre_Intakelow:Treatmentpost_treatment | Fold Change Log2(Fibre_Intakelow:Treatmentpost_treatment/Fibre_Intakelow:Treatmentpre_treatment) | Positive samples | Positive Fibre_Intakelow:Treatmentpre_treatment | Positive Fibre_Intakelow:Treatmentpost_treatment | Positive_Fibre_Intakelow:Treatmentpre_treatment_percent | Positive_Fibre_Intakelow:Treatmentpost_treatment_percent | Mean Fibre_Intakehigh:Treatmentpre_treatment | Median Fibre_Intakehigh:Treatmentpre_treatment | SD Fibre_Intakehigh:Treatmentpre_treatment | Mean Fibre_Intakehigh:Treatmentpost_treatment | Median Fibre_Intakehigh:Treatmentpost_treatment | SD Fibre_Intakehigh:Treatmentpost_treatment | Fold Change Log2(Fibre_Intakehigh:Treatmentpost_treatment/Fibre_Intakehigh:Treatmentpre_treatment) | Positive Fibre_Intakehigh:Treatmentpre_treatment | Positive Fibre_Intakehigh:Treatmentpost_treatment | Positive_Fibre_Intakehigh:Treatmentpre_treatment_percent | Positive_Fibre_Intakehigh:Treatmentpost_treatment_percent |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Shannon | 74 | 0.0028 | 3.2 | 3.2 | 3 | 3.6 | 3.8 | 0.54 | 3.1 | 2.8 | 0.57 | -0.22 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 3.1 | 3 | 0.46 | 3 | 3 | 0.39 | -0.047 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
| Richness | 74 | 0.00058 | 34 | 34 | 28 | 47 | 52 | 16 | 34 | 24 | 16 | -0.47 | 36 / 36 (100%) | 7 / 7 (100%) | 7 / 7 (100%) | 1 | 1 | 30 | 26 | 12 | 30 | 26 | 12 | 0 | 11 / 11 (100%) | 11 / 11 (100%) | 1 | 1 |
Microbial functions were analyzed using supervised and unsupervised multivariate techniques. Functional profiles were ordinated using the unsupervised methods Principal Coordinates Analysis (PCoA), Non-Metric Multidimensional Scaling (NMDS) and Principal Component Analysis (PCA). The supervised methods Adonis and Redundancy analysis (RDA) were used to assess if variance in the functional profiles was significantly associated with the study condition. A guide explaining these methods can be found here. Sparse Partial Least Square Discriminant Analysis (sPLS-DA) from the MixMc package was additionaly used to extract features associated with the condition of interest.
Differentially abundant microbial functions were identified using the univariate methods ANOVA or LMER (linear mixed effect regression) on clr transformed relative abundance data, Fisher's exact test, and Aldex2 (ANOVA-like Differential Expression). Aldex2 was run on read count data. Fisher's exact test was used to test for diffrerences in the presence and absence (detection rate) of microbial functions across study groups.
LMER is used for repeated measures data, using random effects to control for correlation between samples from the same subject. Fixed effects are included for treatment groups, time, and treatment over time, where appropriate. The LMER P values correspond to a nested model test of the significance of including the corresponding fixed effect.
ALDEx2 uses subsampling (Bayesian sampling) to estimate the underlying technical variation. For each subsample instance, center log-ratio transformed data is statistically compared across study groups and computed P values are corrected for multiple testing using the Benjamini–Hochberg procedure. The expected P value (mean P value) is reported, which are those that would likely have been observed if the same samples had been run multiple times. The expected values are reported for both the distribution of P values and for the distribution of Benjamini–Hochberg corrected values.
| Function | Annotation | P lmer study group effect (sqrt) | FDR lmer study group effect (sqrt) | Pbonf lmer study group effect (sqrt) | P lmer time effect (sqrt) | FDR lmer time effect (sqrt) | Pbonf lmer time effect (sqrt) | P lmer treatment effect (interaction) (sqrt) | FDR lmer treatment effect (interaction) (sqrt) | Pbonf lmer treatment effect (interaction) (sqrt) | P lmer study group effect (clr) | FDR lmer study group effect (clr) | Pbonf lmer study group effect (clr) | P lmer time effect (clr) | FDR lmer time effect (clr) | Pbonf lmer time effect (clr) | P lmer treatment effect (interaction) (clr) | FDR lmer treatment effect (interaction) (clr) | Pbonf lmer treatment effect (interaction) (clr) | Mean Pos | Mean Abundance | Median Abundance | Mean Fibre_Intakelow:Treatmentpre_treatment | Median Fibre_Intakelow:Treatmentpre_treatment | SD Fibre_Intakelow:Treatmentpre_treatment | Mean Fibre_Intakelow:Treatmentpost_treatment | Median Fibre_Intakelow:Treatmentpost_treatment | SD Fibre_Intakelow:Treatmentpost_treatment | Fold Change Log2(Fibre_Intakelow:Treatmentpost_treatment/Fibre_Intakelow:Treatmentpre_treatment) | Positive samples | Positive Fibre_Intakelow:Treatmentpre_treatment | Positive Fibre_Intakelow:Treatmentpost_treatment | Positive_Fibre_Intakelow:Treatmentpre_treatment_percent | Positive_Fibre_Intakelow:Treatmentpost_treatment_percent | Mean Fibre_Intakehigh:Treatmentpre_treatment | Median Fibre_Intakehigh:Treatmentpre_treatment | SD Fibre_Intakehigh:Treatmentpre_treatment | Mean Fibre_Intakehigh:Treatmentpost_treatment | Median Fibre_Intakehigh:Treatmentpost_treatment | SD Fibre_Intakehigh:Treatmentpost_treatment | Fold Change Log2(Fibre_Intakehigh:Treatmentpost_treatment/Fibre_Intakehigh:Treatmentpre_treatment) | Positive Fibre_Intakehigh:Treatmentpre_treatment | Positive Fibre_Intakehigh:Treatmentpost_treatment | Positive_Fibre_Intakehigh:Treatmentpre_treatment_percent | Positive_Fibre_Intakehigh:Treatmentpost_treatment_percent |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3.A.1.140.6 | The ATP-binding Cassette (ABC) Superfamily | 0.12 | 0.34 | 1 | 0.56 | 0.76 | 1 | 0.35 | 0.6 | 1 | 0.048 | 0.27 | 1 | 0.32 | 0.83 | 1 | 0.2 | 0.69 | 1 | 0.00022 | 3.6e-05 | 0 | 0 | 0 | 0 | 1.7e-05 | 0 | 4.5e-05 | Inf | 6 / 36 (17%) | 0 / 7 (0%) | 1 / 7 (14%) | 0 | 0.143 | 7e-05 | 0 | 0.00016 | 3.8e-05 | 0 | 1e-04 | -0.88 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 2.A.1.1.92 | The Major Facilitator Superfamily (MFS) | 0.022 | 0.34 | 1 | 0.0092 | 0.22 | 1 | 0.015 | 0.37 | 1 | 0.0059 | 0.16 | 1 | 0.055 | 0.62 | 1 | 0.062 | 0.58 | 1 | 0.00049 | 6.8e-05 | 0 | 0.00021 | 0 | 0.00043 | 0.00013 | 0 | 0.00033 | -0.69 | 5 / 36 (14%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 0 | 0 | 0 | 6.5e-06 | 0 | 2.1e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
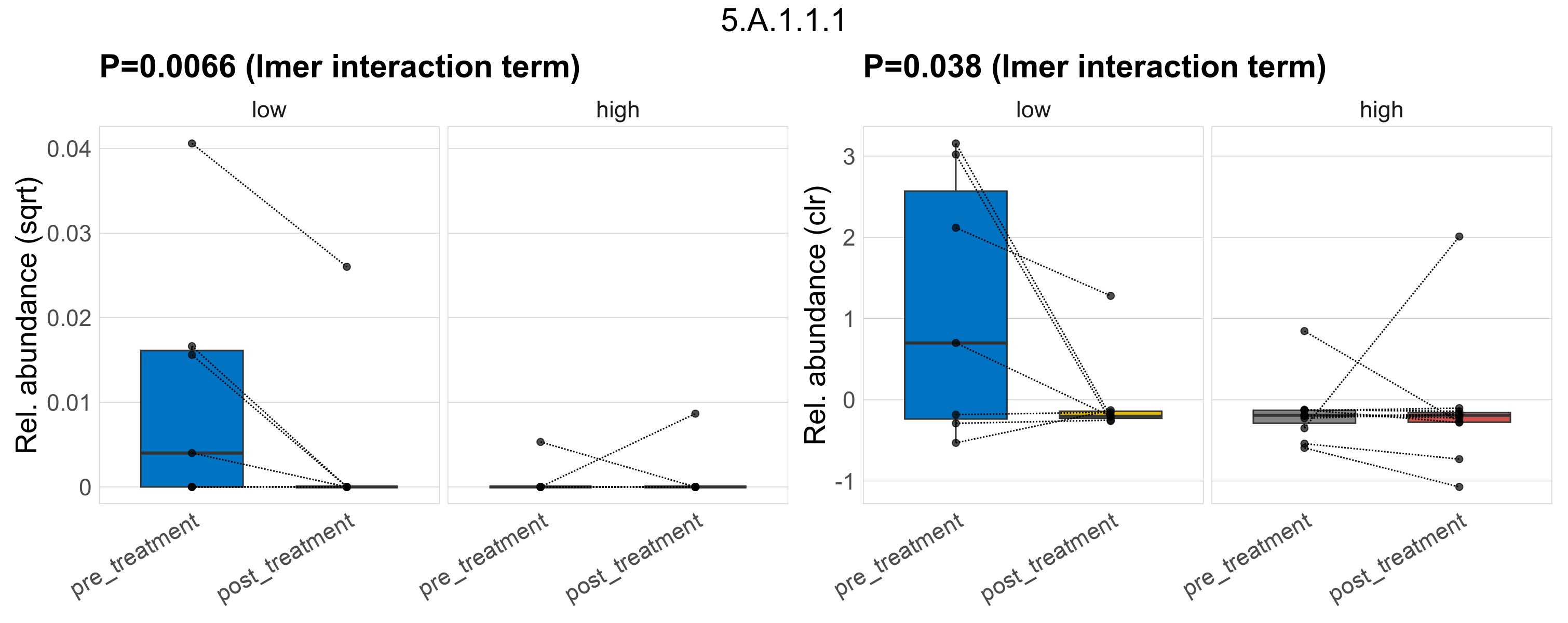
| 5.A.1.1.1 | The Disulfide Bond Oxidoreductase D (DsbD) Family | 0.0076 | 0.34 | 1 | 0.0014 | 0.16 | 0.77 | 0.0066 | 0.37 | 1 | 0.0022 | 0.15 | 1 | 0.012 | 0.5 | 1 | 0.038 | 0.58 | 1 | 0.00042 | 8.2e-05 | 0 | 0.00031 | 1.6e-05 | 6e-04 | 9.7e-05 | 0 | 0.00026 | -1.7 | 7 / 36 (19%) | 4 / 7 (57%) | 1 / 7 (14%) | 0.571 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 6.8e-06 | 0 | 2.3e-05 | 1.4 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.A.1.2.1 | The ATP-binding Cassette (ABC) Superfamily | 0.058 | 0.34 | 1 | 0.14 | 0.44 | 1 | 0.083 | 0.39 | 1 | 0.04 | 0.27 | 1 | 0.13 | 0.66 | 1 | 0.076 | 0.6 | 1 | 0.00036 | 7.1e-05 | 0 | 0.00016 | 0 | 3e-04 | 0.00016 | 0 | 0.00044 | 0 | 7 / 36 (19%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 3e-06 | 0 | 9.9e-06 | 2.1e-05 | 0 | 4.7e-05 | 2.8 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 3.A.1.1.46 | The ATP-binding Cassette (ABC) Superfamily | 0.16 | 0.34 | 1 | 0.11 | 0.4 | 1 | 0.73 | 0.88 | 1 | 0.26 | 0.58 | 1 | 0.25 | 0.77 | 1 | 0.99 | 1 | 1 | 0.00062 | 1e-04 | 0 | 0.00027 | 0 | 0.00066 | 0.00022 | 0 | 0.00059 | -0.3 | 6 / 36 (17%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 2.5e-05 | 0 | 7.2e-05 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 4.A.6.1.11 | The PTS Mannose-Fructose-Sorbose (Man) Family | 0.25 | 0.36 | 1 | 0.13 | 0.42 | 1 | 0.69 | 0.86 | 1 | 0.99 | 0.99 | 1 | 0.27 | 0.78 | 1 | 1 | 1 | 1 | 0.00011 | 1.8e-05 | 0 | 4.8e-05 | 0 | 8.8e-05 | 7.7e-06 | 0 | 2e-05 | -2.6 | 6 / 36 (17%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 2.3e-05 | 0 | 5.1e-05 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 1.B.23.1.22 | The Cyanobacterial Porin (CBP) Family | 0.46 | 0.53 | 1 | 0.32 | 0.61 | 1 | 0.71 | 0.87 | 1 | 0.86 | 0.95 | 1 | 0.55 | 0.91 | 1 | 0.94 | 0.99 | 1 | 7e-04 | 0.00015 | 0 | 0.00035 | 0 | 0.00084 | 0.00017 | 0 | 0.00046 | -1 | 8 / 36 (22%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0.00012 | 0 | 0.00034 | 4.9e-05 | 0 | 0.00013 | -1.3 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 1.A.23.2.1 | The Small Conductance Mechanosensitive Ion Channel (MscS) Family | 0.18 | 0.34 | 1 | 0.59 | 0.78 | 1 | 0.14 | 0.43 | 1 | 0.52 | 0.76 | 1 | 0.75 | 0.94 | 1 | 0.33 | 0.73 | 1 | 0.00041 | 4.6e-05 | 0 | 0.00012 | 0 | 0.00031 | 7.7e-05 | 0 | 2e-04 | -0.64 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 2.6e-05 | 0 | 6.9e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 1.E.34.2.1 | The Putative Actinobacterial Holin-X (Hol-X) Family | 0.56 | 0.61 | 1 | 0.49 | 0.75 | 1 | 0.81 | 0.92 | 1 | 0.31 | 0.62 | 1 | 0.28 | 0.78 | 1 | 0.52 | 0.82 | 1 | 0.00013 | 1.5e-05 | 0 | 1.7e-05 | 0 | 4.5e-05 | 3.9e-05 | 0 | 1e-04 | 1.2 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3e-06 | 0 | 9.9e-06 | 1e-05 | 0 | 3.4e-05 | 1.7 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.B.1.1.2 | The Na+-transporting Carboxylic Acid Decarboxylase (NaT-DC) Family | 0.84 | 0.88 | 1 | 0.81 | 0.91 | 1 | 0.69 | 0.86 | 1 | 0.61 | 0.82 | 1 | 0.91 | 0.98 | 1 | 0.98 | 1 | 1 | 0.00016 | 3.6e-05 | 0 | 2.8e-05 | 0 | 5e-05 | 2.3e-05 | 0 | 5.1e-05 | -0.28 | 8 / 36 (22%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 3.4e-05 | 0 | 9.7e-05 | 5.2e-05 | 0 | 0.00015 | 0.61 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 1.E.24.1.5 | The Bacterophage Dp-1 Holin (Dp-1 Holin) Family | 0.29 | 0.39 | 1 | 0.22 | 0.51 | 1 | 0.2 | 0.48 | 1 | 0.8 | 0.91 | 1 | 0.44 | 0.86 | 1 | 0.41 | 0.76 | 1 | 0.00014 | 1.2e-05 | 0 | 3.2e-05 | 0 | 8.5e-05 | 0 | 0 | 0 | -Inf | 3 / 36 (8.3%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 2.6e-06 | 0 | 8.5e-06 | 1.6e-05 | 0 | 5.3e-05 | 2.6 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
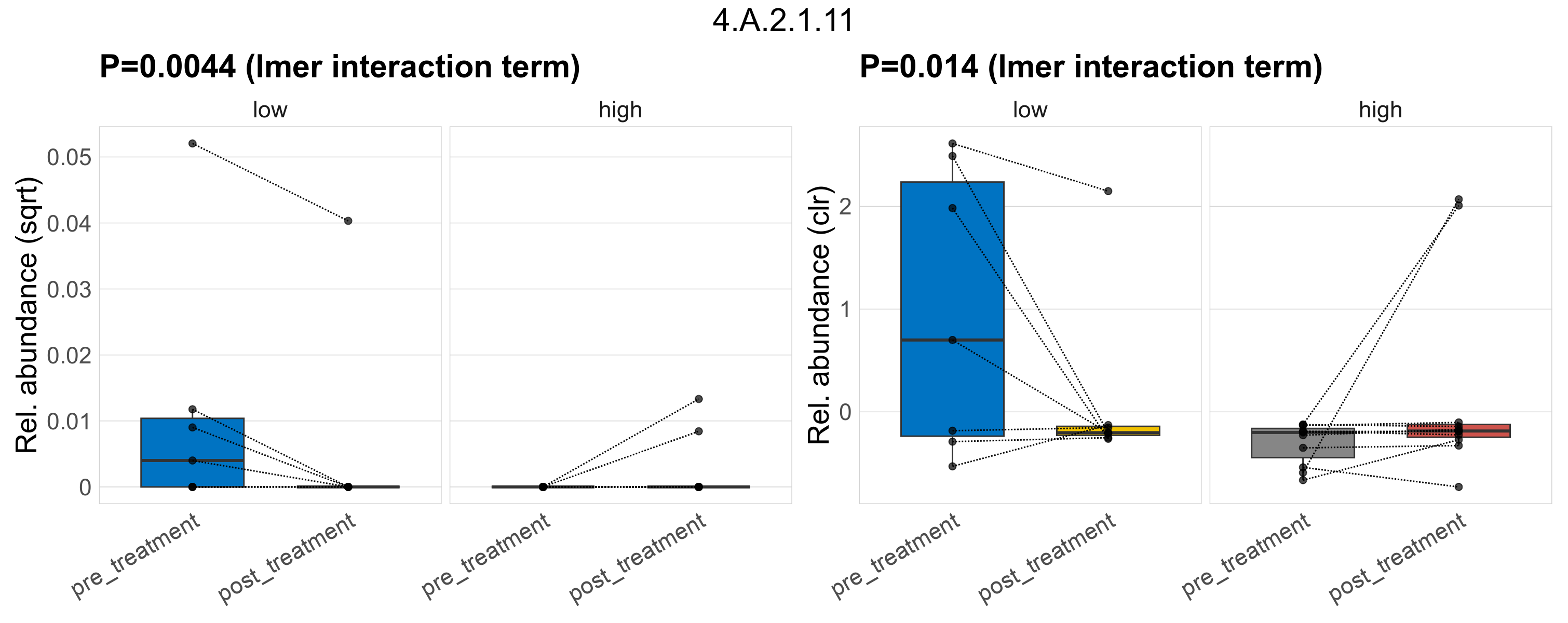
| 4.A.2.1.10 | The PTS Fructose-Mannitol (Fru) Family | 0.18 | 0.34 | 1 | 0.61 | 0.79 | 1 | 0.24 | 0.52 | 1 | 0.34 | 0.63 | 1 | 0.37 | 0.84 | 1 | 0.89 | 0.97 | 1 | 0.00075 | 6.3e-05 | 0 | 0.00017 | 0 | 0.00045 | 0.00015 | 0 | 0.00038 | -0.18 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.A.23.4.3 | The Small Conductance Mechanosensitive Ion Channel (MscS) Family | 0.031 | 0.34 | 1 | 0.0067 | 0.22 | 1 | 0.061 | 0.37 | 1 | 0.038 | 0.27 | 1 | 0.051 | 0.62 | 1 | 0.32 | 0.73 | 1 | 0.00016 | 2.2e-05 | 0 | 8.1e-05 | 0 | 0.00017 | 2.9e-05 | 0 | 7.7e-05 | -1.5 | 5 / 36 (14%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 9.B.102.1.1 | The YedE/YeeE (YedE/YeeE) Family | 0.28 | 0.38 | 1 | 0.35 | 0.63 | 1 | 0.6 | 0.82 | 1 | 0.62 | 0.82 | 1 | 0.88 | 0.97 | 1 | 0.64 | 0.86 | 1 | 2e-04 | 2.2e-05 | 0 | 6.7e-05 | 0 | 0.00018 | 2.9e-05 | 0 | 7.7e-05 | -1.2 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 7.3e-06 | 0 | 2.4e-05 | 2.7e-06 | 0 | 8.9e-06 | -1.4 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.8.1.2 | The Gluconate:H+ Symporter (GntP) Family | 0.17 | 0.34 | 1 | 0.096 | 0.39 | 1 | 0.19 | 0.48 | 1 | 0.66 | 0.85 | 1 | 0.63 | 0.91 | 1 | 0.49 | 0.82 | 1 | 0.00049 | 5.5e-05 | 0 | 0.00019 | 0 | 0.00049 | 8.7e-05 | 0 | 0.00023 | -1.1 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3e-06 | 0 | 9.9e-06 | 2.7e-06 | 0 | 8.9e-06 | -0.15 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.98.1.3 | The Putative Sulfate Exporter (PSE) Family | 0.16 | 0.34 | 1 | 0.45 | 0.72 | 1 | 0.19 | 0.48 | 1 | 0.47 | 0.72 | 1 | 0.68 | 0.92 | 1 | 0.69 | 0.88 | 1 | 0.00049 | 4.1e-05 | 0 | 0.00012 | 0 | 0.00031 | 7.7e-05 | 0 | 2e-04 | -0.64 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 1e-05 | 0 | 3.4e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.85.1.3 | The Aromatic Acid Exporter (ArAE) Family | 0.076 | 0.34 | 1 | 0.038 | 0.28 | 1 | 0.042 | 0.37 | 1 | 0.045 | 0.27 | 1 | 0.29 | 0.78 | 1 | 0.17 | 0.68 | 1 | 0.00097 | 0.00011 | 0 | 0.00037 | 0 | 0.00093 | 0.00017 | 0 | 0.00046 | -1.1 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 8.1e-06 | 0 | 2.7e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 9.B.14.1.3 | The Putative Heme Handling Protein (HHP) Family | 0.097 | 0.34 | 1 | 0.026 | 0.25 | 1 | 0.11 | 0.42 | 1 | 0.23 | 0.54 | 1 | 0.19 | 0.72 | 1 | 0.5 | 0.82 | 1 | 0.00036 | 5.9e-05 | 0 | 0.00019 | 0 | 0.00044 | 9.7e-05 | 0 | 0.00026 | -0.97 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 5.5e-06 | 0 | 1.2e-05 | 5.4e-06 | 0 | 1.8e-05 | -0.026 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 2.A.3.1.14 | The Amino Acid-Polyamine-Organocation (APC) Family | 0.33 | 0.42 | 1 | 0.6 | 0.78 | 1 | 0.74 | 0.89 | 1 | 0.86 | 0.95 | 1 | 0.6 | 0.91 | 1 | 0.7 | 0.88 | 1 | 0.00037 | 6.1e-05 | 0 | 0.00012 | 0 | 0.00026 | 0.00015 | 0 | 0.00038 | 0.32 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.8e-05 | 0 | 7.3e-05 | 2.7e-06 | 0 | 8.9e-06 | -3.4 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 2.A.1.46.6 | The Major Facilitator Superfamily (MFS) | 0.13 | 0.34 | 1 | 0.36 | 0.64 | 1 | 0.47 | 0.72 | 1 | 0.14 | 0.4 | 1 | 0.87 | 0.97 | 1 | 0.72 | 0.88 | 1 | 0.00034 | 2.9e-05 | 0 | 7e-05 | 0 | 0.00018 | 7.7e-05 | 0 | 2e-04 | 0.14 | 3 / 36 (8.3%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 3.A.1.106.3 | The ATP-binding Cassette (ABC) Superfamily | 0.12 | 0.34 | 1 | 0.12 | 0.41 | 1 | 0.44 | 0.69 | 1 | 0.043 | 0.27 | 1 | 0.39 | 0.84 | 1 | 0.53 | 0.82 | 1 | 0.0011 | 0.00089 | 0.00044 | 0.00032 | 0.00023 | 0.00038 | 0.00088 | 0.00036 | 0.001 | 1.5 | 29 / 36 (81%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 0.00095 | 0.00077 | 0.001 | 0.0012 | 0.00098 | 0.0011 | 0.34 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 3.A.1.135.1 | The ATP-binding Cassette (ABC) Superfamily | 0.59 | 0.65 | 1 | 0.39 | 0.67 | 1 | 0.89 | 0.96 | 1 | 0.21 | 0.52 | 1 | 0.28 | 0.78 | 1 | 0.66 | 0.87 | 1 | 0.00017 | 1.8e-05 | 0 | 0 | 0 | 0 | 2.3e-05 | 0 | 6.1e-05 | Inf | 4 / 36 (11%) | 0 / 7 (0%) | 1 / 7 (14%) | 0 | 0.143 | 1.2e-05 | 0 | 3.9e-05 | 3.4e-05 | 0 | 7.8e-05 | 1.5 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 2.A.103.1.1 | The Bacterial Murein Precursor Exporter (MPE) Family | 0.18 | 0.34 | 1 | 0.13 | 0.43 | 1 | 0.23 | 0.52 | 1 | 0.6 | 0.82 | 1 | 0.051 | 0.62 | 1 | 0.1 | 0.65 | 1 | 0.00042 | 3.5e-05 | 0 | 8.4e-05 | 0 | 0.00022 | 9.5e-05 | 0 | 2e-04 | 0.18 | 3 / 36 (8.3%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 1.S.2.1.1 | The Bacterial Microcompartment Shell/Pore-forming Protein-2 (BMC-SP2) Family | 0.44 | 0.52 | 1 | 0.93 | 0.97 | 1 | 0.43 | 0.69 | 1 | 0.4 | 0.67 | 1 | 0.57 | 0.91 | 1 | 0.19 | 0.68 | 1 | 0.00031 | 4.4e-05 | 0 | 1e-04 | 0 | 0.00027 | 9.7e-05 | 0 | 0.00026 | -0.044 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 1.4e-05 | 0 | 3.7e-05 | 2.7e-06 | 0 | 8.9e-06 | -2.4 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 1.A.35.4.1 | The CorA Metal Ion Transporter (MIT) Family | 0.046 | 0.34 | 1 | 0.024 | 0.24 | 1 | 0.03 | 0.37 | 1 | 0.036 | 0.27 | 1 | 0.11 | 0.65 | 1 | 0.083 | 0.61 | 1 | 0.00034 | 6.6e-05 | 0 | 0.00024 | 0 | 0.00057 | 7.7e-05 | 0 | 2e-04 | -1.6 | 7 / 36 (19%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1.3e-05 | 0 | 3.5e-05 | 2.3 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 3.A.1.107.3 | The ATP-binding Cassette (ABC) Superfamily | 0.2 | 0.34 | 1 | 0.35 | 0.64 | 1 | 0.33 | 0.59 | 1 | 0.67 | 0.86 | 1 | 0.68 | 0.92 | 1 | 0.9 | 0.97 | 1 | 0.00056 | 6.2e-05 | 0 | 0.00019 | 0 | 0.00049 | 0.00012 | 0 | 0.00031 | -0.66 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 8.1e-06 | 0 | 2.7e-05 | 1.6 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.55.3.1 | The Metal Ion (Mn2+-iron) Transporter (Nramp) Family | 0.47 | 0.54 | 1 | 0.58 | 0.77 | 1 | 0.96 | 0.99 | 1 | 0.66 | 0.85 | 1 | 0.63 | 0.91 | 1 | 0.98 | 1 | 1 | 0.00016 | 3e-05 | 0 | 3.6e-05 | 0 | 8.8e-05 | 8.7e-05 | 0 | 0.00023 | 1.3 | 7 / 36 (19%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 5.5e-06 | 0 | 1.2e-05 | 1.6e-05 | 0 | 3.7e-05 | 1.5 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 9.B.143.4.2 | The 6 TMS DUF1275/Pf06912 (DUF1275) Family | 0.59 | 0.65 | 1 | 0.64 | 0.8 | 1 | 0.97 | 0.99 | 1 | 0.16 | 0.43 | 1 | 0.34 | 0.83 | 1 | 0.55 | 0.83 | 1 | 0.00013 | 1.1e-05 | 0 | 0 | 0 | 0 | 4.4e-06 | 0 | 1.2e-05 | Inf | 3 / 36 (8.3%) | 0 / 7 (0%) | 1 / 7 (14%) | 0 | 0.143 | 7.3e-06 | 0 | 2.4e-05 | 2.6e-05 | 0 | 8.6e-05 | 1.8 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 9.B.154.1.1 | The Putative Holin-2 (PH-2) Family | 0.52 | 0.58 | 1 | 0.38 | 0.65 | 1 | 0.045 | 0.37 | 1 | 0.85 | 0.94 | 1 | 0.75 | 0.94 | 1 | 0.067 | 0.58 | 1 | 0.0013 | 0.00028 | 0 | 0.00037 | 0 | 0.00098 | 4e-05 | 0 | 0.00011 | -3.2 | 8 / 36 (22%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 5e-05 | 0 | 0.00016 | 0.00061 | 0 | 0.0014 | 3.6 | 2 / 11 (18%) | 4 / 11 (36%) | 0.182 | 0.364 |
| 2.A.6.2.13 | The Resistance-Nodulation-Cell Division (RND) Superfamily | 0.067 | 0.34 | 1 | 0.016 | 0.23 | 1 | 0.083 | 0.39 | 1 | 0.03 | 0.27 | 1 | 0.088 | 0.62 | 1 | 0.28 | 0.73 | 1 | 0.00094 | 0.00016 | 0 | 0.00047 | 0 | 0.0011 | 0.00032 | 0 | 0.00084 | -0.55 | 6 / 36 (17%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 1.1e-05 | 0 | 3.6e-05 | 2.7e-06 | 0 | 8.9e-06 | -2 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.A.3.6.2 | The P-type ATPase (P-ATPase) Superfamily | 0.094 | 0.34 | 1 | 0.14 | 0.44 | 1 | 0.13 | 0.43 | 1 | 0.036 | 0.27 | 1 | 0.3 | 0.8 | 1 | 0.24 | 0.7 | 1 | 0.00075 | 8.3e-05 | 0 | 0.00022 | 0 | 0.00053 | 2e-04 | 0 | 0.00054 | -0.14 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.76.1.3 | The Resistance to Homoserine/Threonine (RhtB) Family | 0.45 | 0.52 | 1 | 0.66 | 0.81 | 1 | 0.72 | 0.88 | 1 | 0.82 | 0.93 | 1 | 0.64 | 0.92 | 1 | 0.72 | 0.88 | 1 | 0.00014 | 2.3e-05 | 0 | 1.9e-05 | 0 | 4.4e-05 | 5.8e-05 | 0 | 0.00015 | 1.6 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 3e-06 | 0 | 9.9e-06 | 2.3e-05 | 0 | 5.2e-05 | 2.9 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 1.B.14.2.14 | The Outer Membrane Receptor (OMR) Family | 0.08 | 0.34 | 1 | 0.17 | 0.46 | 1 | 0.11 | 0.43 | 1 | 0.05 | 0.28 | 1 | 0.37 | 0.84 | 1 | 0.23 | 0.7 | 1 | 0.00058 | 6.4e-05 | 0 | 0.00017 | 0 | 4e-04 | 0.00015 | 0 | 0.00038 | -0.18 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 8.1e-06 | 0 | 2.7e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.A.12.3.1 | The Intracellular Chloride Channel (CLIC) Family | 0.33 | 0.42 | 1 | 0.54 | 0.76 | 1 | 0.69 | 0.86 | 1 | 0.68 | 0.86 | 1 | 0.8 | 0.94 | 1 | 0.72 | 0.88 | 1 | 0.00028 | 3.2e-05 | 0 | 8.4e-05 | 0 | 0.00022 | 4.8e-05 | 0 | 0.00013 | -0.81 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 1.1e-05 | 0 | 3.6e-05 | 8.1e-06 | 0 | 2.7e-05 | -0.44 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 1.B.38.4.1 | The Treponema Porin Major Surface Protein (TP-MSP) Family | 0.21 | 0.35 | 1 | 0.27 | 0.55 | 1 | 0.35 | 0.6 | 1 | 0.92 | 0.97 | 1 | 0.52 | 0.87 | 1 | 0.62 | 0.85 | 1 | 1e-04 | 1.4e-05 | 0 | 3.2e-05 | 0 | 5.7e-05 | 8.8e-06 | 0 | 2.3e-05 | -1.9 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 8.9e-06 | 0 | 3e-05 | 1.2e-05 | 0 | 4e-05 | 0.43 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.2.3.1 | The Glycoside-Pentoside-Hexuronide (GPH):Cation Symporter Family | 0.28 | 0.38 | 1 | 0.76 | 0.88 | 1 | 0.28 | 0.54 | 1 | 0.91 | 0.97 | 1 | 0.5 | 0.87 | 1 | 0.84 | 0.93 | 1 | 2e-04 | 2.8e-05 | 0 | 6.7e-05 | 0 | 0.00018 | 5.8e-05 | 0 | 0.00015 | -0.21 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 9.5e-06 | 0 | 2.3e-05 | 1.9 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 1.A.34.1.2 | The Bacillus Gap Junction-like Channel-forming Complex (GJ-CC) Family | 0.31 | 0.4 | 1 | 0.7 | 0.85 | 1 | 0.27 | 0.53 | 1 | 0.88 | 0.96 | 1 | 0.46 | 0.87 | 1 | 0.18 | 0.68 | 1 | 0.00053 | 4.4e-05 | 0 | 1e-04 | 0 | 0.00027 | 0.00012 | 0 | 0.00031 | 0.26 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 6e-06 | 0 | 2e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.A.1.17.3 | The Major Facilitator Superfamily (MFS) | 0.18 | 0.34 | 1 | 0.68 | 0.83 | 1 | 0.26 | 0.53 | 1 | 0.48 | 0.73 | 1 | 0.58 | 0.91 | 1 | 0.77 | 0.9 | 1 | 0.00054 | 4.5e-05 | 0 | 0.00012 | 0 | 0.00031 | 9.7e-05 | 0 | 0.00026 | -0.31 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 1e-05 | 0 | 3.4e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.C.1.1.1 | The TonB-ExbB-ExbD/TolA-TolQ-TolR Outer Membrane Receptor Energizers and Stabilizers (TonB/TolA) Family | 0.16 | 0.34 | 1 | 0.87 | 0.95 | 1 | 0.68 | 0.86 | 1 | 0.25 | 0.56 | 1 | 0.61 | 0.91 | 1 | 0.86 | 0.94 | 1 | 0.00017 | 1.9e-05 | 0 | 3.6e-05 | 0 | 8.8e-05 | 5.8e-05 | 0 | 0.00015 | 0.69 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 3.A.1.2.4 | The ATP-binding Cassette (ABC) Superfamily | 0.022 | 0.34 | 1 | 0.00083 | 0.16 | 0.47 | 0.01 | 0.37 | 1 | 0.0012 | 0.15 | 0.69 | 0.0047 | 0.36 | 1 | 0.049 | 0.58 | 1 | 0.00068 | 0.00017 | 0 | 0.00054 | 8.1e-05 | 0.0012 | 3e-04 | 0 | 0.00079 | -0.85 | 9 / 36 (25%) | 5 / 7 (71%) | 1 / 7 (14%) | 0.714 | 0.143 | 1.4e-05 | 0 | 3.7e-05 | 8.1e-06 | 0 | 2.7e-05 | -0.79 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 2.A.7.3.6 | The Drug/Metabolite Transporter (DMT) Superfamily | 0.27 | 0.38 | 1 | 0.6 | 0.78 | 1 | 0.24 | 0.52 | 1 | 0.99 | 0.99 | 1 | 0.1 | 0.64 | 1 | 0.015 | 0.46 | 1 | 0.00043 | 3.6e-05 | 0 | 8.4e-05 | 0 | 0.00022 | 9.7e-05 | 0 | 0.00026 | 0.21 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 1.B.21.1.1 | The OmpG Porin (OmpG) Family | 0.26 | 0.37 | 1 | 0.66 | 0.82 | 1 | 0.64 | 0.84 | 1 | 0.83 | 0.93 | 1 | 0.48 | 0.87 | 1 | 0.2 | 0.69 | 1 | 3e-04 | 2.5e-05 | 0 | 6.7e-05 | 0 | 0.00018 | 5.8e-05 | 0 | 0.00015 | -0.21 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3e-06 | 0 | 9.9e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.A.49.5.3 | The Chloride Carrier/Channel (ClC) Family | 0.057 | 0.34 | 1 | 0.13 | 0.42 | 1 | 0.24 | 0.52 | 1 | 0.045 | 0.27 | 1 | 0.12 | 0.66 | 1 | 0.26 | 0.72 | 1 | 0.00024 | 4.1e-05 | 0 | 1e-04 | 0 | 0.00018 | 9.7e-05 | 0 | 0.00026 | -0.044 | 6 / 36 (17%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 3e-06 | 0 | 9.9e-06 | 2.7e-06 | 0 | 8.9e-06 | -0.15 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.A.1.122.15 | The ATP-binding Cassette (ABC) Superfamily | 0.063 | 0.34 | 1 | 0.0091 | 0.22 | 1 | 0.036 | 0.37 | 1 | 0.019 | 0.26 | 1 | 0.05 | 0.62 | 1 | 0.14 | 0.65 | 1 | 0.00049 | 8.2e-05 | 0 | 0.00023 | 0 | 0.00053 | 0.00018 | 0 | 0.00049 | -0.35 | 6 / 36 (17%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 3e-06 | 0 | 9.9e-06 | 2.7e-06 | 0 | 8.9e-06 | -0.15 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 9.B.146.1.7 | The Putative Undecaprenyl-phosphate N-Acetylglucosaminyl Transferase (MurG) Family | 0.054 | 0.34 | 1 | 0.16 | 0.46 | 1 | 0.19 | 0.48 | 1 | 0.029 | 0.27 | 1 | 0.24 | 0.77 | 1 | 0.24 | 0.7 | 1 | 6e-04 | 6.7e-05 | 0 | 0.00019 | 0 | 0.00036 | 0.00015 | 0 | 0.00041 | -0.34 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 4.A.1.1.2 | The PTS Glucose-Glucoside (Glc) Family | 0.14 | 0.34 | 1 | 0.51 | 0.75 | 1 | 0.76 | 0.9 | 1 | 0.46 | 0.72 | 1 | 0.087 | 0.62 | 1 | 0.23 | 0.7 | 1 | 0.00035 | 6.8e-05 | 0 | 0.00019 | 0 | 0.00049 | 0.00014 | 0 | 0.00027 | -0.44 | 7 / 36 (19%) | 2 / 7 (29%) | 3 / 7 (43%) | 0.286 | 0.429 | 2.6e-06 | 0 | 8.5e-06 | 1.1e-05 | 0 | 3.6e-05 | 2.1 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.1.20.2 | The Major Facilitator Superfamily (MFS) | 0.16 | 0.34 | 1 | 0.38 | 0.65 | 1 | 0.17 | 0.46 | 1 | 0.47 | 0.73 | 1 | 0.68 | 0.92 | 1 | 0.63 | 0.85 | 1 | 0.00034 | 2.8e-05 | 0 | 8.4e-05 | 0 | 0.00022 | 4.8e-05 | 0 | 0.00013 | -0.81 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 8.1e-06 | 0 | 2.7e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 3.A.1.4.1 | The ATP-binding Cassette (ABC) Superfamily | 0.19 | 0.34 | 1 | 0.6 | 0.78 | 1 | 0.52 | 0.75 | 1 | 0.65 | 0.85 | 1 | 0.65 | 0.92 | 1 | 0.84 | 0.93 | 1 | 0.0011 | 0.00025 | 0 | 0.00073 | 0 | 0.0019 | 0.00048 | 0 | 0.0012 | -0.6 | 8 / 36 (22%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 1.5e-05 | 0 | 3.3e-05 | 3.4e-05 | 0 | 7.8e-05 | 1.2 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 9.B.126.1.1 | The Putative Lipid IV Exporter (YhjD) Family | 0.16 | 0.34 | 1 | 0.038 | 0.28 | 1 | 0.23 | 0.51 | 1 | 0.47 | 0.73 | 1 | 0.34 | 0.83 | 1 | 0.98 | 1 | 1 | 0.00025 | 4.2e-05 | 0 | 0.00012 | 0 | 0.00031 | 7.7e-05 | 0 | 2e-04 | -0.64 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 5.5e-06 | 0 | 1.2e-05 | 2.7e-06 | 0 | 8.9e-06 | -1 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 8.A.46.1.1 | The Glycan-binding Protein (SusD) Family | 0.89 | 0.92 | 1 | 0.76 | 0.88 | 1 | 0.61 | 0.82 | 1 | 0.36 | 0.64 | 1 | 0.5 | 0.87 | 1 | 0.45 | 0.79 | 1 | 7.5e-05 | 1e-05 | 0 | 8.1e-06 | 0 | 2.1e-05 | 8.3e-06 | 0 | 1.4e-05 | 0.035 | 5 / 36 (14%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 1.8e-05 | 0 | 5.9e-05 | 6e-06 | 0 | 2e-05 | -1.6 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 1.B.46.1.1 | The Outer Membrane LolAB Lipoprotein Insertion Apparatus (LolAB) Family | 0.47 | 0.54 | 1 | 0.76 | 0.88 | 1 | 0.56 | 0.78 | 1 | 0.43 | 0.69 | 1 | 0.66 | 0.92 | 1 | 0.21 | 0.69 | 1 | 0.00026 | 2.2e-05 | 0 | 5e-05 | 0 | 0.00013 | 3.9e-05 | 0 | 1e-04 | -0.36 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 1.5e-05 | 0 | 4.9e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 1.B.14.1.9 | The Outer Membrane Receptor (OMR) Family | 0.28 | 0.38 | 1 | 0.87 | 0.94 | 1 | 0.65 | 0.84 | 1 | 0.88 | 0.96 | 1 | 0.58 | 0.91 | 1 | 0.9 | 0.97 | 1 | 0.00054 | 7.5e-05 | 0 | 0.00017 | 0 | 0.00045 | 0.00018 | 0 | 0.00049 | 0.082 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 5.1e-06 | 0 | 1.7e-05 | 1.6e-05 | 0 | 3.7e-05 | 1.6 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 4.A.1.2.7 | The PTS Glucose-Glucoside (Glc) Family | 0.15 | 0.34 | 1 | 0.82 | 0.91 | 1 | 0.92 | 0.97 | 1 | 0.079 | 0.32 | 1 | 0.77 | 0.94 | 1 | 0.56 | 0.83 | 1 | 0.00042 | 4.6e-05 | 0 | 7.9e-05 | 0 | 0.00018 | 0.00015 | 0 | 0.00041 | 0.93 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 3.A.1.139.2 | The ATP-binding Cassette (ABC) Superfamily | 0.73 | 0.79 | 1 | 0.55 | 0.76 | 1 | 0.45 | 0.7 | 1 | 0.29 | 0.61 | 1 | 0.54 | 0.89 | 1 | 0.37 | 0.74 | 1 | 0.00027 | 3.8e-05 | 0 | 5e-05 | 0 | 0.00013 | 9.7e-05 | 0 | 0.00026 | 0.96 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 1.7e-05 | 0 | 4e-05 | 1.3e-05 | 0 | 4.3e-05 | -0.39 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 2.A.1.21.22 | The Major Facilitator Superfamily (MFS) | 0.87 | 0.9 | 1 | 0.26 | 0.55 | 1 | 0.97 | 0.99 | 1 | 0.62 | 0.82 | 1 | 0.077 | 0.62 | 1 | 0.56 | 0.83 | 1 | 0.00019 | 7.5e-05 | 0 | 5.8e-05 | 0 | 0.00013 | 8.9e-05 | 2.7e-05 | 0.00013 | 0.62 | 14 / 36 (39%) | 2 / 7 (29%) | 4 / 7 (57%) | 0.286 | 0.571 | 3.8e-05 | 0 | 7.2e-05 | 0.00011 | 0 | 0.00027 | 1.5 | 3 / 11 (27%) | 5 / 11 (45%) | 0.273 | 0.455 |
| 2.A.17.1.5 | The Proton-dependent Oligopeptide Transporter (POT/PTR) Family | 0.22 | 0.35 | 1 | 0.37 | 0.65 | 1 | 0.47 | 0.72 | 1 | 0.67 | 0.85 | 1 | 0.25 | 0.77 | 1 | 0.32 | 0.73 | 1 | 0.00021 | 3.6e-05 | 0 | 7.2e-05 | 0 | 0.00018 | 9.4e-05 | 0 | 2e-04 | 0.38 | 6 / 36 (17%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 6e-06 | 0 | 2e-05 | 5.4e-06 | 0 | 1.8e-05 | -0.15 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.1.25.2 | The Major Facilitator Superfamily (MFS) | 0.084 | 0.34 | 1 | 0.27 | 0.56 | 1 | 0.18 | 0.48 | 1 | 0.017 | 0.25 | 1 | 0.23 | 0.76 | 1 | 0.16 | 0.67 | 1 | 0.00046 | 6.4e-05 | 0 | 0.00015 | 0 | 0.00035 | 0.00017 | 0 | 0.00046 | 0.18 | 5 / 36 (14%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.6.4.1 | The Resistance-Nodulation-Cell Division (RND) Superfamily | 0.13 | 0.34 | 1 | 0.65 | 0.81 | 1 | 0.6 | 0.82 | 1 | 0.19 | 0.5 | 1 | 0.65 | 0.92 | 1 | 0.69 | 0.88 | 1 | 0.00043 | 0.00011 | 0 | 0.00022 | 0 | 0.00048 | 0.00025 | 0 | 0.00061 | 0.18 | 9 / 36 (25%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 1.7e-05 | 0 | 4.9e-05 | 3.4e-05 | 0 | 1e-04 | 1 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 9.B.1.1.6 | The Integral Membrane CAAX Protease (CAAX Protease) Family | 0.028 | 0.34 | 1 | 0.021 | 0.23 | 1 | 0.021 | 0.37 | 1 | 0.012 | 0.22 | 1 | 0.087 | 0.62 | 1 | 0.056 | 0.58 | 1 | 0.00026 | 3.6e-05 | 0 | 0.00013 | 0 | 0.00031 | 3.9e-05 | 0 | 1e-04 | -1.7 | 5 / 36 (14%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 0 | 0 | 0 | 1.1e-05 | 0 | 3.6e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.1.14.33 | The Major Facilitator Superfamily (MFS) | 0.16 | 0.34 | 1 | 0.24 | 0.53 | 1 | 0.14 | 0.43 | 1 | 0.3 | 0.62 | 1 | 0.34 | 0.83 | 1 | 0.99 | 1 | 1 | 0.00061 | 5.1e-05 | 0 | 0.00015 | 0 | 4e-04 | 0.00011 | 0 | 0.00028 | -0.45 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.A.35.1.1 | The CorA Metal Ion Transporter (MIT) Family | 0.74 | 0.79 | 1 | 0.59 | 0.78 | 1 | 0.31 | 0.58 | 1 | 0.34 | 0.63 | 1 | 0.65 | 0.92 | 1 | 0.25 | 0.7 | 1 | 2e-04 | 3.4e-05 | 0 | 3.6e-05 | 0 | 8.8e-05 | 9.7e-05 | 0 | 0.00026 | 1.4 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.5e-05 | 0 | 7.2e-05 | 2.7e-06 | 0 | 8.9e-06 | -3.2 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 2.A.21.3.19 | The Solute:Sodium Symporter (SSS) Family | 0.066 | 0.34 | 1 | 0.94 | 0.98 | 1 | 0.24 | 0.52 | 1 | 0.12 | 0.38 | 1 | 0.83 | 0.95 | 1 | 0.55 | 0.83 | 1 | 0.00076 | 0.00038 | 1.6e-05 | 6e-04 | 0.00022 | 0.00092 | 0.00055 | 0.00012 | 0.00079 | -0.13 | 18 / 36 (50%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 7.7e-05 | 0 | 0.00017 | 0.00044 | 0 | 0.00077 | 2.5 | 3 / 11 (27%) | 5 / 11 (45%) | 0.273 | 0.455 |
| 2.A.1.1.42 | The Major Facilitator Superfamily (MFS) | 0.25 | 0.36 | 1 | 0.18 | 0.48 | 1 | 0.86 | 0.94 | 1 | 0.11 | 0.36 | 1 | 0.18 | 0.72 | 1 | 0.97 | 1 | 1 | 0.0011 | 0.00084 | 0.00068 | 0.00033 | 0.00034 | 0.00035 | 0.00085 | 0.00072 | 0.00087 | 1.4 | 27 / 36 (75%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.00086 | 0.00042 | 0.0013 | 0.0011 | 0.00091 | 0.00096 | 0.36 | 9 / 11 (82%) | 10 / 11 (91%) | 0.818 | 0.909 |
| 3.A.1.11.7 | The ATP-binding Cassette (ABC) Superfamily | 0.3 | 0.4 | 1 | 0.19 | 0.49 | 1 | 0.39 | 0.64 | 1 | 0.15 | 0.42 | 1 | 0.12 | 0.66 | 1 | 0.37 | 0.75 | 1 | 0.00027 | 6e-05 | 0 | 0 | 0 | 0 | 8.2e-05 | 0 | 0.00015 | Inf | 8 / 36 (22%) | 0 / 7 (0%) | 2 / 7 (29%) | 0 | 0.286 | 8.8e-05 | 0 | 0.00027 | 5.6e-05 | 0 | 1e-04 | -0.65 | 2 / 11 (18%) | 4 / 11 (36%) | 0.182 | 0.364 |
| 3.A.10.1.5 | The H+. Na+-translocating Pyrophosphatase (M+-PPase) Family | 0.29 | 0.38 | 1 | 0.28 | 0.57 | 1 | 0.83 | 0.92 | 1 | 0.53 | 0.76 | 1 | 0.92 | 0.98 | 1 | 0.35 | 0.74 | 1 | 0.00019 | 1.6e-05 | 0 | 5e-05 | 0 | 0.00013 | 1.9e-05 | 0 | 5.1e-05 | -1.4 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 6.6e-06 | 0 | 2.2e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.A.23.1.8 | The Dicarboxylate/Amino Acid:Cation (Na+ or H+) Symporter (DAACS) Family | 0.14 | 0.34 | 1 | 0.36 | 0.64 | 1 | 0.8 | 0.91 | 1 | 0.32 | 0.63 | 1 | 0.42 | 0.86 | 1 | 0.97 | 1 | 1 | 0.00056 | 6.3e-05 | 0 | 0.00015 | 0 | 0.00026 | 0.00014 | 0 | 0.00036 | -0.1 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.2e-05 | 0 | 7.3e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 8.A.1.1.3 | The Membrane Fusion Protein (MFP) Family | 0.14 | 0.34 | 1 | 0.92 | 0.96 | 1 | 0.91 | 0.96 | 1 | 0.45 | 0.71 | 1 | 0.43 | 0.86 | 1 | 0.62 | 0.85 | 1 | 0.00021 | 3.5e-05 | 0 | 8.9e-05 | 0 | 0.00022 | 7.5e-05 | 0 | 0.00015 | -0.25 | 6 / 36 (17%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 2.6e-06 | 0 | 8.5e-06 | 8.1e-06 | 0 | 2.7e-05 | 1.6 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.53.3.11 | The Sulfate Permease (SulP) Family | 0.3 | 0.4 | 1 | 0.99 | 1 | 1 | 0.75 | 0.9 | 1 | 0.88 | 0.95 | 1 | 0.58 | 0.91 | 1 | 0.8 | 0.92 | 1 | 0.00027 | 3e-05 | 0 | 6.7e-05 | 0 | 0.00018 | 6.8e-05 | 0 | 0.00018 | 0.021 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3e-06 | 0 | 9.9e-06 | 1e-05 | 0 | 3.4e-05 | 1.7 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 8.A.42.1.6 | The small hydrophilic YodL domain/protein (YodL) Family | 0.94 | 0.95 | 1 | 0.63 | 0.8 | 1 | 0.99 | 0.99 | 1 | 0.9 | 0.97 | 1 | 0.36 | 0.84 | 1 | 0.59 | 0.85 | 1 | 0.00059 | 0.00031 | 0.00019 | 0.00023 | 0.00027 | 0.00024 | 0.00028 | 0.00025 | 0.00029 | 0.28 | 19 / 36 (53%) | 4 / 7 (57%) | 5 / 7 (71%) | 0.571 | 0.714 | 3e-04 | 0 | 0.00053 | 4e-04 | 0 | 0.00057 | 0.42 | 5 / 11 (45%) | 5 / 11 (45%) | 0.455 | 0.455 |
| 9.B.307.1.2 | The FtsH AAA(+) Zinc Metaloprotease (FtsH) Family | 0.16 | 0.34 | 1 | 0.66 | 0.82 | 1 | 0.14 | 0.43 | 1 | 0.19 | 0.5 | 1 | 0.99 | 1 | 1 | 0.23 | 0.7 | 1 | 0.00052 | 7.2e-05 | 0 | 0.00017 | 0 | 0.00044 | 0.00017 | 0 | 0.00046 | 0 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 1.6e-05 | 0 | 3.7e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 2.A.1.15.13 | The Major Facilitator Superfamily (MFS) | 0.0073 | 0.34 | 1 | 0.0029 | 0.22 | 1 | 0.0056 | 0.35 | 1 | 0.00038 | 0.11 | 0.22 | 0.0098 | 0.46 | 1 | 0.015 | 0.46 | 1 | 0.00022 | 3.6e-05 | 0 | 0.00012 | 1.6e-05 | 0.00021 | 5.8e-05 | 0 | 0.00015 | -1 | 6 / 36 (17%) | 4 / 7 (57%) | 1 / 7 (14%) | 0.571 | 0.143 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 3.A.1.3.25 | The ATP-binding Cassette (ABC) Superfamily | 0.19 | 0.34 | 1 | 0.79 | 0.9 | 1 | 0.79 | 0.91 | 1 | 0.07 | 0.3 | 1 | 1 | 1 | 1 | 0.69 | 0.88 | 1 | 0.00013 | 2.1e-05 | 0 | 2.3e-06 | 0 | 6.1e-06 | 0 | 0 | 0 | -Inf | 6 / 36 (17%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 4.7e-05 | 0 | 0.00012 | 2e-05 | 0 | 4.9e-05 | -1.2 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 3.A.1.2.27 | The ATP-binding Cassette (ABC) Superfamily | 0.14 | 0.34 | 1 | 0.092 | 0.39 | 1 | 0.25 | 0.52 | 1 | 0.47 | 0.73 | 1 | 0.23 | 0.76 | 1 | 0.53 | 0.82 | 1 | 0.00047 | 0.00012 | 0 | 0.00032 | 0 | 0.00066 | 0.00019 | 0 | 0.00051 | -0.75 | 9 / 36 (25%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 2.9e-05 | 0 | 5.9e-05 | 2.6e-05 | 0 | 6.9e-05 | -0.16 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 2.A.33.1.5 | The NhaA Na+:H+ Antiporter (NhaA) Family | 0.11 | 0.34 | 1 | 0.023 | 0.24 | 1 | 0.032 | 0.37 | 1 | 0.024 | 0.27 | 1 | 0.096 | 0.64 | 1 | 0.063 | 0.58 | 1 | 0.00059 | 0.00044 | 0.00042 | 0.00022 | 0.00011 | 0.00025 | 0.00063 | 0.00024 | 0.00076 | 1.5 | 27 / 36 (75%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.00047 | 0.00048 | 0.00032 | 0.00044 | 0.00044 | 0.00034 | -0.095 | 10 / 11 (91%) | 9 / 11 (82%) | 0.909 | 0.818 |
| 2.C.1.1.3 | The TonB-ExbB-ExbD/TolA-TolQ-TolR Outer Membrane Receptor Energizers and Stabilizers (TonB/TolA) Family | 0.15 | 0.34 | 1 | 0.62 | 0.79 | 1 | 0.47 | 0.72 | 1 | 0.35 | 0.64 | 1 | 0.3 | 0.8 | 1 | 0.21 | 0.69 | 1 | 4e-04 | 5.5e-05 | 0 | 0.00014 | 0 | 0.00035 | 0.00014 | 0 | 0.00033 | 0 | 5 / 36 (14%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 2.6e-06 | 0 | 8.5e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.A.3.1.4 | The Amino Acid-Polyamine-Organocation (APC) Family | 0.23 | 0.35 | 1 | 0.047 | 0.29 | 1 | 0.44 | 0.69 | 1 | 0.5 | 0.75 | 1 | 0.051 | 0.62 | 1 | 0.48 | 0.81 | 1 | 0.00021 | 4.2e-05 | 0 | 5.3e-05 | 0 | 0.00013 | 0.00014 | 0 | 3e-04 | 1.4 | 7 / 36 (19%) | 2 / 7 (29%) | 3 / 7 (43%) | 0.286 | 0.429 | 0 | 0 | 0 | 1.5e-05 | 0 | 3.3e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 3.A.1.13.1 | The ATP-binding Cassette (ABC) Superfamily | 0.077 | 0.34 | 1 | 0.023 | 0.24 | 1 | 0.068 | 0.37 | 1 | 0.038 | 0.27 | 1 | 0.28 | 0.78 | 1 | 0.47 | 0.81 | 1 | 0.00065 | 5.4e-05 | 0 | 0.00016 | 0 | 4e-04 | 0.00012 | 0 | 0.00031 | -0.42 | 3 / 36 (8.3%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 3.A.1.5.1 | The ATP-binding Cassette (ABC) Superfamily | 0.16 | 0.34 | 1 | 0.037 | 0.28 | 1 | 0.13 | 0.43 | 1 | 0.27 | 0.6 | 1 | 0.06 | 0.62 | 1 | 0.17 | 0.68 | 1 | 0.00033 | 7.2e-05 | 0 | 0.00016 | 0 | 0.00035 | 0.00015 | 0 | 0.00038 | -0.093 | 8 / 36 (22%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 2.5e-05 | 0 | 7.2e-05 | 1.6e-05 | 0 | 3.7e-05 | -0.64 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
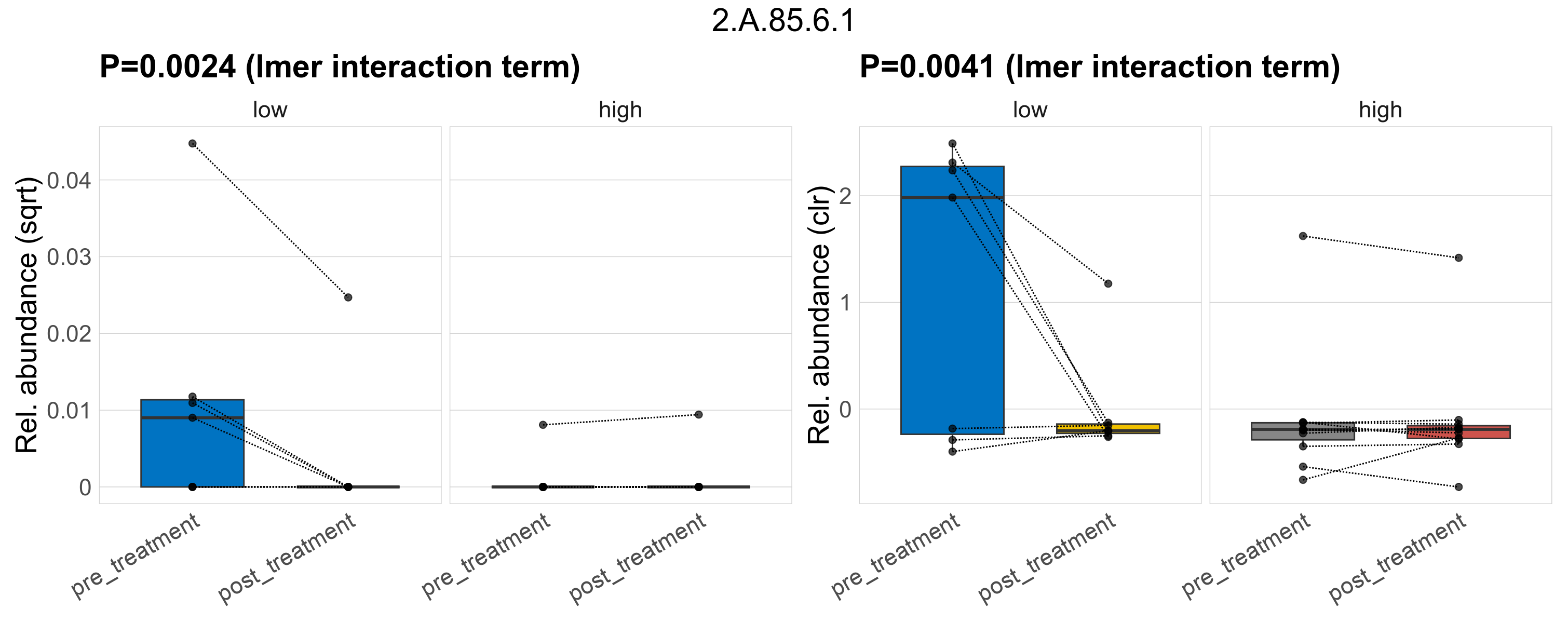
| 2.A.85.7.1 | The Aromatic Acid Exporter (ArAE) Family | 0.12 | 0.34 | 1 | 0.084 | 0.39 | 1 | 0.42 | 0.67 | 1 | 0.26 | 0.59 | 1 | 0.38 | 0.84 | 1 | 0.99 | 1 | 1 | 0.00056 | 6.2e-05 | 0 | 0.00019 | 0 | 0.00044 | 0.00012 | 0 | 0.00031 | -0.66 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 1.1e-05 | 0 | 3.6e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.A.40.1.1 | The Nucleobase/Ascorbate Transporter (NAT) or Nucleobase:Cation Symporter-2 (NCS2) Family | 0.051 | 0.34 | 1 | 0.027 | 0.25 | 1 | 0.057 | 0.37 | 1 | 0.081 | 0.32 | 1 | 0.095 | 0.64 | 1 | 0.16 | 0.67 | 1 | 0.00031 | 7e-05 | 0 | 0.00022 | 0 | 0.00048 | 9.7e-05 | 0 | 0.00026 | -1.2 | 8 / 36 (22%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 9.9e-06 | 0 | 2.5e-05 | 1.6e-05 | 0 | 3.7e-05 | 0.69 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 1.A.30.2.2 | The H+- or Na+-translocating Bacterial Flagellar Motor/ExbBD Outer Membrane Transport Energizer (Mot/Exb) Superfamily | 0.046 | 0.34 | 1 | 0.16 | 0.46 | 1 | 0.27 | 0.53 | 1 | 0.0083 | 0.19 | 1 | 0.13 | 0.66 | 1 | 0.27 | 0.72 | 1 | 0.00042 | 4.6e-05 | 0 | 0.00011 | 0 | 0.00022 | 0.00013 | 0 | 0.00033 | 0.24 | 4 / 36 (11%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 2.A.4.1.4 | The Cation Diffusion Facilitator (CDF) Family | 0.053 | 0.34 | 1 | 0.066 | 0.36 | 1 | 0.27 | 0.53 | 1 | 0.036 | 0.27 | 1 | 0.14 | 0.66 | 1 | 0.49 | 0.82 | 1 | 0.00024 | 3.3e-05 | 0 | 8.8e-05 | 0 | 0.00017 | 7.7e-05 | 0 | 2e-04 | -0.19 | 5 / 36 (14%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 3e-06 | 0 | 9.9e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 9.B.62.1.4 | The Copper Resistance (CopD) Family | 0.34 | 0.43 | 1 | 0.95 | 0.98 | 1 | 0.9 | 0.96 | 1 | 0.5 | 0.75 | 1 | 0.41 | 0.85 | 1 | 0.32 | 0.73 | 1 | 0.00019 | 2.2e-05 | 0 | 5e-05 | 0 | 0.00013 | 4.8e-05 | 0 | 0.00013 | -0.059 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 5.1e-06 | 0 | 1.7e-05 | 2.7e-06 | 0 | 8.9e-06 | -0.92 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 1.B.11.2.1 | The Outer Membrane Fimbrial Usher Porin (FUP) Family | 0.064 | 0.34 | 1 | 0.055 | 0.31 | 1 | 0.13 | 0.43 | 1 | 0.03 | 0.27 | 1 | 0.23 | 0.76 | 1 | 0.39 | 0.75 | 1 | 0.001 | 8.7e-05 | 0 | 0.00025 | 0 | 0.00057 | 0.00019 | 0 | 0.00051 | -0.4 | 3 / 36 (8.3%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 2.A.28.3.4 | The Bile Acid:Na+ Symporter (BASS) Family | 0.26 | 0.37 | 1 | 0.55 | 0.76 | 1 | 0.56 | 0.78 | 1 | 0.92 | 0.97 | 1 | 0.68 | 0.92 | 1 | 0.76 | 0.9 | 1 | 0.00033 | 3.7e-05 | 0 | 1e-04 | 0 | 0.00027 | 6.8e-05 | 0 | 0.00018 | -0.56 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 5.1e-06 | 0 | 1.7e-05 | 8.1e-06 | 0 | 2.7e-05 | 0.67 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.3.13.1 | The Amino Acid-Polyamine-Organocation (APC) Family | 0.06 | 0.34 | 1 | 0.13 | 0.42 | 1 | 0.1 | 0.42 | 1 | 0.047 | 0.27 | 1 | 0.33 | 0.83 | 1 | 0.23 | 0.7 | 1 | 0.00033 | 3.7e-05 | 0 | 1e-04 | 0 | 0.00022 | 7.7e-05 | 0 | 2e-04 | -0.38 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.42.2.3 | The Hydroxy/Aromatic Amino Acid Permease (HAAAP) Family | 0.18 | 0.34 | 1 | 0.76 | 0.88 | 1 | 0.3 | 0.57 | 1 | 0.39 | 0.67 | 1 | 0.35 | 0.84 | 1 | 0.92 | 0.98 | 1 | 0.00054 | 4.5e-05 | 0 | 0.00012 | 0 | 0.00031 | 0.00011 | 0 | 0.00028 | -0.13 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
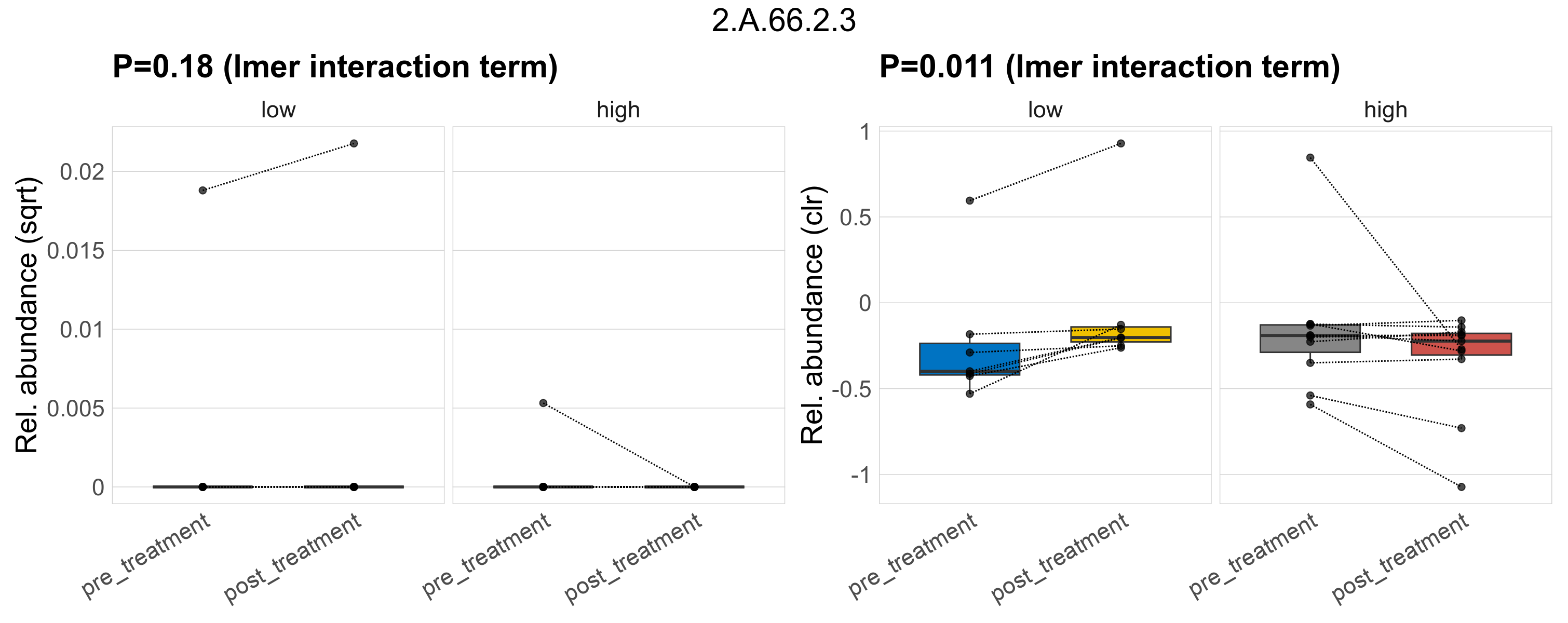
| 2.A.66.2.3 | The Multidrug/Oligosaccharidyl-lipid/Polysaccharide (MOP) Flippase Superfamily | 0.32 | 0.41 | 1 | 0.41 | 0.69 | 1 | 0.18 | 0.48 | 1 | 0.68 | 0.86 | 1 | 0.069 | 0.62 | 1 | 0.011 | 0.46 | 1 | 0.00029 | 2.4e-05 | 0 | 5e-05 | 0 | 0.00013 | 6.8e-05 | 0 | 0.00018 | 0.44 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 1.B.48.2.5 | The Curli Fiber Subunit Porin. CgsA. CsgG (CsgG) Family | 0.84 | 0.88 | 1 | 0.89 | 0.96 | 1 | 0.098 | 0.42 | 1 | 0.34 | 0.63 | 1 | 0.84 | 0.96 | 1 | 0.2 | 0.69 | 1 | 0.00018 | 4e-05 | 0 | 8.1e-06 | 0 | 2.1e-05 | 4.4e-06 | 0 | 1.2e-05 | -0.88 | 8 / 36 (22%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 1.4e-05 | 0 | 3.1e-05 | 0.00011 | 0 | 0.00021 | 3 | 2 / 11 (18%) | 4 / 11 (36%) | 0.182 | 0.364 |
| 8.A.1.6.1 | The Membrane Fusion Protein (MFP) Family | 0.12 | 0.34 | 1 | 0.55 | 0.76 | 1 | 0.64 | 0.83 | 1 | 0.055 | 0.29 | 1 | 0.49 | 0.87 | 1 | 0.67 | 0.88 | 1 | 0.00085 | 7.1e-05 | 0 | 0.00016 | 0 | 4e-04 | 2e-04 | 0 | 0.00054 | 0.32 | 3 / 36 (8.3%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 9.B.65.1.1 | The Putative Transporter (YhgE) Family | 0.19 | 0.34 | 1 | 0.11 | 0.4 | 1 | 0.45 | 0.71 | 1 | 0.75 | 0.89 | 1 | 0.59 | 0.91 | 1 | 0.72 | 0.88 | 1 | 0.00056 | 9.4e-05 | 0 | 0.00031 | 0 | 8e-04 | 0.00015 | 0 | 0.00038 | -1 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 1.6e-05 | 0 | 3.9e-05 | 2.7e-06 | 0 | 8.9e-06 | -2.6 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 1.A.62.2.3 | The Homotrimeric Cation Channel (TRIC) Family | 0.11 | 0.34 | 1 | 0.14 | 0.44 | 1 | 0.12 | 0.43 | 1 | 0.41 | 0.68 | 1 | 0.91 | 0.98 | 1 | 0.5 | 0.82 | 1 | 0.00021 | 1.8e-05 | 0 | 6.7e-05 | 0 | 0.00018 | 1.9e-05 | 0 | 5.1e-05 | -1.8 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.1.2.9 | The Major Facilitator Superfamily (MFS) | 0.13 | 0.34 | 1 | 0.52 | 0.75 | 1 | 0.2 | 0.48 | 1 | 0.33 | 0.63 | 1 | 0.97 | 0.99 | 1 | 0.48 | 0.81 | 1 | 0.00021 | 2.3e-05 | 0 | 5.3e-05 | 0 | 0.00013 | 4.8e-05 | 0 | 0.00013 | -0.14 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 1e-05 | 0 | 3.4e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 9.A.47.2.2 | The Tight Adherence (Pilus) Biogenesis Apparatus (TABA) Family | 0.2 | 0.34 | 1 | 0.25 | 0.53 | 1 | 0.54 | 0.77 | 1 | 0.028 | 0.27 | 1 | 0.29 | 0.78 | 1 | 0.19 | 0.68 | 1 | 0.0021 | 0.0017 | 0.0013 | 0.00099 | 0.0013 | 0.00099 | 0.0016 | 0.0017 | 0.0017 | 0.69 | 29 / 36 (81%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.0018 | 0.0013 | 0.0022 | 0.002 | 0.0013 | 0.0015 | 0.15 | 11 / 11 (100%) | 10 / 11 (91%) | 1 | 0.909 |
| 3.A.10.1.10 | The H+. Na+-translocating Pyrophosphatase (M+-PPase) Family | 0.94 | 0.95 | 1 | 0.61 | 0.79 | 1 | 0.49 | 0.73 | 1 | 0.53 | 0.76 | 1 | 0.89 | 0.98 | 1 | 0.86 | 0.94 | 1 | 0.00084 | 0.00052 | 1e-04 | 0.00043 | 0.00028 | 0.00073 | 0.00023 | 0.00011 | 0.00035 | -0.9 | 22 / 36 (61%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.00041 | 9.7e-05 | 0.00069 | 0.00085 | 2.9e-05 | 0.0017 | 1.1 | 8 / 11 (73%) | 6 / 11 (55%) | 0.727 | 0.545 |
| 2.A.13.1.1 | The C4-Dicarboxylate Uptake (Dcu) Family | 0.17 | 0.34 | 1 | 0.094 | 0.39 | 1 | 0.3 | 0.57 | 1 | 0.53 | 0.76 | 1 | 0.29 | 0.78 | 1 | 0.61 | 0.85 | 1 | 0.00029 | 6.5e-05 | 0 | 2e-04 | 0 | 0.00048 | 7.7e-05 | 0 | 2e-04 | -1.4 | 8 / 36 (22%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 2.7e-05 | 0 | 7.3e-05 | 9.5e-06 | 0 | 2.3e-05 | -1.5 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 2.A.37.1.1 | The Monovalent Cation:Proton Antiporter-2 (CPA2) Family | 0.073 | 0.34 | 1 | 0.091 | 0.39 | 1 | 0.16 | 0.46 | 1 | 0.12 | 0.38 | 1 | 0.37 | 0.84 | 1 | 0.4 | 0.75 | 1 | 0.00047 | 1e-04 | 0 | 0.00032 | 0 | 0.00075 | 0.00015 | 0 | 0.00038 | -1.1 | 8 / 36 (22%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 2.2e-05 | 0 | 7.3e-05 | 1.5e-05 | 0 | 3.3e-05 | -0.55 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 3.A.1.7.1 | The ATP-binding Cassette (ABC) Superfamily | 0.2 | 0.34 | 1 | 0.64 | 0.8 | 1 | 0.99 | 0.99 | 1 | 0.51 | 0.75 | 1 | 0.4 | 0.85 | 1 | 0.69 | 0.88 | 1 | 6e-04 | 0.00013 | 0 | 0.00032 | 0 | 0.00084 | 0.00033 | 0 | 0.00084 | 0.044 | 8 / 36 (22%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 5.5e-06 | 0 | 1.2e-05 | 1.5e-05 | 0 | 3.3e-05 | 1.4 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 1.A.14.2.2 | The Calcium Transporter A (CaTA) (formerly the Testis-Enhanced Gene Transfer (TEGT) Family | 0.37 | 0.45 | 1 | 0.95 | 0.98 | 1 | 0.38 | 0.63 | 1 | 0.67 | 0.85 | 1 | 0.54 | 0.89 | 1 | 0.19 | 0.68 | 1 | 0.00044 | 3.7e-05 | 0 | 8.4e-05 | 0 | 0.00022 | 8.7e-05 | 0 | 0.00023 | 0.051 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 1.1e-05 | 0 | 3.6e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 1.A.43.3.3 | The Camphor Resistance or Fluoride Exporter (Fluc) Family | 0.23 | 0.35 | 1 | 0.76 | 0.88 | 1 | 0.89 | 0.96 | 1 | 0.069 | 0.3 | 1 | 0.76 | 0.94 | 1 | 0.9 | 0.97 | 1 | 0.00044 | 3e-04 | 0.00018 | 0.00019 | 0.00011 | 0.00021 | 0.00019 | 0 | 0.00025 | 0 | 25 / 36 (69%) | 4 / 7 (57%) | 3 / 7 (43%) | 0.571 | 0.429 | 0.00037 | 0.00028 | 0.00042 | 0.00039 | 0.00022 | 0.00038 | 0.076 | 10 / 11 (91%) | 8 / 11 (73%) | 0.909 | 0.727 |
| 2.A.3.1.7 | The Amino Acid-Polyamine-Organocation (APC) Family | 0.23 | 0.36 | 1 | 0.43 | 0.7 | 1 | 1 | 1 | 1 | 0.76 | 0.89 | 1 | 0.8 | 0.94 | 1 | 0.35 | 0.74 | 1 | 0.00044 | 8.6e-05 | 0 | 0.00025 | 0 | 0.00062 | 0.00013 | 0 | 3e-04 | -0.94 | 7 / 36 (19%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 3.6e-05 | 0 | 0.00011 | 5.4e-06 | 0 | 1.8e-05 | -2.7 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
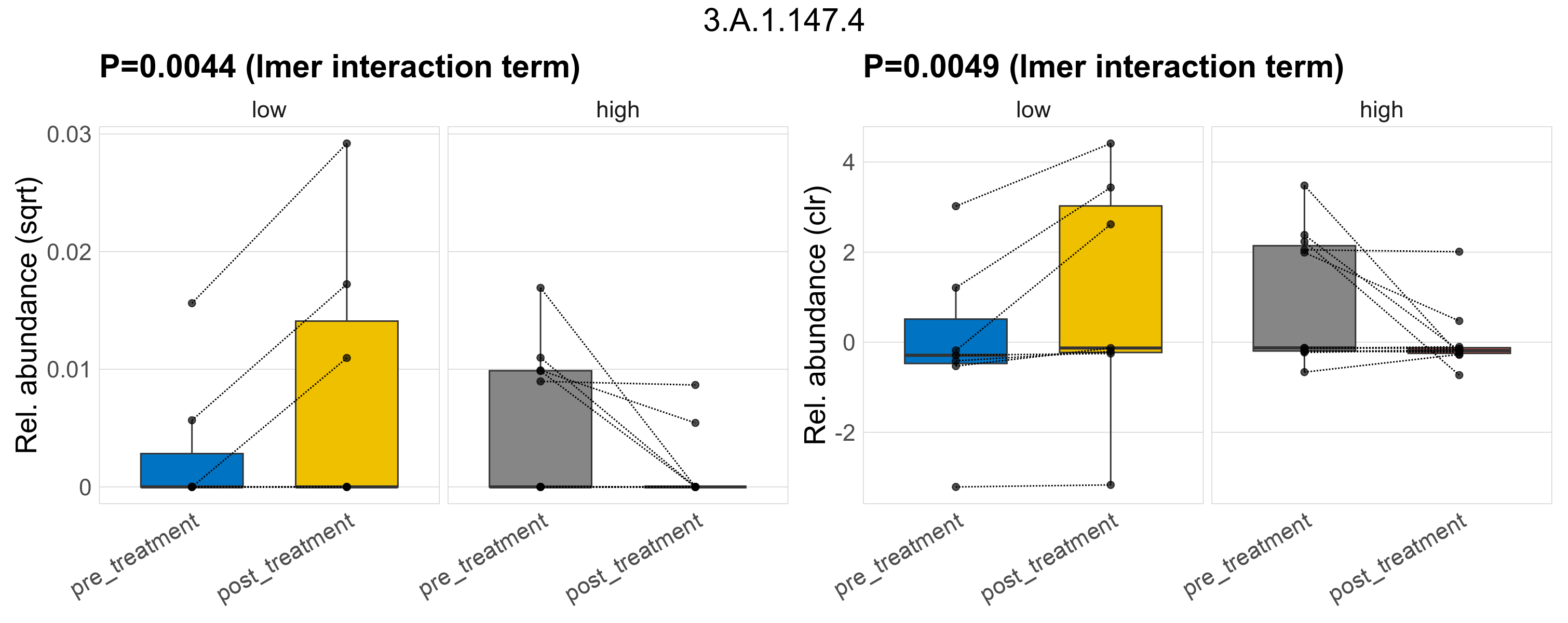
| 3.A.1.147.4 | The ATP-binding Cassette (ABC) Superfamily | 0.5 | 0.56 | 1 | 0.029 | 0.25 | 1 | 0.0044 | 0.35 | 1 | 0.19 | 0.49 | 1 | 0.048 | 0.62 | 1 | 0.0049 | 0.46 | 1 | 0.00019 | 6.5e-05 | 0 | 3.9e-05 | 0 | 9.1e-05 | 0.00018 | 0 | 0.00032 | 2.2 | 12 / 36 (33%) | 2 / 7 (29%) | 3 / 7 (43%) | 0.286 | 0.429 | 6.2e-05 | 0 | 8.9e-05 | 9.5e-06 | 0 | 2.3e-05 | -2.7 | 5 / 11 (45%) | 2 / 11 (18%) | 0.455 | 0.182 |
| 4.A.2.1.24 | The PTS Fructose-Mannitol (Fru) Family | 0.092 | 0.34 | 1 | 0.081 | 0.38 | 1 | 0.066 | 0.37 | 1 | 0.11 | 0.36 | 1 | 0.56 | 0.91 | 1 | 0.24 | 0.7 | 1 | 0.00053 | 5.9e-05 | 0 | 2e-04 | 0 | 0.00053 | 8.7e-05 | 0 | 0.00023 | -1.2 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 8.1e-06 | 0 | 2.7e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.45.2.5 | The Arsenite-Antimonite (ArsB) Efflux Family | 0.33 | 0.42 | 1 | 0.45 | 0.72 | 1 | 0.97 | 0.99 | 1 | 0.72 | 0.87 | 1 | 0.91 | 0.98 | 1 | 0.5 | 0.82 | 1 | 0.00021 | 3.5e-05 | 0 | 8.6e-05 | 0 | 0.00022 | 6.8e-05 | 0 | 0.00018 | -0.34 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 1.4e-05 | 0 | 3.7e-05 | 2.7e-06 | 0 | 8.9e-06 | -2.4 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 2.A.3.7.4 | The Amino Acid-Polyamine-Organocation (APC) Family | 0.26 | 0.37 | 1 | 0.87 | 0.94 | 1 | 0.94 | 0.98 | 1 | 0.83 | 0.94 | 1 | 0.37 | 0.84 | 1 | 0.54 | 0.83 | 1 | 0.00045 | 5e-05 | 0 | 0.00012 | 0 | 0.00031 | 0.00013 | 0 | 0.00033 | 0.12 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 5.4e-06 | 0 | 1.8e-05 | 1.1 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.A.1.122.30 | The ATP-binding Cassette (ABC) Superfamily | 0.39 | 0.47 | 1 | 0.42 | 0.69 | 1 | 0.84 | 0.92 | 1 | 0.075 | 0.31 | 1 | 0.2 | 0.74 | 1 | 0.51 | 0.82 | 1 | 0.0022 | 0.0017 | 0.0013 | 0.0012 | 0.00094 | 0.0013 | 0.0017 | 0.0019 | 0.0019 | 0.5 | 28 / 36 (78%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.0018 | 0.0013 | 0.0027 | 0.002 | 0.0014 | 0.0017 | 0.15 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 2.A.47.4.8 | The Divalent Anion:Na+ Symporter (DASS) Family | 0.093 | 0.34 | 1 | 0.074 | 0.37 | 1 | 0.17 | 0.47 | 1 | 0.096 | 0.34 | 1 | 0.21 | 0.74 | 1 | 0.39 | 0.75 | 1 | 0.00044 | 7.4e-05 | 0 | 2e-04 | 0 | 0.00048 | 0.00015 | 0 | 0.00041 | -0.42 | 6 / 36 (17%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 7.3e-06 | 0 | 2.4e-05 | 5.4e-06 | 0 | 1.8e-05 | -0.43 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.66.2.16 | The Multidrug/Oligosaccharidyl-lipid/Polysaccharide (MOP) Flippase Superfamily | 0.14 | 0.34 | 1 | 0.22 | 0.51 | 1 | 0.054 | 0.37 | 1 | 0.044 | 0.27 | 1 | 0.14 | 0.66 | 1 | 0.026 | 0.57 | 1 | 3e-04 | 4.2e-05 | 0 | 0 | 0 | 0 | 8.9e-05 | 0 | 0.00023 | Inf | 5 / 36 (14%) | 0 / 7 (0%) | 1 / 7 (14%) | 0 | 0.143 | 7.9e-05 | 0 | 0.00019 | 2.7e-06 | 0 | 8.8e-06 | -4.9 | 3 / 11 (27%) | 1 / 11 (9.1%) | 0.273 | 0.0909 |
| 3.A.1.6.1 | The ATP-binding Cassette (ABC) Superfamily | 0.19 | 0.34 | 1 | 0.2 | 0.49 | 1 | 0.66 | 0.84 | 1 | 0.38 | 0.65 | 1 | 0.07 | 0.62 | 1 | 0.35 | 0.74 | 1 | 0.0011 | 0.00012 | 0 | 0.00029 | 0 | 0.00076 | 0.00031 | 0 | 0.00076 | 0.096 | 4 / 36 (11%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 0 | 0 | 0 | 8.1e-06 | 0 | 2.7e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.A.43.3.1 | The Camphor Resistance or Fluoride Exporter (Fluc) Family | 0.5 | 0.56 | 1 | 0.82 | 0.91 | 1 | 0.82 | 0.92 | 1 | 0.13 | 0.39 | 1 | 0.52 | 0.87 | 1 | 0.72 | 0.88 | 1 | 0.00059 | 0.00044 | 0.00041 | 0.00043 | 0.00014 | 0.00052 | 0.00044 | 0.00059 | 0.00044 | 0.033 | 27 / 36 (75%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.00048 | 0.00042 | 0.00044 | 0.00042 | 4e-04 | 0.00032 | -0.19 | 9 / 11 (82%) | 10 / 11 (91%) | 0.818 | 0.909 |
| 2.A.2.3.4 | The Glycoside-Pentoside-Hexuronide (GPH):Cation Symporter Family | 0.12 | 0.34 | 1 | 0.18 | 0.47 | 1 | 0.1 | 0.42 | 1 | 0.27 | 0.6 | 1 | 0.48 | 0.87 | 1 | 0.23 | 0.7 | 1 | 0.00037 | 7.2e-05 | 0 | 2e-04 | 0 | 0.00048 | 0.00013 | 0 | 0.00033 | -0.62 | 7 / 36 (19%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 7.7e-06 | 0 | 2.6e-05 | 2e-05 | 0 | 3.9e-05 | 1.4 | 1 / 11 (9.1%) | 3 / 11 (27%) | 0.0909 | 0.273 |
| 2.A.23.1.7 | The Dicarboxylate/Amino Acid:Cation (Na+ or H+) Symporter (DAACS) Family | 0.068 | 0.34 | 1 | 0.72 | 0.86 | 1 | 0.78 | 0.91 | 1 | 0.035 | 0.27 | 1 | 0.82 | 0.95 | 1 | 0.7 | 0.88 | 1 | 0.00039 | 5.4e-05 | 0 | 0.00011 | 0 | 0.00022 | 0.00017 | 0 | 4e-04 | 0.63 | 5 / 36 (14%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 3.D.4.3.2 | The Proton-translocating Cytochrome Oxidase (COX) Superfamily | 0.22 | 0.35 | 1 | 0.49 | 0.75 | 1 | 0.56 | 0.78 | 1 | 0.62 | 0.82 | 1 | 0.94 | 0.98 | 1 | 0.98 | 1 | 1 | 0.00062 | 8.6e-05 | 0 | 0.00022 | 0 | 0.00058 | 0.00018 | 0 | 0.00049 | -0.29 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 1.1e-05 | 0 | 3.6e-05 | 1.3e-05 | 0 | 4.5e-05 | 0.24 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
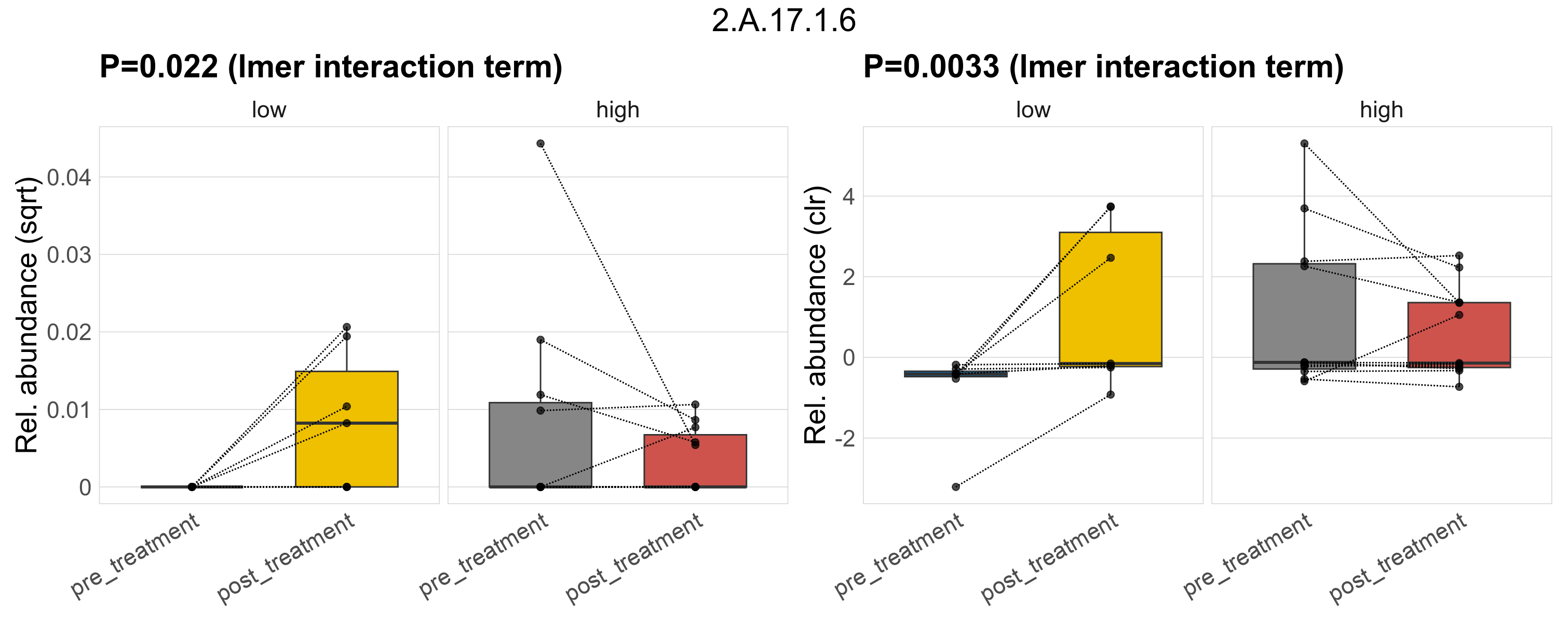
| 2.A.17.1.6 | The Proton-dependent Oligopeptide Transporter (POT/PTR) Family | 0.065 | 0.34 | 1 | 0.047 | 0.29 | 1 | 0.022 | 0.37 | 1 | 0.02 | 0.26 | 1 | 0.0023 | 0.31 | 1 | 0.0033 | 0.46 | 1 | 3e-04 | 0.00011 | 0 | 0 | 0 | 0 | 0.00014 | 6.8e-05 | 0.00018 | Inf | 13 / 36 (36%) | 0 / 7 (0%) | 4 / 7 (57%) | 0 | 0.571 | 0.00023 | 0 | 0.00058 | 2.8e-05 | 0 | 3.9e-05 | -3 | 4 / 11 (36%) | 5 / 11 (45%) | 0.364 | 0.455 |
| 3.D.4.5.1 | The Proton-translocating Cytochrome Oxidase (COX) Superfamily | 0.1 | 0.34 | 1 | 0.027 | 0.25 | 1 | 0.11 | 0.42 | 1 | 0.23 | 0.54 | 1 | 0.23 | 0.76 | 1 | 0.62 | 0.85 | 1 | 0.00056 | 0.00015 | 0 | 0.00048 | 0 | 0.0012 | 0.00026 | 0 | 0.00063 | -0.88 | 10 / 36 (28%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 1.7e-05 | 0 | 3.7e-05 | 1.6e-05 | 0 | 3.7e-05 | -0.087 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 2.A.1.7.17 | The Major Facilitator Superfamily (MFS) | 0.077 | 0.34 | 1 | 0.032 | 0.27 | 1 | 0.054 | 0.37 | 1 | 0.24 | 0.56 | 1 | 0.074 | 0.62 | 1 | 0.14 | 0.65 | 1 | 0.00028 | 1e-04 | 0 | 0.00019 | 0.00014 | 0.00022 | 3.3e-05 | 0 | 5.6e-05 | -2.5 | 13 / 36 (36%) | 5 / 7 (71%) | 2 / 7 (29%) | 0.714 | 0.286 | 6.4e-05 | 0 | 0.00013 | 0.00012 | 0 | 0.00032 | 0.91 | 3 / 11 (27%) | 3 / 11 (27%) | 0.273 | 0.273 |
| 9.B.211.1.2 | The 10 TMS Putative Permease (10PP) Family | 0.12 | 0.34 | 1 | 0.45 | 0.72 | 1 | 0.34 | 0.6 | 1 | 0.059 | 0.29 | 1 | 0.63 | 0.91 | 1 | 0.21 | 0.69 | 1 | 0.00024 | 0.00019 | 0.00018 | 0.00017 | 0.00014 | 0.00018 | 0.00013 | 6.2e-05 | 0.00017 | -0.39 | 28 / 36 (78%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 0.00026 | 0.00024 | 0.00013 | 0.00016 | 7.5e-05 | 2e-04 | -0.7 | 11 / 11 (100%) | 8 / 11 (73%) | 1 | 0.727 |
| 2.A.42.2.5 | The Hydroxy/Aromatic Amino Acid Permease (HAAAP) Family | 0.091 | 0.34 | 1 | 0.1 | 0.39 | 1 | 0.19 | 0.48 | 1 | 0.045 | 0.27 | 1 | 0.37 | 0.84 | 1 | 0.56 | 0.83 | 1 | 0.00066 | 5.5e-05 | 0 | 0.00015 | 0 | 0.00035 | 0.00014 | 0 | 0.00036 | -0.1 | 3 / 36 (8.3%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 9.B.209.1.1 | The DUF2633 (DUF2633) Family | 0.15 | 0.34 | 1 | 0.9 | 0.96 | 1 | 0.92 | 0.97 | 1 | 0.18 | 0.49 | 1 | 0.62 | 0.91 | 1 | 0.54 | 0.83 | 1 | 3e-04 | 2.5e-05 | 0 | 5.3e-05 | 0 | 0.00013 | 7.7e-05 | 0 | 2e-04 | 0.54 | 3 / 36 (8.3%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 9.B.160.1.16 | The Colicin V Production (CvpA) Family | 0.11 | 0.34 | 1 | 0.27 | 0.56 | 1 | 0.13 | 0.43 | 1 | 0.13 | 0.39 | 1 | 0.95 | 0.98 | 1 | 0.51 | 0.82 | 1 | 0.00018 | 2e-05 | 0 | 5.3e-05 | 0 | 0.00013 | 4.8e-05 | 0 | 0.00013 | -0.14 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 5.A.3.4.1 | The Prokaryotic Molybdopterin-containing Oxidoreductase (PMO) Family | 0.09 | 0.34 | 1 | 0.29 | 0.58 | 1 | 0.16 | 0.46 | 1 | 0.091 | 0.34 | 1 | 0.58 | 0.91 | 1 | 0.35 | 0.74 | 1 | 0.00059 | 0.00013 | 0 | 0.00033 | 0 | 0.00079 | 0.00029 | 0 | 0.00071 | -0.19 | 8 / 36 (22%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 8.9e-06 | 0 | 3e-05 | 2.6e-05 | 0 | 6.9e-05 | 1.5 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 5.A.3.4.3 | The Prokaryotic Molybdopterin-containing Oxidoreductase (PMO) Family | 0.6 | 0.66 | 1 | 0.34 | 0.63 | 1 | 0.69 | 0.86 | 1 | 0.62 | 0.82 | 1 | 0.58 | 0.91 | 1 | 0.96 | 1 | 1 | 0.00026 | 5.9e-05 | 0 | 5.3e-05 | 0 | 0.00013 | 0.00019 | 0 | 0.00051 | 1.8 | 8 / 36 (22%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 1.2e-05 | 0 | 2.8e-05 | 2.3e-05 | 0 | 4.1e-05 | 0.94 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 4.A.6.1.4 | The PTS Mannose-Fructose-Sorbose (Man) Family | 0.071 | 0.34 | 1 | 0.9 | 0.96 | 1 | 0.36 | 0.6 | 1 | 0.14 | 0.4 | 1 | 0.5 | 0.87 | 1 | 0.75 | 0.89 | 1 | 0.00026 | 3.7e-05 | 0 | 8.7e-05 | 0 | 0.00018 | 8.5e-05 | 0 | 0.00018 | -0.034 | 5 / 36 (14%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0 | 0 | 0 | 1.1e-05 | 0 | 3.6e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
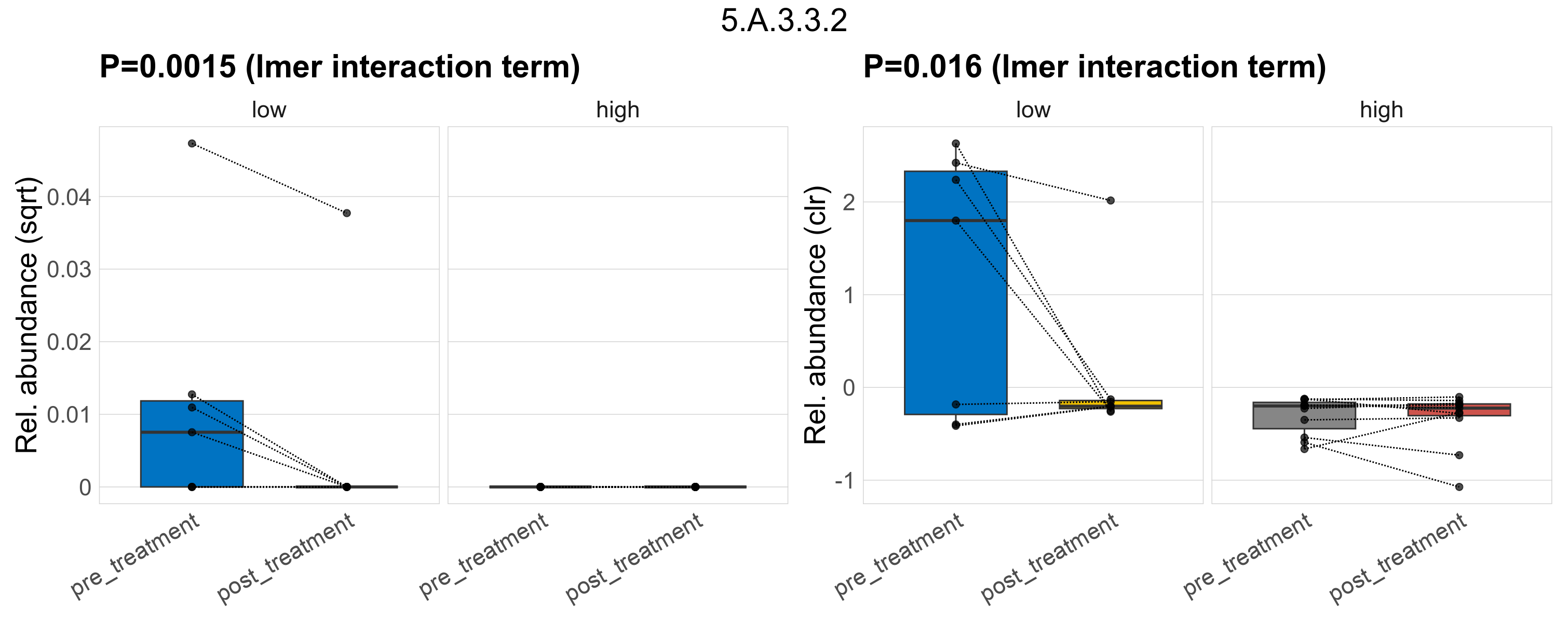
| 8.A.3.3.2 | The Cytoplasmic Membrane-Periplasmic Auxiliary-1 (MPA1) Protein with Cytoplasmic (C) Domain (MPA1-C or MPA1+C) Family | 0.49 | 0.56 | 1 | 0.59 | 0.78 | 1 | 0.51 | 0.75 | 1 | 0.44 | 0.71 | 1 | 0.81 | 0.95 | 1 | 0.16 | 0.67 | 1 | 0.00083 | 9.2e-05 | 0 | 0.00024 | 0 | 0.00062 | 0.00015 | 0 | 0.00041 | -0.68 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 5.2e-05 | 0 | 0.00015 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 2.A.3.1.1 | The Amino Acid-Polyamine-Organocation (APC) Family | 0.043 | 0.34 | 1 | 0.0066 | 0.22 | 1 | 0.063 | 0.37 | 1 | 0.026 | 0.27 | 1 | 0.035 | 0.62 | 1 | 0.29 | 0.73 | 1 | 0.00039 | 5.4e-05 | 0 | 0.00019 | 0 | 0.00044 | 7.7e-05 | 0 | 2e-04 | -1.3 | 5 / 36 (14%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 5.1e-06 | 0 | 1.7e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 3.D.1.9.1 | The H+ or Na+-translocating NADH Dehydrogenase (NDH) Family | 0.94 | 0.95 | 1 | 0.73 | 0.87 | 1 | 0.58 | 0.79 | 1 | 0.46 | 0.72 | 1 | 0.59 | 0.91 | 1 | 0.42 | 0.77 | 1 | 0.00013 | 2.5e-05 | 0 | 1.6e-05 | 0 | 3.1e-05 | 3.4e-05 | 0 | 7.9e-05 | 1.1 | 7 / 36 (19%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 2.9e-05 | 0 | 7.4e-05 | 2.2e-05 | 0 | 7.1e-05 | -0.4 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 8.A.5.1.5 | The Voltage-gated K+ Channel β-subunit (Kvβ) Family | 0.038 | 0.34 | 1 | 0.076 | 0.38 | 1 | 0.036 | 0.37 | 1 | 0.0069 | 0.17 | 1 | 0.11 | 0.64 | 1 | 0.045 | 0.58 | 1 | 0.00034 | 5.7e-05 | 0 | 0.00015 | 0 | 3e-04 | 0.00013 | 0 | 0.00033 | -0.21 | 6 / 36 (17%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 0 | 0 | 0 | 1.3e-05 | 0 | 3.5e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 2.A.121.1.1 | The Sulfate Transporter (CysZ) Family | 0.12 | 0.34 | 1 | 0.71 | 0.86 | 1 | 0.26 | 0.52 | 1 | 0.15 | 0.42 | 1 | 0.64 | 0.92 | 1 | 0.2 | 0.69 | 1 | 0.00032 | 4.4e-05 | 0 | 8.4e-05 | 0 | 0.00018 | 0.00012 | 0 | 0.00031 | 0.51 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 1.7e-05 | 0 | 3.9e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 8.A.2.1.3 | The Secretin Auxiliary Lipoprotein (SAL) Family | 0.21 | 0.35 | 1 | 0.19 | 0.49 | 1 | 0.61 | 0.82 | 1 | 0.6 | 0.82 | 1 | 0.89 | 0.98 | 1 | 0.35 | 0.74 | 1 | 0.00017 | 1.4e-05 | 0 | 5e-05 | 0 | 0.00013 | 1.9e-05 | 0 | 5.1e-05 | -1.4 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3e-06 | 0 | 9.9e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 1.B.33.2.4 | The Outer Membrane Protein Insertion Porin (Bam Complex) (OmpIP) Family | 0.036 | 0.34 | 1 | 0.04 | 0.28 | 1 | 0.039 | 0.37 | 1 | 0.043 | 0.27 | 1 | 0.19 | 0.72 | 1 | 0.12 | 0.65 | 1 | 0.00034 | 3.8e-05 | 0 | 0.00014 | 0 | 0.00031 | 3.9e-05 | 0 | 1e-04 | -1.8 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 1.1e-05 | 0 | 3.6e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.3.1.24 | The Amino Acid-Polyamine-Organocation (APC) Family | 0.22 | 0.35 | 1 | 0.079 | 0.38 | 1 | 0.056 | 0.37 | 1 | 0.71 | 0.87 | 1 | 0.45 | 0.86 | 1 | 0.24 | 0.7 | 1 | 0.00021 | 3.5e-05 | 0 | 8.9e-05 | 0 | 0.00022 | 4.8e-05 | 0 | 0.00013 | -0.89 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 1.1e-05 | 0 | 3.6e-05 | 1.6e-05 | 0 | 3.7e-05 | 0.54 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 3.A.1.120.6 | The ATP-binding Cassette (ABC) Superfamily | 0.32 | 0.41 | 1 | 0.89 | 0.96 | 1 | 0.66 | 0.84 | 1 | 0.8 | 0.92 | 1 | 0.59 | 0.91 | 1 | 0.82 | 0.93 | 1 | 0.00034 | 5.6e-05 | 0 | 0.00013 | 0 | 0.00036 | 0.00013 | 0 | 0.00033 | 0 | 6 / 36 (17%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 5.5e-06 | 0 | 1.2e-05 | 1.3e-05 | 0 | 3.3e-05 | 1.2 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 9.B.34.1.2 | The Kinase/Phosphatase/Cyclic-GMP Synthase/Cyclic di-GMP Hydrolase (KPSH) Family | 0.37 | 0.45 | 1 | 0.16 | 0.46 | 1 | 0.65 | 0.84 | 1 | 0.79 | 0.9 | 1 | 0.18 | 0.72 | 1 | 1 | 1 | 1 | 4e-04 | 4.5e-05 | 0 | 5e-05 | 0 | 0.00013 | 0.00016 | 0 | 0.00044 | 1.7 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 9.5e-06 | 0 | 2.3e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 2.A.19.1.1 | The Ca2+:Cation Antiporter (CaCA) Family | 0.14 | 0.34 | 1 | 0.53 | 0.76 | 1 | 0.46 | 0.71 | 1 | 0.28 | 0.6 | 1 | 0.58 | 0.91 | 1 | 0.54 | 0.83 | 1 | 0.00023 | 3.2e-05 | 0 | 6.8e-05 | 0 | 0.00013 | 7.7e-05 | 0 | 2e-04 | 0.18 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1e-05 | 0 | 3.4e-05 | 1.9 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 8.A.46.2.1 | The Glycan-binding Protein (SusD) Family | 0.49 | 0.56 | 1 | 0.34 | 0.63 | 1 | 0.46 | 0.71 | 1 | 0.13 | 0.39 | 1 | 0.23 | 0.76 | 1 | 0.31 | 0.73 | 1 | 6.5e-05 | 5.4e-06 | 0 | 0 | 0 | 0 | 8.8e-06 | 0 | 2.3e-05 | Inf | 3 / 36 (8.3%) | 0 / 7 (0%) | 1 / 7 (14%) | 0 | 0.143 | 6e-06 | 0 | 2e-05 | 6e-06 | 0 | 2e-05 | 0 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 1.B.17.3.10 | The Outer Membrane Factor (OMF) Family | 0.068 | 0.34 | 1 | 0.14 | 0.44 | 1 | 0.055 | 0.37 | 1 | 0.062 | 0.3 | 1 | 0.38 | 0.84 | 1 | 0.13 | 0.65 | 1 | 0.00036 | 5e-05 | 0 | 0.00014 | 0 | 0.00031 | 9.7e-05 | 0 | 0.00026 | -0.53 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 1.3e-05 | 0 | 3.5e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 2.A.1.2.65 | The Major Facilitator Superfamily (MFS) | 0.21 | 0.35 | 1 | 0.86 | 0.94 | 1 | 0.78 | 0.91 | 1 | 0.9 | 0.97 | 1 | 0.19 | 0.72 | 1 | 0.53 | 0.82 | 1 | 0.00025 | 4.1e-05 | 0 | 0.00012 | 0 | 0.00031 | 7.3e-05 | 0 | 0.00015 | -0.72 | 6 / 36 (17%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 3e-06 | 0 | 9.9e-06 | 9.5e-06 | 0 | 2.3e-05 | 1.7 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 2.A.1.2.15 | The Major Facilitator Superfamily (MFS) | 0.2 | 0.34 | 1 | 0.096 | 0.39 | 1 | 0.2 | 0.48 | 1 | 0.93 | 0.97 | 1 | 0.6 | 0.91 | 1 | 0.47 | 0.81 | 1 | 0.00028 | 3.1e-05 | 0 | 1e-04 | 0 | 0.00027 | 4.8e-05 | 0 | 0.00013 | -1.1 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3e-06 | 0 | 9.9e-06 | 2.7e-06 | 0 | 8.9e-06 | -0.15 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.1.1.2 | The Major Facilitator Superfamily (MFS) | 0.041 | 0.34 | 1 | 0.016 | 0.23 | 1 | 0.052 | 0.37 | 1 | 0.021 | 0.26 | 1 | 0.11 | 0.64 | 1 | 0.24 | 0.7 | 1 | 0.00078 | 6.5e-05 | 0 | 0.00026 | 0 | 0.00062 | 7.7e-05 | 0 | 2e-04 | -1.8 | 3 / 36 (8.3%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 1.E.14.1.4 | The CidA/LrgA Holin (CidA/LrgA Holin) Family | 0.91 | 0.93 | 1 | 0.23 | 0.51 | 1 | 0.05 | 0.37 | 1 | 0.02 | 0.26 | 1 | 0.19 | 0.72 | 1 | 0.013 | 0.46 | 1 | 0.00015 | 2e-05 | 0 | 1.7e-05 | 0 | 4.5e-05 | 6.8e-05 | 0 | 0.00018 | 2 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 1.2e-05 | 0 | 2.3e-05 | 0 | 0 | 0 | -Inf | 3 / 11 (27%) | 0 / 11 (0%) | 0.273 | 0 |
| 3.A.1.105.15 | The ATP-binding Cassette (ABC) Superfamily | 0.17 | 0.34 | 1 | 0.15 | 0.44 | 1 | 0.83 | 0.92 | 1 | 0.71 | 0.87 | 1 | 0.77 | 0.94 | 1 | 0.33 | 0.73 | 1 | 0.00034 | 8.5e-05 | 0 | 0.00026 | 0 | 0.00062 | 0.00013 | 0 | 3e-04 | -1 | 9 / 36 (25%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 2.4e-05 | 0 | 4.3e-05 | 5.4e-06 | 0 | 1.8e-05 | -2.2 | 3 / 11 (27%) | 1 / 11 (9.1%) | 0.273 | 0.0909 |
| 1.B.6.2.1 | The OmpA-OmpF Porin (OOP) Family | 0.055 | 0.34 | 1 | 0.15 | 0.45 | 1 | 0.26 | 0.52 | 1 | 0.085 | 0.33 | 1 | 0.42 | 0.86 | 1 | 0.61 | 0.85 | 1 | 2e-04 | 1.6e-05 | 0 | 4.5e-05 | 0 | 8.9e-05 | 3.9e-05 | 0 | 1e-04 | -0.21 | 3 / 36 (8.3%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 4.A.6.1.3 | The PTS Mannose-Fructose-Sorbose (Man) Family | 0.078 | 0.34 | 1 | 0.47 | 0.73 | 1 | 0.57 | 0.79 | 1 | 0.065 | 0.3 | 1 | 0.47 | 0.87 | 1 | 0.64 | 0.86 | 1 | 0.00037 | 3.1e-05 | 0 | 7e-05 | 0 | 0.00014 | 8.7e-05 | 0 | 0.00023 | 0.31 | 3 / 36 (8.3%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 2.A.40.7.7 | The Nucleobase/Ascorbate Transporter (NAT) or Nucleobase:Cation Symporter-2 (NCS2) Family | 0.26 | 0.37 | 1 | 0.45 | 0.72 | 1 | 0.56 | 0.78 | 1 | 0.45 | 0.71 | 1 | 0.61 | 0.91 | 1 | 0.6 | 0.85 | 1 | 0.00043 | 5.9e-05 | 0 | 1e-04 | 0 | 0.00027 | 0.00019 | 0 | 0.00051 | 0.93 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 2.7e-06 | 0 | 8.9e-06 | 0.054 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 1.A.30.2.1 | The H+- or Na+-translocating Bacterial Flagellar Motor/ExbBD Outer Membrane Transport Energizer (Mot/Exb) Superfamily | 0.018 | 0.34 | 1 | 0.0094 | 0.22 | 1 | 0.021 | 0.37 | 1 | 0.0053 | 0.16 | 1 | 0.054 | 0.62 | 1 | 0.074 | 0.6 | 1 | 0.00035 | 4.8e-05 | 0 | 0.00018 | 0 | 0.00035 | 6.8e-05 | 0 | 0.00018 | -1.4 | 5 / 36 (14%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.86.1.4 | The Autoinducer-2 Exporter (AI-2E) Family (Formerly the PerM Family. TC #9.B.22) | 0.27 | 0.37 | 1 | 0.98 | 1 | 1 | 0.4 | 0.65 | 1 | 0.87 | 0.95 | 1 | 0.14 | 0.66 | 1 | 0.021 | 0.52 | 1 | 0.00032 | 2.7e-05 | 0 | 6.7e-05 | 0 | 0.00018 | 6.8e-05 | 0 | 0.00018 | 0.021 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 1.B.17.3.5 | The Outer Membrane Factor (OMF) Family | 0.022 | 0.34 | 1 | 0.091 | 0.39 | 1 | 0.25 | 0.52 | 1 | 0.015 | 0.23 | 1 | 0.065 | 0.62 | 1 | 0.27 | 0.72 | 1 | 0.00028 | 3.9e-05 | 0 | 0.00011 | 0 | 0.00015 | 8.7e-05 | 0 | 0.00023 | -0.34 | 5 / 36 (14%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 5.A.3.4.2 | The Prokaryotic Molybdopterin-containing Oxidoreductase (PMO) Family | 0.13 | 0.34 | 1 | 0.17 | 0.47 | 1 | 0.062 | 0.37 | 1 | 0.17 | 0.45 | 1 | 0.4 | 0.84 | 1 | 0.14 | 0.65 | 1 | 0.00059 | 9.8e-05 | 0 | 0.00025 | 0 | 0.00062 | 0.00021 | 0 | 0.00056 | -0.25 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 3e-06 | 0 | 9.9e-06 | 2.1e-05 | 0 | 4.7e-05 | 2.8 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 4.A.1.1.3 | The PTS Glucose-Glucoside (Glc) Family | 0.5 | 0.56 | 1 | 0.44 | 0.72 | 1 | 0.32 | 0.59 | 1 | 0.7 | 0.87 | 1 | 0.95 | 0.98 | 1 | 0.79 | 0.91 | 1 | 0.00026 | 5e-05 | 0 | 8.6e-05 | 0 | 0.00022 | 7.7e-05 | 0 | 2e-04 | -0.16 | 7 / 36 (19%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.5e-05 | 0 | 7.2e-05 | 3.6e-05 | 0 | 1e-04 | 0.53 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 3.A.1.21.1 | The ATP-binding Cassette (ABC) Superfamily | 0.19 | 0.34 | 1 | 0.72 | 0.87 | 1 | 0.51 | 0.75 | 1 | 0.22 | 0.52 | 1 | 0.82 | 0.95 | 1 | 0.35 | 0.74 | 1 | 5e-04 | 7e-05 | 0 | 0.00014 | 0 | 0.00036 | 2e-04 | 0 | 0.00054 | 0.51 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 1.3e-05 | 0 | 3.5e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 9.B.55.1.1 | The 4 TMS DUF307/YccF (DUF307) Family | 0.16 | 0.34 | 1 | 0.13 | 0.42 | 1 | 0.37 | 0.63 | 1 | 0.87 | 0.95 | 1 | 0.9 | 0.98 | 1 | 0.44 | 0.79 | 1 | 0.00031 | 2.6e-05 | 0 | 1e-04 | 0 | 0.00027 | 2.9e-05 | 0 | 7.7e-05 | -1.8 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.A.1.1.1 | The Major Facilitator Superfamily (MFS) | 0.32 | 0.41 | 1 | 0.76 | 0.88 | 1 | 0.96 | 0.99 | 1 | 0.94 | 0.98 | 1 | 0.61 | 0.91 | 1 | 0.74 | 0.89 | 1 | 0.00093 | 1e-04 | 0 | 0.00022 | 0 | 0.00058 | 0.00026 | 0 | 0.00069 | 0.24 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 1e-05 | 0 | 3.4e-05 | 2.1e-05 | 0 | 6.8e-05 | 1.1 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.1.3.17 | The Major Facilitator Superfamily (MFS) | 0.15 | 0.34 | 1 | 0.33 | 0.62 | 1 | 0.21 | 0.49 | 1 | 0.3 | 0.62 | 1 | 0.58 | 0.91 | 1 | 0.37 | 0.74 | 1 | 0.00031 | 5.2e-05 | 0 | 0.00013 | 0 | 0.00031 | 0.00011 | 0 | 0.00028 | -0.24 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 5.1e-06 | 0 | 1.7e-05 | 1.6e-05 | 0 | 3.7e-05 | 1.6 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 2.A.7.22.1 | The Drug/Metabolite Transporter (DMT) Superfamily | 0.11 | 0.34 | 1 | 0.22 | 0.51 | 1 | 0.1 | 0.42 | 1 | 0.22 | 0.52 | 1 | 0.75 | 0.94 | 1 | 0.34 | 0.73 | 1 | 0.00046 | 5.1e-05 | 0 | 0.00015 | 0 | 4e-04 | 7.7e-05 | 0 | 2e-04 | -0.96 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.1e-05 | 0 | 6.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 8.A.1.6.2 | The Membrane Fusion Protein (MFP) Family | 0.8 | 0.84 | 1 | 0.18 | 0.48 | 1 | 0.082 | 0.39 | 1 | 0.11 | 0.37 | 1 | 0.053 | 0.62 | 1 | 0.011 | 0.46 | 1 | 0.00023 | 3.2e-05 | 0 | 1.7e-05 | 0 | 4.5e-05 | 7.7e-05 | 0 | 2e-04 | 2.2 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3.5e-05 | 0 | 0.00011 | 1e-05 | 0 | 3.4e-05 | -1.8 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 1.A.16.3.1 | The Formate-Nitrite Transporter (FNT) Family | 0.15 | 0.34 | 1 | 0.29 | 0.58 | 1 | 0.091 | 0.41 | 1 | 0.39 | 0.66 | 1 | 0.8 | 0.94 | 1 | 0.33 | 0.73 | 1 | 0.00044 | 4.8e-05 | 0 | 0.00015 | 0 | 4e-04 | 7.7e-05 | 0 | 2e-04 | -0.96 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 1.3e-05 | 0 | 3.5e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 2.A.20.1.1 | The Inorganic Phosphate Transporter (PiT) Family | 0.23 | 0.35 | 1 | 0.51 | 0.75 | 1 | 0.73 | 0.88 | 1 | 0.82 | 0.93 | 1 | 0.51 | 0.87 | 1 | 0.39 | 0.75 | 1 | 6e-04 | 6.7e-05 | 0 | 0.00019 | 0 | 0.00049 | 0.00015 | 0 | 0.00038 | -0.34 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 5.1e-06 | 0 | 1.7e-05 | 2.7e-06 | 0 | 8.9e-06 | -0.92 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 1.B.1.1.1 | The General Bacterial Porin (GBP) Family | 0.22 | 0.35 | 1 | 0.43 | 0.7 | 1 | 0.18 | 0.48 | 1 | 0.86 | 0.95 | 1 | 0.86 | 0.96 | 1 | 0.49 | 0.82 | 1 | 3e-04 | 4.1e-05 | 0 | 0.00012 | 0 | 0.00031 | 5.8e-05 | 0 | 0.00015 | -1 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 2.1e-05 | 0 | 4.7e-05 | 3 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 1.A.77.3.11 | The Mg2+/Ca2+ Uniporter (MCU) Family | 0.18 | 0.34 | 1 | 0.6 | 0.78 | 1 | 0.24 | 0.52 | 1 | 0.45 | 0.71 | 1 | 0.41 | 0.85 | 1 | 0.85 | 0.94 | 1 | 0.00037 | 3.1e-05 | 0 | 8.4e-05 | 0 | 0.00022 | 6.8e-05 | 0 | 0.00018 | -0.3 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 3.A.1.2.2 | The ATP-binding Cassette (ABC) Superfamily | 0.11 | 0.34 | 1 | 0.18 | 0.47 | 1 | 0.044 | 0.37 | 1 | 0.11 | 0.37 | 1 | 0.59 | 0.91 | 1 | 0.12 | 0.65 | 1 | 0.00081 | 0.00011 | 0 | 0.00033 | 0 | 0.00084 | 2e-04 | 0 | 0.00054 | -0.72 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.9e-05 | 0 | 6.8e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 3.A.2.1.1 | The H+- or Na+-translocating F-type. V-type and A-type ATPase (F-ATPase) Superfamily | 0.058 | 0.34 | 1 | 0.04 | 0.28 | 1 | 0.059 | 0.37 | 1 | 0.01 | 0.22 | 1 | 0.016 | 0.5 | 1 | 0.034 | 0.58 | 1 | 6e-04 | 0.00023 | 0 | 0.00054 | 8.1e-05 | 0.0012 | 0.00058 | 0 | 0.0015 | 0.1 | 14 / 36 (39%) | 6 / 7 (86%) | 2 / 7 (29%) | 0.857 | 0.286 | 1.9e-05 | 0 | 3.6e-05 | 3.1e-05 | 0 | 5.5e-05 | 0.71 | 3 / 11 (27%) | 3 / 11 (27%) | 0.273 | 0.273 |
| 3.A.1.3.26 | The ATP-binding Cassette (ABC) Superfamily | 0.11 | 0.34 | 1 | 0.32 | 0.61 | 1 | 0.39 | 0.64 | 1 | 0.14 | 0.4 | 1 | 0.98 | 0.99 | 1 | 0.99 | 1 | 1 | 0.00076 | 0.00015 | 0 | 0.00042 | 0 | 0.0011 | 0.00032 | 0 | 0.00079 | -0.39 | 7 / 36 (19%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 7.7e-06 | 0 | 2.6e-05 | 1.1e-05 | 0 | 3.6e-05 | 0.51 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 1.B.14.1.1 | The Outer Membrane Receptor (OMR) Family | 0.14 | 0.34 | 1 | 0.63 | 0.8 | 1 | 0.99 | 0.99 | 1 | 0.094 | 0.34 | 1 | 0.34 | 0.83 | 1 | 0.82 | 0.93 | 1 | 0.00057 | 7.9e-05 | 0 | 0.00016 | 0 | 0.00035 | 0.00024 | 0 | 0.00064 | 0.58 | 5 / 36 (14%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 6e-06 | 0 | 2e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.A.7.26.1 | The Drug/Metabolite Transporter (DMT) Superfamily | 0.27 | 0.38 | 1 | 0.56 | 0.77 | 1 | 0.43 | 0.69 | 1 | 0.76 | 0.89 | 1 | 0.75 | 0.94 | 1 | 1 | 1 | 1 | 0.00017 | 1.9e-05 | 0 | 5e-05 | 0 | 0.00013 | 2.9e-05 | 0 | 7.7e-05 | -0.79 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1e-05 | 0 | 3.4e-05 | 1.9 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.1.33.2 | The Major Facilitator Superfamily (MFS) | 0.85 | 0.88 | 1 | 0.66 | 0.82 | 1 | 0.94 | 0.98 | 1 | 0.76 | 0.89 | 1 | 0.62 | 0.91 | 1 | 0.97 | 1 | 1 | 0.00035 | 8.9e-05 | 0 | 6.4e-05 | 0 | 0.00013 | 0.00012 | 0 | 0.00028 | 0.91 | 9 / 36 (25%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 6.2e-05 | 0 | 0.00015 | 0.00011 | 0 | 0.00029 | 0.83 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 1.A.23.4.5 | The Small Conductance Mechanosensitive Ion Channel (MscS) Family | 0.033 | 0.34 | 1 | 0.067 | 0.36 | 1 | 0.069 | 0.37 | 1 | 0.031 | 0.27 | 1 | 0.18 | 0.72 | 1 | 0.16 | 0.67 | 1 | 0.00023 | 5.8e-05 | 0 | 0.00017 | 1.6e-05 | 0.00035 | 9.1e-05 | 0 | 0.00023 | -0.9 | 9 / 36 (25%) | 4 / 7 (57%) | 2 / 7 (29%) | 0.571 | 0.286 | 7.7e-06 | 0 | 2.6e-05 | 1.8e-05 | 0 | 4.5e-05 | 1.2 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 1.B.25.1.5 | The Outer Membrane Porin (Opr) Family | 0.18 | 0.34 | 1 | 0.45 | 0.72 | 1 | 0.99 | 0.99 | 1 | 0.41 | 0.68 | 1 | 0.57 | 0.91 | 1 | 0.76 | 0.9 | 1 | 0.00031 | 3.5e-05 | 0 | 7.9e-05 | 0 | 0.00018 | 8.7e-05 | 0 | 0.00023 | 0.14 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 7.7e-06 | 0 | 2.6e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 3.A.1.25.6 | The ATP-binding Cassette (ABC) Superfamily | 0.21 | 0.35 | 1 | 0.55 | 0.76 | 1 | 0.21 | 0.48 | 1 | 0.25 | 0.56 | 1 | 0.62 | 0.91 | 1 | 0.39 | 0.75 | 1 | 0.00068 | 0.00044 | 2e-04 | 2e-04 | 8.1e-05 | 0.00034 | 0.00046 | 0.00024 | 8e-04 | 1.2 | 23 / 36 (64%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 0.00079 | 0.00053 | 0.0016 | 0.00022 | 0.00022 | 0.00022 | -1.8 | 7 / 11 (64%) | 7 / 11 (64%) | 0.636 | 0.636 |
| 3.A.1.129.1 | The ATP-binding Cassette (ABC) Superfamily | 0.25 | 0.36 | 1 | 0.77 | 0.88 | 1 | 0.39 | 0.64 | 1 | 0.71 | 0.87 | 1 | 0.44 | 0.86 | 1 | 0.83 | 0.93 | 1 | 0.00056 | 0.00012 | 0 | 0.00031 | 0 | 8e-04 | 0.00027 | 0 | 0.00069 | -0.2 | 8 / 36 (22%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 2.2e-05 | 0 | 7.3e-05 | 2e-05 | 0 | 3.9e-05 | -0.14 | 1 / 11 (9.1%) | 3 / 11 (27%) | 0.0909 | 0.273 |
| 2.A.1.2.60 | The Major Facilitator Superfamily (MFS) | 0.08 | 0.34 | 1 | 0.079 | 0.38 | 1 | 0.33 | 0.59 | 1 | 0.19 | 0.5 | 1 | 0.68 | 0.92 | 1 | 0.55 | 0.83 | 1 | 0.00021 | 4.8e-05 | 0 | 0.00015 | 0 | 0.00035 | 8.3e-05 | 0 | 0.00018 | -0.85 | 8 / 36 (22%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 5.5e-06 | 0 | 1.2e-05 | 2.7e-06 | 0 | 8.9e-06 | -1 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 1.B.14.14.1 | The Outer Membrane Receptor (OMR) Family | 0.41 | 0.49 | 1 | 0.57 | 0.77 | 1 | 0.72 | 0.88 | 1 | 0.15 | 0.41 | 1 | 0.77 | 0.94 | 1 | 0.64 | 0.86 | 1 | 0.00013 | 1.4e-05 | 0 | 8.1e-06 | 0 | 2.1e-05 | 0 | 0 | 0 | -Inf | 4 / 36 (11%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 3.9e-05 | 0 | 0.00011 | 3e-06 | 0 | 1e-05 | -3.7 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 9.B.226.1.8 | The Uncharacterized 9 or 10 TMS Protein (U-TMP) Family | 0.46 | 0.53 | 1 | 0.51 | 0.75 | 1 | 0.85 | 0.93 | 1 | 0.19 | 0.49 | 1 | 0.79 | 0.94 | 1 | 0.96 | 1 | 1 | 0.00089 | 0.00071 | 0.00044 | 0.00044 | 0.00023 | 0.00054 | 0.00068 | 0.00096 | 0.00065 | 0.63 | 29 / 36 (81%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 0.00081 | 0.00049 | 0.0015 | 0.00082 | 0.00044 | 0.00079 | 0.018 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 9.B.22.1.8 | The Leukotoxin Secretion Morphogenesis Protein C (MorC) Family | 0.017 | 0.34 | 1 | 0.2 | 0.49 | 1 | 0.1 | 0.42 | 1 | 0.093 | 0.34 | 1 | 0.59 | 0.91 | 1 | 0.25 | 0.71 | 1 | 0.0013 | 0.00033 | 0 | 0.00087 | 0 | 0.0011 | 0.00049 | 0 | 0.00089 | -0.83 | 9 / 36 (25%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 2.2e-05 | 0 | 7.3e-05 | 0.00019 | 0 | 0.00053 | 3.1 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 1.B.75.1.3 | The DUF481 Putative Beta Barrel Porin (DUF481) Family | 0.26 | 0.37 | 1 | 0.22 | 0.51 | 1 | 0.34 | 0.6 | 1 | 0.99 | 0.99 | 1 | 0.062 | 0.62 | 1 | 0.12 | 0.65 | 1 | 0.00026 | 3.6e-05 | 0 | 8.4e-05 | 0 | 0.00022 | 9.4e-05 | 0 | 2e-04 | 0.16 | 5 / 36 (14%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 2.6e-06 | 0 | 8.5e-06 | 2.7e-06 | 0 | 8.9e-06 | 0.054 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.40.4.3 | The Nucleobase/Ascorbate Transporter (NAT) or Nucleobase:Cation Symporter-2 (NCS2) Family | 0.12 | 0.34 | 1 | 0.58 | 0.77 | 1 | 0.98 | 0.99 | 1 | 0.29 | 0.6 | 1 | 0.34 | 0.83 | 1 | 0.75 | 0.89 | 1 | 0.00028 | 3.9e-05 | 0 | 8.9e-05 | 0 | 0.00022 | 9.8e-05 | 0 | 0.00018 | 0.14 | 5 / 36 (14%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0 | 0 | 0 | 1e-05 | 0 | 3.4e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.1.4.3 | The Major Facilitator Superfamily (MFS) | 0.25 | 0.36 | 1 | 0.83 | 0.92 | 1 | 0.53 | 0.76 | 1 | 0.61 | 0.82 | 1 | 0.79 | 0.94 | 1 | 0.54 | 0.82 | 1 | 0.00027 | 4.6e-05 | 0 | 8.6e-05 | 0 | 0.00022 | 0.00013 | 0 | 0.00033 | 0.6 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 3e-06 | 0 | 9.9e-06 | 1.2e-05 | 0 | 2.6e-05 | 2 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 5.A.3.1.1 | The Prokaryotic Molybdopterin-containing Oxidoreductase (PMO) Family | 0.24 | 0.36 | 1 | 0.61 | 0.79 | 1 | 0.34 | 0.6 | 1 | 0.54 | 0.77 | 1 | 0.78 | 0.94 | 1 | 0.4 | 0.75 | 1 | 0.00036 | 8e-05 | 0 | 0.00015 | 0 | 0.00035 | 0.00023 | 0 | 0.00056 | 0.62 | 8 / 36 (22%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 1.6e-05 | 0 | 3.6e-05 | 2.7e-06 | 0 | 8.9e-06 | -2.6 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 9.B.174.1.2 | The Two Tunnel Gated C-terminal Processing Protease (CTP) Family | 0.33 | 0.42 | 1 | 0.39 | 0.67 | 1 | 0.72 | 0.88 | 1 | 0.92 | 0.97 | 1 | 0.38 | 0.84 | 1 | 0.65 | 0.86 | 1 | 5e-04 | 5.6e-05 | 0 | 1e-04 | 0 | 0.00027 | 0.00016 | 0 | 0.00044 | 0.68 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1e-05 | 0 | 3.4e-05 | 1.9 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.17.1.2 | The Proton-dependent Oligopeptide Transporter (POT/PTR) Family | 0.16 | 0.34 | 1 | 0.37 | 0.65 | 1 | 0.092 | 0.41 | 1 | 0.54 | 0.77 | 1 | 0.85 | 0.96 | 1 | 0.27 | 0.72 | 1 | 0.00026 | 2.9e-05 | 0 | 8.4e-05 | 0 | 0.00022 | 3.9e-05 | 0 | 1e-04 | -1.1 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 1.6e-05 | 0 | 3.7e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 3.A.1.125.1 | The ATP-binding Cassette (ABC) Superfamily | 0.041 | 0.34 | 1 | 0.0052 | 0.22 | 1 | 0.031 | 0.37 | 1 | 0.073 | 0.31 | 1 | 0.035 | 0.62 | 1 | 0.15 | 0.66 | 1 | 0.00043 | 0.00012 | 0 | 0.00042 | 1.6e-05 | 0.00091 | 0.00014 | 0 | 0.00036 | -1.6 | 10 / 36 (28%) | 4 / 7 (57%) | 1 / 7 (14%) | 0.571 | 0.143 | 1.9e-05 | 0 | 3.8e-05 | 1.7e-05 | 0 | 4e-05 | -0.16 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 3.A.1.5.42 | The ATP-binding Cassette (ABC) Superfamily | 0.4 | 0.47 | 1 | 0.82 | 0.91 | 1 | 0.4 | 0.65 | 1 | 0.68 | 0.86 | 1 | 0.63 | 0.91 | 1 | 0.24 | 0.7 | 1 | 0.00033 | 7.4e-05 | 0 | 0.00016 | 0 | 4e-04 | 0.00018 | 0 | 0.00046 | 0.17 | 8 / 36 (22%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 1.9e-05 | 0 | 3.8e-05 | 1.1e-05 | 0 | 3.6e-05 | -0.79 | 3 / 11 (27%) | 1 / 11 (9.1%) | 0.273 | 0.0909 |
| 9.B.186.1.1 | The Putative Lipoprotein Supressor of a ts bamD mutant. YiaD (YiaD) Family | 0.26 | 0.37 | 1 | 0.2 | 0.49 | 1 | 0.68 | 0.85 | 1 | 0.97 | 0.99 | 1 | 0.077 | 0.62 | 1 | 0.39 | 0.75 | 1 | 0.00018 | 2e-05 | 0 | 3.4e-05 | 0 | 8.9e-05 | 5.4e-05 | 0 | 1e-04 | 0.67 | 4 / 36 (11%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 0 | 0 | 0 | 1e-05 | 0 | 3.4e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.109.1.7 | The Tellurium Ion Resistance (TerC) Family | 0.53 | 0.6 | 1 | 0.2 | 0.49 | 1 | 0.087 | 0.4 | 1 | 0.46 | 0.72 | 1 | 0.31 | 0.8 | 1 | 0.052 | 0.58 | 1 | 0.00062 | 6.9e-05 | 0 | 1e-04 | 0 | 0.00027 | 0.00023 | 0 | 0.00061 | 1.2 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 1.4e-05 | 0 | 3.7e-05 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 1.B.11.2.2 | The Outer Membrane Fimbrial Usher Porin (FUP) Family | 0.09 | 0.34 | 1 | 0.12 | 0.42 | 1 | 0.037 | 0.37 | 1 | 0.86 | 0.95 | 1 | 0.22 | 0.74 | 1 | 0.058 | 0.58 | 1 | 7.4e-05 | 8.2e-06 | 0 | 1.9e-05 | 0 | 4.5e-05 | 0 | 0 | 0 | -Inf | 4 / 36 (11%) | 2 / 7 (29%) | 0 / 7 (0%) | 0.286 | 0 | 0 | 0 | 0 | 1.5e-05 | 0 | 3.3e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 2.A.1.52.2 | The Major Facilitator Superfamily (MFS) | 0.25 | 0.36 | 1 | 0.2 | 0.49 | 1 | 0.83 | 0.92 | 1 | 0.72 | 0.87 | 1 | 0.78 | 0.94 | 1 | 0.16 | 0.67 | 1 | 0.00037 | 4.1e-05 | 0 | 0.00013 | 0 | 0.00036 | 6.8e-05 | 0 | 0.00018 | -0.93 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 5.5e-06 | 0 | 1.2e-05 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 9.B.168.1.2 | The DUF3078 Putative Porin (DUF3078) Family | 0.17 | 0.34 | 1 | 0.37 | 0.65 | 1 | 0.16 | 0.46 | 1 | 0.056 | 0.29 | 1 | 0.25 | 0.77 | 1 | 0.13 | 0.65 | 1 | 8.6e-05 | 9.6e-06 | 0 | 0 | 0 | 0 | 1.3e-05 | 0 | 3.5e-05 | Inf | 4 / 36 (11%) | 0 / 7 (0%) | 1 / 7 (14%) | 0 | 0.143 | 2e-05 | 0 | 4.5e-05 | 3e-06 | 0 | 1e-05 | -2.7 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 2.A.8.1.3 | The Gluconate:H+ Symporter (GntP) Family | 0.13 | 0.34 | 1 | 0.44 | 0.71 | 1 | 0.54 | 0.76 | 1 | 0.13 | 0.39 | 1 | 0.43 | 0.86 | 1 | 0.6 | 0.85 | 1 | 0.00037 | 5.1e-05 | 0 | 0.00012 | 0 | 0.00026 | 0.00014 | 0 | 0.00036 | 0.22 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 2.7e-06 | 0 | 8.9e-06 | 0.054 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.C.1.4.3 | The TonB-ExbB-ExbD/TolA-TolQ-TolR Outer Membrane Receptor Energizers and Stabilizers (TonB/TolA) Family | 0.81 | 0.85 | 1 | 0.53 | 0.76 | 1 | 0.16 | 0.45 | 1 | 0.55 | 0.78 | 1 | 0.69 | 0.93 | 1 | 0.28 | 0.73 | 1 | 0.00019 | 2.6e-05 | 0 | 1.6e-05 | 0 | 4.3e-05 | 0 | 0 | 0 | -Inf | 5 / 36 (14%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 1.1e-05 | 0 | 3.6e-05 | 6.5e-05 | 0 | 0.00014 | 2.6 | 1 / 11 (9.1%) | 3 / 11 (27%) | 0.0909 | 0.273 |
| 2.A.14.1.2 | The Lactate Permease (LctP) Family | 0.17 | 0.34 | 1 | 0.51 | 0.75 | 1 | 0.21 | 0.48 | 1 | 0.37 | 0.65 | 1 | 0.52 | 0.87 | 1 | 0.74 | 0.89 | 1 | 0.00065 | 5.5e-05 | 0 | 0.00015 | 0 | 4e-04 | 0.00012 | 0 | 0.00031 | -0.32 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 8.1e-06 | 0 | 2.7e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.21.3.21 | The Solute:Sodium Symporter (SSS) Family | 0.064 | 0.34 | 1 | 0.0076 | 0.22 | 1 | 0.031 | 0.37 | 1 | 0.0095 | 0.21 | 1 | 0.06 | 0.62 | 1 | 0.16 | 0.67 | 1 | 0.00085 | 9.4e-05 | 0 | 0.00027 | 0 | 0.00066 | 0.00021 | 0 | 0.00056 | -0.36 | 4 / 36 (11%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 8.A.1.7.1 | The Membrane Fusion Protein (MFP) Family | 0.14 | 0.34 | 1 | 0.054 | 0.31 | 1 | 0.28 | 0.54 | 1 | 0.32 | 0.63 | 1 | 0.7 | 0.93 | 1 | 0.71 | 0.88 | 1 | 0.00043 | 4.8e-05 | 0 | 0.00015 | 0 | 4e-04 | 8.7e-05 | 0 | 0.00023 | -0.79 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 3e-06 | 0 | 9.9e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 1.A.1.5.25 | The Voltage-gated Ion Channel (VIC) Superfamily | 0.58 | 0.64 | 1 | 0.51 | 0.75 | 1 | 0.52 | 0.75 | 1 | 0.32 | 0.63 | 1 | 0.46 | 0.87 | 1 | 0.36 | 0.74 | 1 | 0.00067 | 0.00048 | 0.00039 | 0.00032 | 4e-04 | 0.00032 | 0.00044 | 0.00036 | 0.00046 | 0.46 | 26 / 36 (72%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.00051 | 0.00037 | 0.00081 | 0.00059 | 0.00057 | 0.00045 | 0.21 | 8 / 11 (73%) | 10 / 11 (91%) | 0.727 | 0.909 |
| 1.B.6.1.21 | The OmpA-OmpF Porin (OOP) Family | 0.055 | 0.34 | 1 | 0.023 | 0.24 | 1 | 0.067 | 0.37 | 1 | 0.037 | 0.27 | 1 | 0.25 | 0.77 | 1 | 0.44 | 0.79 | 1 | 0.00036 | 3e-05 | 0 | 9.6e-05 | 0 | 0.00022 | 5.8e-05 | 0 | 0.00015 | -0.73 | 3 / 36 (8.3%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 3.A.1.1.3 | The ATP-binding Cassette (ABC) Superfamily | 0.61 | 0.66 | 1 | 0.13 | 0.42 | 1 | 0.12 | 0.43 | 1 | 0.28 | 0.6 | 1 | 0.029 | 0.62 | 1 | 0.0079 | 0.46 | 1 | 0.00029 | 4e-05 | 0 | 5e-05 | 0 | 0.00013 | 0.00014 | 0 | 0.00036 | 1.5 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 8.5e-06 | 0 | 2.1e-05 | 5.4e-06 | 0 | 1.8e-05 | -0.65 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 1.B.76.1.8 | The Copper Resistance Putative Porin (CopB) Family | 0.22 | 0.35 | 1 | 0.16 | 0.45 | 1 | 0.63 | 0.83 | 1 | 0.85 | 0.94 | 1 | 0.9 | 0.98 | 1 | 0.19 | 0.68 | 1 | 6e-04 | 6.7e-05 | 0 | 0.00024 | 0 | 0.00062 | 9.7e-05 | 0 | 0.00026 | -1.3 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 8.1e-06 | 0 | 1.9e-05 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 2.A.8.1.7 | The Gluconate:H+ Symporter (GntP) Family | 0.1 | 0.34 | 1 | 0.1 | 0.39 | 1 | 0.19 | 0.48 | 1 | 0.099 | 0.35 | 1 | 0.3 | 0.8 | 1 | 0.47 | 0.81 | 1 | 0.00047 | 6.6e-05 | 0 | 0.00019 | 0 | 0.00044 | 0.00015 | 0 | 0.00038 | -0.34 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 2.7e-06 | 0 | 8.9e-06 | 0.054 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.64.1.1 | The Twin Arginine Targeting (Tat) Family | 0.2 | 0.34 | 1 | 0.7 | 0.85 | 1 | 0.45 | 0.71 | 1 | 0.45 | 0.71 | 1 | 1 | 1 | 1 | 0.78 | 0.91 | 1 | 0.00062 | 8.6e-05 | 0 | 2e-04 | 0 | 0.00053 | 2e-04 | 0 | 0.00054 | 0 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 3e-06 | 0 | 9.9e-06 | 2.1e-05 | 0 | 6.8e-05 | 2.8 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.A.1.27.3 | The ATP-binding Cassette (ABC) Superfamily | 0.12 | 0.34 | 1 | 0.4 | 0.68 | 1 | 0.7 | 0.86 | 1 | 0.28 | 0.6 | 1 | 0.76 | 0.94 | 1 | 0.82 | 0.93 | 1 | 0.00083 | 0.00021 | 0 | 0.00056 | 0 | 0.0014 | 0.00044 | 0 | 0.0011 | -0.35 | 9 / 36 (25%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 1.8e-05 | 0 | 3.5e-05 | 1.9e-05 | 0 | 6.2e-05 | 0.078 | 3 / 11 (27%) | 1 / 11 (9.1%) | 0.273 | 0.0909 |
| 2.A.47.3.2 | The Divalent Anion:Na+ Symporter (DASS) Family | 0.038 | 0.34 | 1 | 0.032 | 0.27 | 1 | 0.031 | 0.37 | 1 | 0.042 | 0.27 | 1 | 0.13 | 0.66 | 1 | 0.086 | 0.61 | 1 | 0.00036 | 6.9e-05 | 0 | 0.00024 | 0 | 0.00053 | 7.7e-05 | 0 | 2e-04 | -1.6 | 7 / 36 (19%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 2.3e-05 | 0 | 6.8e-05 | 3.1 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 9.B.118.1.1 | The 11 or 12 TMS YhfT (YhfT) Family | 0.024 | 0.34 | 1 | 0.018 | 0.23 | 1 | 0.02 | 0.37 | 1 | 0.012 | 0.22 | 1 | 0.062 | 0.62 | 1 | 0.051 | 0.58 | 1 | 0.00036 | 6.9e-05 | 0 | 0.00022 | 0 | 0.00043 | 0.00011 | 0 | 0.00028 | -1 | 7 / 36 (19%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1.6e-05 | 0 | 3.7e-05 | 2.6 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 4.A.6.1.7 | The PTS Mannose-Fructose-Sorbose (Man) Family | 0.23 | 0.35 | 1 | 0.32 | 0.61 | 1 | 0.17 | 0.46 | 1 | 0.067 | 0.3 | 1 | 0.066 | 0.62 | 1 | 0.064 | 0.58 | 1 | 0.00039 | 0.00027 | 0.00012 | 2e-04 | 0 | 4e-04 | 0.00026 | 0.00013 | 0.00029 | 0.38 | 25 / 36 (69%) | 3 / 7 (43%) | 6 / 7 (86%) | 0.429 | 0.857 | 0.00042 | 0.00019 | 0.00079 | 0.00017 | 8.8e-05 | 0.00021 | -1.3 | 8 / 11 (73%) | 8 / 11 (73%) | 0.727 | 0.727 |
| 2.A.1.7.18 | The Major Facilitator Superfamily (MFS) | 0.48 | 0.55 | 1 | 0.91 | 0.96 | 1 | 0.66 | 0.84 | 1 | 0.79 | 0.9 | 1 | 0.9 | 0.98 | 1 | 0.7 | 0.88 | 1 | 0.00023 | 1e-04 | 0 | 9.5e-05 | 3.2e-05 | 0.00012 | 0.00013 | 2.7e-05 | 0.00024 | 0.45 | 16 / 36 (44%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 6.7e-05 | 0 | 0.00013 | 0.00012 | 0 | 0.00029 | 0.84 | 3 / 11 (27%) | 5 / 11 (45%) | 0.273 | 0.455 |
| 2.A.8.1.4 | The Gluconate:H+ Symporter (GntP) Family | 0.28 | 0.38 | 1 | 0.99 | 1 | 1 | 0.15 | 0.44 | 1 | 0.89 | 0.96 | 1 | 0.69 | 0.93 | 1 | 0.23 | 0.7 | 1 | 3e-04 | 5.8e-05 | 0 | 0.00012 | 0 | 0.00031 | 0.00012 | 0 | 0.00031 | 0 | 7 / 36 (19%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 3.9e-05 | 0 | 7e-05 | 3.9 | 1 / 11 (9.1%) | 4 / 11 (36%) | 0.0909 | 0.364 |
| 2.A.69.4.6 | The Auxin Efflux Carrier (AEC) Family | 0.41 | 0.49 | 1 | 0.75 | 0.88 | 1 | 0.91 | 0.96 | 1 | 0.15 | 0.41 | 1 | 0.95 | 0.98 | 1 | 0.71 | 0.88 | 1 | 0.00094 | 0.00076 | 0.00057 | 0.00051 | 0.00034 | 0.00057 | 0.00068 | 0.00036 | 0.00076 | 0.42 | 29 / 36 (81%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 0.00084 | 0.00052 | 0.0011 | 0.00089 | 0.00091 | 0.00075 | 0.083 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 2.A.102.3.1 | The 4-Toluene Sulfonate Uptake Permease (TSUP) Family | 0.72 | 0.77 | 1 | 0.36 | 0.65 | 1 | 0.15 | 0.45 | 1 | 0.018 | 0.26 | 1 | 0.047 | 0.62 | 1 | 0.0084 | 0.46 | 1 | 8.2e-05 | 1.1e-05 | 0 | 1.7e-05 | 0 | 4.5e-05 | 2.9e-05 | 0 | 7.7e-05 | 0.77 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 5.5e-06 | 0 | 1.2e-05 | 2.7e-06 | 0 | 8.9e-06 | -1 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 2.A.8.1.8 | The Gluconate:H+ Symporter (GntP) Family | 0.2 | 0.34 | 1 | 0.2 | 0.49 | 1 | 0.62 | 0.83 | 1 | 0.85 | 0.94 | 1 | 0.69 | 0.93 | 1 | 0.31 | 0.73 | 1 | 0.00044 | 3.7e-05 | 0 | 0.00012 | 0 | 0.00031 | 6.8e-05 | 0 | 0.00018 | -0.82 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3e-06 | 0 | 9.9e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 9.B.121.1.1 | The AsmA (AsmA) Family | 0.24 | 0.36 | 1 | 0.3 | 0.59 | 1 | 0.76 | 0.9 | 1 | 0.51 | 0.75 | 1 | 0.42 | 0.86 | 1 | 0.9 | 0.97 | 1 | 0.00043 | 8.4e-05 | 0 | 2e-04 | 0 | 0.00048 | 0.00016 | 0 | 0.00044 | -0.32 | 7 / 36 (19%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 3.5e-05 | 0 | 0.00011 | 8.1e-06 | 0 | 2.7e-05 | -2.1 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 3.A.1.23.1 | The ATP-binding Cassette (ABC) Superfamily | 0.86 | 0.89 | 1 | 0.87 | 0.94 | 1 | 0.41 | 0.67 | 1 | 0.9 | 0.97 | 1 | 0.72 | 0.94 | 1 | 0.75 | 0.89 | 1 | 0.00049 | 0.00029 | 7.3e-05 | 0.00025 | 0.00014 | 0.00037 | 0.00025 | 0.00014 | 3e-04 | 0 | 21 / 36 (58%) | 6 / 7 (86%) | 4 / 7 (57%) | 0.857 | 0.571 | 0.00053 | 0 | 0.001 | 9.1e-05 | 2.9e-05 | 0.00013 | -2.5 | 5 / 11 (45%) | 6 / 11 (55%) | 0.455 | 0.545 |
| 4.B.1.1.1 | The Nicotinamide Ribonucleoside (NR) Uptake Permease (PnuC) Family | 0.11 | 0.34 | 1 | 0.09 | 0.39 | 1 | 0.12 | 0.43 | 1 | 0.27 | 0.6 | 1 | 0.58 | 0.91 | 1 | 0.56 | 0.83 | 1 | 0.00054 | 7.5e-05 | 0 | 0.00026 | 0 | 0.00067 | 0.00011 | 0 | 0.00028 | -1.2 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 3e-06 | 0 | 9.9e-06 | 1e-05 | 0 | 3.4e-05 | 1.7 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 9.B.157.1.3 | The Cell Shape-determining MreBCD (MreBCD) Family | 0.026 | 0.34 | 1 | 0.0048 | 0.22 | 1 | 0.0034 | 0.35 | 1 | 0.0017 | 0.15 | 0.97 | 0.023 | 0.61 | 1 | 0.012 | 0.46 | 1 | 0.00048 | 9.4e-05 | 0 | 0.00029 | 3.2e-05 | 0.00065 | 0.00017 | 0 | 0.00046 | -0.77 | 7 / 36 (19%) | 4 / 7 (57%) | 1 / 7 (14%) | 0.571 | 0.143 | 0 | 0 | 0 | 1.6e-05 | 0 | 4.3e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 1.B.35.2.1 | The Oligogalacturonate-specific Porin (KdgM) Family | 0.068 | 0.34 | 1 | 0.039 | 0.28 | 1 | 0.099 | 0.42 | 1 | 0.058 | 0.29 | 1 | 0.23 | 0.76 | 1 | 0.44 | 0.79 | 1 | 0.00051 | 4.3e-05 | 0 | 0.00017 | 0 | 0.00044 | 4.8e-05 | 0 | 0.00013 | -1.8 | 3 / 36 (8.3%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 2.A.1.57.3 | The Major Facilitator Superfamily (MFS) | 0.47 | 0.54 | 1 | 0.72 | 0.87 | 1 | 0.87 | 0.94 | 1 | 0.71 | 0.87 | 1 | 0.81 | 0.95 | 1 | 0.6 | 0.85 | 1 | 0.00059 | 6.6e-05 | 0 | 0.00015 | 0 | 4e-04 | 0.00011 | 0 | 0.00028 | -0.45 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 4.4e-05 | 0 | 0.00015 | 6.8e-06 | 0 | 2.3e-05 | -2.7 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.D.11.1.1 | The Periplasmic Nitrate Reductase Complex (NAP) Complex Family | 0.054 | 0.34 | 1 | 0.011 | 0.23 | 1 | 0.048 | 0.37 | 1 | 0.019 | 0.26 | 1 | 0.062 | 0.62 | 1 | 0.17 | 0.68 | 1 | 0.00094 | 0.00018 | 0 | 0.00055 | 2.8e-05 | 0.0013 | 0.00037 | 0 | 0.00097 | -0.57 | 7 / 36 (19%) | 4 / 7 (57%) | 1 / 7 (14%) | 0.571 | 0.143 | 1.1e-05 | 0 | 3.6e-05 | 6.8e-06 | 0 | 2.3e-05 | -0.69 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.40.3.3 | The Nucleobase/Ascorbate Transporter (NAT) or Nucleobase:Cation Symporter-2 (NCS2) Family | 0.099 | 0.34 | 1 | 0.84 | 0.93 | 1 | 0.76 | 0.9 | 1 | 0.11 | 0.36 | 1 | 0.67 | 0.92 | 1 | 0.94 | 0.99 | 1 | 0.00053 | 7.3e-05 | 0 | 0.00017 | 0 | 4e-04 | 0.00019 | 0 | 0.00046 | 0.16 | 5 / 36 (14%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0 | 0 | 0 | 1e-05 | 0 | 3.4e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.B.33.1.3 | The Outer Membrane Protein Insertion Porin (Bam Complex) (OmpIP) Family | 0.18 | 0.34 | 1 | 0.35 | 0.64 | 1 | 0.17 | 0.46 | 1 | 0.48 | 0.73 | 1 | 0.58 | 0.91 | 1 | 0.29 | 0.73 | 1 | 0.00064 | 0.00025 | 0 | 6e-04 | 0 | 0.0015 | 0.00058 | 0 | 0.0015 | -0.049 | 14 / 36 (39%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 2.2e-05 | 0 | 3.7e-05 | 3.9e-05 | 0 | 5.3e-05 | 0.83 | 4 / 11 (36%) | 5 / 11 (45%) | 0.364 | 0.455 |
| 3.A.1.17.1 | The ATP-binding Cassette (ABC) Superfamily | 0.028 | 0.34 | 1 | 0.025 | 0.25 | 1 | 0.027 | 0.37 | 1 | 0.029 | 0.27 | 1 | 0.14 | 0.66 | 1 | 0.067 | 0.58 | 1 | 0.00052 | 7.2e-05 | 0 | 0.00029 | 0 | 0.00066 | 5.8e-05 | 0 | 0.00015 | -2.3 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 1.3e-05 | 0 | 3.5e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 2.A.21.1.1 | The Solute:Sodium Symporter (SSS) Family | 0.077 | 0.34 | 1 | 0.46 | 0.73 | 1 | 0.32 | 0.59 | 1 | 0.83 | 0.93 | 1 | 0.82 | 0.95 | 1 | 0.58 | 0.84 | 1 | 6.6e-05 | 7.3e-06 | 0 | 1.9e-05 | 0 | 4.5e-05 | 9.7e-06 | 0 | 2.6e-05 | -0.97 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 4.A.6.1.5 | The PTS Mannose-Fructose-Sorbose (Man) Family | 0.2 | 0.34 | 1 | 0.97 | 0.99 | 1 | 0.91 | 0.96 | 1 | 0.26 | 0.6 | 1 | 0.9 | 0.98 | 1 | 0.86 | 0.94 | 1 | 0.00058 | 8.1e-05 | 0 | 0.00017 | 0 | 0.00044 | 0.00023 | 0 | 0.00061 | 0.44 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 5.4e-06 | 0 | 1.8e-05 | 1.1 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.7.3.22 | The Drug/Metabolite Transporter (DMT) Superfamily | 0.39 | 0.47 | 1 | 0.17 | 0.46 | 1 | 0.12 | 0.43 | 1 | 0.68 | 0.86 | 1 | 0.19 | 0.72 | 1 | 0.087 | 0.61 | 1 | 0.00038 | 3.1e-05 | 0 | 5e-05 | 0 | 0.00013 | 0.00011 | 0 | 0.00028 | 1.1 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3e-06 | 0 | 9.9e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.A.42.2.1 | The Hydroxy/Aromatic Amino Acid Permease (HAAAP) Family | 0.14 | 0.34 | 1 | 0.15 | 0.44 | 1 | 0.19 | 0.48 | 1 | 0.56 | 0.79 | 1 | 0.95 | 0.98 | 1 | 0.83 | 0.93 | 1 | 0.00079 | 8.8e-05 | 0 | 0.00034 | 0 | 0.00089 | 9.7e-05 | 0 | 0.00026 | -1.8 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3e-06 | 0 | 9.9e-06 | 1e-05 | 0 | 3.4e-05 | 1.7 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.6.2.1 | The Resistance-Nodulation-Cell Division (RND) Superfamily | 0.16 | 0.34 | 1 | 0.038 | 0.28 | 1 | 0.25 | 0.52 | 1 | 0.27 | 0.6 | 1 | 0.061 | 0.62 | 1 | 0.35 | 0.74 | 1 | 0.00056 | 0.00016 | 0 | 0.00039 | 3.2e-05 | 0.00092 | 0.00033 | 0 | 0.00087 | -0.24 | 10 / 36 (28%) | 4 / 7 (57%) | 1 / 7 (14%) | 0.571 | 0.143 | 3.8e-05 | 0 | 7.6e-05 | 1.8e-05 | 0 | 4.1e-05 | -1.1 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 1.B.55.3.1 | The Poly Acetyl Glucosamine Porin (PgaA) Family | 0.19 | 0.34 | 1 | 0.14 | 0.44 | 1 | 0.26 | 0.52 | 1 | 0.56 | 0.79 | 1 | 0.48 | 0.87 | 1 | 0.63 | 0.85 | 1 | 0.00071 | 0.00014 | 0 | 0.00039 | 0 | 0.00097 | 0.00026 | 0 | 0.00069 | -0.58 | 7 / 36 (19%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.1e-05 | 0 | 5.1e-05 | 1.8e-05 | 0 | 4.1e-05 | -0.22 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 2.A.58.2.1 | The Phosphate:Na+ Symporter (PNaS) Family | 0.11 | 0.34 | 1 | 0.25 | 0.54 | 1 | 0.3 | 0.57 | 1 | 0.056 | 0.29 | 1 | 0.25 | 0.77 | 1 | 0.34 | 0.73 | 1 | 0.00038 | 6.3e-05 | 0 | 0.00015 | 0 | 0.00035 | 0.00016 | 0 | 0.00044 | 0.093 | 6 / 36 (17%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 5.4e-06 | 0 | 1.8e-05 | 1.1 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 9.B.124.2.2 | The DUF805 or PF05656 (DUF805) Family | 0.16 | 0.34 | 1 | 0.31 | 0.6 | 1 | 0.15 | 0.45 | 1 | 0.39 | 0.66 | 1 | 0.36 | 0.84 | 1 | 0.97 | 1 | 1 | 0.00034 | 2.8e-05 | 0 | 8.4e-05 | 0 | 0.00022 | 5.8e-05 | 0 | 0.00015 | -0.53 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.B.14.1.4 | The Outer Membrane Receptor (OMR) Family | 0.1 | 0.34 | 1 | 0.11 | 0.4 | 1 | 0.091 | 0.41 | 1 | 0.18 | 0.49 | 1 | 0.4 | 0.84 | 1 | 0.27 | 0.73 | 1 | 0.00047 | 7.9e-05 | 0 | 0.00024 | 0 | 0.00057 | 0.00014 | 0 | 0.00036 | -0.78 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 5.1e-06 | 0 | 1.7e-05 | 1.6e-05 | 0 | 3.7e-05 | 1.6 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 2.A.27.1.1 | The Glutamate:Na+ Symporter (ESS) Family | 0.3 | 0.4 | 1 | 0.5 | 0.75 | 1 | 0.96 | 0.99 | 1 | 0.59 | 0.81 | 1 | 0.72 | 0.93 | 1 | 0.67 | 0.88 | 1 | 0.00035 | 5.8e-05 | 0 | 8.6e-05 | 0 | 0.00022 | 0.00018 | 0 | 0.00049 | 1.1 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1.6e-05 | 0 | 3.7e-05 | 2.6 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 2.A.1.2.62 | The Major Facilitator Superfamily (MFS) | 0.25 | 0.36 | 1 | 0.54 | 0.76 | 1 | 0.77 | 0.9 | 1 | 0.95 | 0.98 | 1 | 0.56 | 0.91 | 1 | 0.18 | 0.68 | 1 | 0.00074 | 6.1e-05 | 0 | 0.00017 | 0 | 0.00045 | 0.00014 | 0 | 0.00036 | -0.28 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 7.3e-06 | 0 | 2.4e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 9.B.75.2.1 | The Ethanol Utilization/Transport (Eut) Protein Family | 0.079 | 0.34 | 1 | 0.48 | 0.73 | 1 | 0.32 | 0.59 | 1 | 0.093 | 0.34 | 1 | 0.53 | 0.89 | 1 | 0.33 | 0.73 | 1 | 0.00021 | 2.3e-05 | 0 | 5.1e-05 | 0 | 9.3e-05 | 5.8e-05 | 0 | 0.00015 | 0.19 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 3.A.1.15.5 | The ATP-binding Cassette (ABC) Superfamily | 0.13 | 0.34 | 1 | 0.24 | 0.53 | 1 | 0.052 | 0.37 | 1 | 0.18 | 0.49 | 1 | 0.86 | 0.96 | 1 | 0.18 | 0.68 | 1 | 0.00061 | 8.5e-05 | 0 | 0.00024 | 0 | 0.00062 | 0.00017 | 0 | 0.00046 | -0.5 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 1.5e-05 | 0 | 3.3e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 8.A.3.2.3 | The Cytoplasmic Membrane-Periplasmic Auxiliary-1 (MPA1) Protein with Cytoplasmic (C) Domain (MPA1-C or MPA1+C) Family | 0.32 | 0.41 | 1 | 0.24 | 0.53 | 1 | 0.14 | 0.43 | 1 | 0.2 | 0.51 | 1 | 0.14 | 0.66 | 1 | 0.13 | 0.65 | 1 | 0.00019 | 7.3e-05 | 0 | 3.7e-05 | 0 | 9e-05 | 1e-04 | 2.7e-05 | 0.00013 | 1.4 | 14 / 36 (39%) | 2 / 7 (29%) | 4 / 7 (57%) | 0.286 | 0.571 | 0.00012 | 0 | 0.00021 | 3.4e-05 | 0 | 5.1e-05 | -1.8 | 4 / 11 (36%) | 4 / 11 (36%) | 0.364 | 0.364 |
| 3.A.1.2.17 | The ATP-binding Cassette (ABC) Superfamily | 0.43 | 0.51 | 1 | 0.84 | 0.92 | 1 | 0.82 | 0.92 | 1 | 0.23 | 0.54 | 1 | 0.97 | 0.99 | 1 | 0.79 | 0.91 | 1 | 0.00018 | 3e-05 | 0 | 1.2e-05 | 0 | 3.1e-05 | 3.9e-06 | 0 | 1e-05 | -1.6 | 6 / 36 (17%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 6.7e-05 | 0 | 0.00019 | 2e-05 | 0 | 4.6e-05 | -1.7 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 3.A.1.2.3 | The ATP-binding Cassette (ABC) Superfamily | 0.078 | 0.34 | 1 | 0.0094 | 0.22 | 1 | 0.041 | 0.37 | 1 | 0.093 | 0.34 | 1 | 0.073 | 0.62 | 1 | 0.2 | 0.69 | 1 | 0.00065 | 0.00015 | 0 | 0.00048 | 0 | 0.0012 | 0.00023 | 0 | 0.00061 | -1.1 | 8 / 36 (22%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 1.1e-05 | 0 | 2.5e-05 | 9.2e-06 | 0 | 2.2e-05 | -0.26 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 2.A.3.2.8 | The Amino Acid-Polyamine-Organocation (APC) Family | 0.19 | 0.34 | 1 | 0.14 | 0.44 | 1 | 0.42 | 0.67 | 1 | 0.51 | 0.75 | 1 | 0.055 | 0.62 | 1 | 0.21 | 0.69 | 1 | 0.00045 | 5e-05 | 0 | 0.00012 | 0 | 0.00031 | 0.00013 | 0 | 3e-04 | 0.12 | 4 / 36 (11%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 4.D.2.4.3 | The Glycosyl Transferase 2 (GT2) Family | 0.21 | 0.35 | 1 | 0.62 | 0.79 | 1 | 0.5 | 0.74 | 1 | 0.54 | 0.77 | 1 | 0.7 | 0.93 | 1 | 0.65 | 0.86 | 1 | 0.00024 | 3.3e-05 | 0 | 6.2e-05 | 0 | 0.00013 | 6.8e-05 | 0 | 0.00018 | 0.13 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 6e-06 | 0 | 2e-05 | 2.1e-05 | 0 | 6.8e-05 | 1.8 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 5.A.3.2.1 | The Prokaryotic Molybdopterin-containing Oxidoreductase (PMO) Family | 0.078 | 0.34 | 1 | 0.54 | 0.76 | 1 | 0.38 | 0.63 | 1 | 0.1 | 0.35 | 1 | 0.51 | 0.87 | 1 | 0.34 | 0.74 | 1 | 0.00037 | 4.1e-05 | 0 | 9e-05 | 0 | 0.00016 | 0.00011 | 0 | 0.00028 | 0.29 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 1e-05 | 0 | 3.4e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.B.14.6.13 | The Outer Membrane Receptor (OMR) Family | 0.9 | 0.93 | 1 | 0.64 | 0.8 | 1 | 0.66 | 0.84 | 1 | 0.39 | 0.66 | 1 | 0.92 | 0.98 | 1 | 0.4 | 0.75 | 1 | 9.3e-05 | 7.8e-06 | 0 | 1.2e-05 | 0 | 3.2e-05 | 4.4e-06 | 0 | 1.2e-05 | -1.4 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 1.5e-05 | 0 | 4.9e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.A.42.1.2 | The Hydroxy/Aromatic Amino Acid Permease (HAAAP) Family | 0.074 | 0.34 | 1 | 0.046 | 0.29 | 1 | 0.045 | 0.37 | 1 | 0.035 | 0.27 | 1 | 0.26 | 0.77 | 1 | 0.17 | 0.68 | 1 | 7e-04 | 7.8e-05 | 0 | 0.00024 | 0 | 0.00057 | 0.00015 | 0 | 0.00041 | -0.68 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.8.1.5 | The Gluconate:H+ Symporter (GntP) Family | 0.19 | 0.34 | 1 | 0.058 | 0.33 | 1 | 0.13 | 0.43 | 1 | 0.44 | 0.71 | 1 | 0.015 | 0.5 | 1 | 0.034 | 0.58 | 1 | 0.00049 | 4.1e-05 | 0 | 1e-04 | 0 | 0.00027 | 0.00011 | 0 | 0.00028 | 0.14 | 3 / 36 (8.3%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 2.A.17.1.4 | The Proton-dependent Oligopeptide Transporter (POT/PTR) Family | 0.52 | 0.58 | 1 | 0.2 | 0.49 | 1 | 0.63 | 0.83 | 1 | 0.92 | 0.97 | 1 | 0.36 | 0.84 | 1 | 0.72 | 0.88 | 1 | 0.00032 | 6.3e-05 | 0 | 3.6e-05 | 0 | 8.8e-05 | 0.00025 | 0 | 0.00067 | 2.8 | 7 / 36 (19%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1.9e-05 | 0 | 3.8e-05 | 2.9 | 1 / 11 (9.1%) | 3 / 11 (27%) | 0.0909 | 0.273 |
| 2.A.115.2.9 | The Novobiocin Exporter (NbcE) Family | 0.67 | 0.73 | 1 | 0.41 | 0.69 | 1 | 0.058 | 0.37 | 1 | 0.62 | 0.82 | 1 | 0.81 | 0.95 | 1 | 0.13 | 0.65 | 1 | 0.00031 | 5.9e-05 | 0 | 6.9e-05 | 0 | 0.00018 | 1.3e-05 | 0 | 3.5e-05 | -2.4 | 7 / 36 (19%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 1.7e-05 | 0 | 4e-05 | 0.00013 | 0 | 0.00028 | 2.9 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 1.B.23.1.16 | The Cyanobacterial Porin (CBP) Family | 0.83 | 0.87 | 1 | 0.57 | 0.77 | 1 | 0.26 | 0.52 | 1 | 0.28 | 0.6 | 1 | 0.3 | 0.8 | 1 | 0.2 | 0.69 | 1 | 0.00047 | 9.1e-05 | 0 | 1e-04 | 0 | 0.00027 | 0.00012 | 0 | 0.00023 | 0.26 | 7 / 36 (19%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 0.00014 | 0 | 0.00039 | 1.8e-05 | 0 | 4.5e-05 | -3 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 2.A.7.3.57 | The Drug/Metabolite Transporter (DMT) Superfamily | 0.93 | 0.95 | 1 | 0.54 | 0.76 | 1 | 0.83 | 0.92 | 1 | 0.44 | 0.7 | 1 | 0.37 | 0.84 | 1 | 0.76 | 0.9 | 1 | 0.00029 | 6.4e-05 | 0 | 2.4e-05 | 0 | 6.4e-05 | 5.3e-05 | 0 | 9.8e-05 | 1.1 | 8 / 36 (22%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 2.6e-05 | 0 | 5.8e-05 | 0.00013 | 0 | 0.00039 | 2.3 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 4.A.2.1.1 | The PTS Fructose-Mannitol (Fru) Family | 0.2 | 0.34 | 1 | 0.2 | 0.49 | 1 | 0.62 | 0.83 | 1 | 0.76 | 0.89 | 1 | 0.74 | 0.94 | 1 | 0.26 | 0.72 | 1 | 0.0013 | 0.00011 | 0 | 0.00035 | 0 | 0.00094 | 2e-04 | 0 | 0.00054 | -0.81 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 8.8e-06 | 0 | 2.9e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.A.1.6.4 | The Major Facilitator Superfamily (MFS) | 0.17 | 0.34 | 1 | 0.011 | 0.23 | 1 | 0.01 | 0.37 | 1 | 0.34 | 0.63 | 1 | 0.086 | 0.62 | 1 | 0.072 | 0.59 | 1 | 0.00036 | 7e-05 | 0 | 0.00017 | 0 | 4e-04 | 0.00011 | 0 | 0.00028 | -0.63 | 7 / 36 (19%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 2.2e-05 | 0 | 7.3e-05 | 3.4e-05 | 0 | 1e-04 | 0.63 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 9.B.27.2.2 | The DedA or YdjX-Z (DedA) Family | 0.25 | 0.36 | 1 | 0.54 | 0.76 | 1 | 0.77 | 0.91 | 1 | 0.94 | 0.98 | 1 | 0.18 | 0.72 | 1 | 0.81 | 0.92 | 1 | 0.00021 | 1.8e-05 | 0 | 3.4e-05 | 0 | 8.9e-05 | 4.8e-05 | 0 | 0.00013 | 0.5 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.B.10.1.1 | The Nucleoside-specific Channel-forming Outer Membrane Porin (Tsx) Family | 0.18 | 0.34 | 1 | 0.77 | 0.88 | 1 | 0.48 | 0.72 | 1 | 0.38 | 0.66 | 1 | 0.7 | 0.93 | 1 | 0.42 | 0.77 | 1 | 0.00018 | 3e-05 | 0 | 5.1e-05 | 0 | 9.3e-05 | 7.7e-05 | 0 | 2e-04 | 0.59 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 3e-06 | 0 | 9.9e-06 | 1.3e-05 | 0 | 3.5e-05 | 2.1 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 1.B.3.1.3 | The Sugar Porin (SP) Family | 0.072 | 0.34 | 1 | 0.0097 | 0.22 | 1 | 0.076 | 0.38 | 1 | 0.023 | 0.27 | 1 | 0.1 | 0.64 | 1 | 0.39 | 0.75 | 1 | 0.00077 | 0.00011 | 0 | 0.00031 | 0 | 0.00075 | 0.00023 | 0 | 0.00061 | -0.43 | 5 / 36 (14%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 3e-06 | 0 | 9.9e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.A.1.12.1 | The Major Facilitator Superfamily (MFS) | 0.056 | 0.34 | 1 | 0.12 | 0.41 | 1 | 0.22 | 0.5 | 1 | 0.031 | 0.27 | 1 | 0.23 | 0.76 | 1 | 0.4 | 0.75 | 1 | 0.00065 | 5.4e-05 | 0 | 0.00015 | 0 | 0.00031 | 0.00013 | 0 | 0.00033 | -0.21 | 3 / 36 (8.3%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 1.E.19.1.2 | The Clostridium difficile TcdE Holin (TcdE Holin) Family | 0.21 | 0.35 | 1 | 1 | 1 | 1 | 0.22 | 0.5 | 1 | 0.064 | 0.3 | 1 | 0.43 | 0.86 | 1 | 0.11 | 0.65 | 1 | 0.00016 | 1.4e-05 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 3 / 36 (8.3%) | 0 / 7 (0%) | 0 / 7 (0%) | 0 | 0 | 3.2e-05 | 0 | 9.5e-05 | 1.3e-05 | 0 | 4.3e-05 | -1.3 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 1.B.14.7.1 | The Outer Membrane Receptor (OMR) Family | 0.23 | 0.35 | 1 | 0.81 | 0.91 | 1 | 0.9 | 0.96 | 1 | 0.57 | 0.79 | 1 | 0.78 | 0.94 | 1 | 0.83 | 0.93 | 1 | 0.00074 | 0.00016 | 0 | 0.00036 | 0 | 0.00088 | 0.00043 | 0 | 0.0011 | 0.26 | 8 / 36 (22%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 1.8e-05 | 0 | 4e-05 | 2.1e-05 | 0 | 4.7e-05 | 0.22 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 4.A.5.1.3 | The PTS Galactitol (Gat) Family | 0.072 | 0.34 | 1 | 0.014 | 0.23 | 1 | 0.048 | 0.37 | 1 | 0.032 | 0.27 | 1 | 0.17 | 0.72 | 1 | 0.33 | 0.73 | 1 | 0.0015 | 0.00013 | 0 | 0.00042 | 0 | 0.0011 | 0.00023 | 0 | 0.00061 | -0.87 | 3 / 36 (8.3%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 9.B.14.1.17 | The Putative Heme Handling Protein (HHP) Family | 0.92 | 0.94 | 1 | 0.57 | 0.77 | 1 | 0.7 | 0.87 | 1 | 0.99 | 0.99 | 1 | 0.61 | 0.91 | 1 | 0.39 | 0.75 | 1 | 0.00047 | 0.00021 | 0 | 0.00013 | 1.6e-05 | 0.00016 | 8.1e-05 | 0 | 0.00014 | -0.68 | 16 / 36 (44%) | 4 / 7 (57%) | 3 / 7 (43%) | 0.571 | 0.429 | 0.00033 | 0 | 0.00076 | 0.00021 | 5.9e-05 | 0.00057 | -0.65 | 3 / 11 (27%) | 6 / 11 (55%) | 0.273 | 0.545 |
| 4.C.1.1.6 | The Fatty Acid Transporter (FAT) Family | 0.08 | 0.34 | 1 | 0.27 | 0.55 | 1 | 0.27 | 0.53 | 1 | 0.08 | 0.32 | 1 | 0.28 | 0.78 | 1 | 0.31 | 0.73 | 1 | 0.00029 | 4.8e-05 | 0 | 0.00011 | 0 | 0.00022 | 0.00012 | 0 | 0.00031 | 0.13 | 6 / 36 (17%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1e-05 | 0 | 3.4e-05 | 1.9 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.A.1.121.6 | The ATP-binding Cassette (ABC) Superfamily | 0.21 | 0.35 | 1 | 0.89 | 0.96 | 1 | 0.69 | 0.86 | 1 | 0.1 | 0.35 | 1 | 0.7 | 0.93 | 1 | 0.46 | 0.8 | 1 | 0.00022 | 9.1e-05 | 0 | 4.1e-05 | 0 | 8.9e-05 | 4.8e-05 | 0 | 8.6e-05 | 0.23 | 15 / 36 (42%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0.00012 | 9.4e-05 | 0.00017 | 0.00012 | 0 | 0.00022 | 0 | 6 / 11 (55%) | 5 / 11 (45%) | 0.545 | 0.455 |
| 2.A.7.3.26 | The Drug/Metabolite Transporter (DMT) Superfamily | 0.11 | 0.34 | 1 | 0.99 | 1 | 1 | 0.81 | 0.91 | 1 | 0.032 | 0.27 | 1 | 0.5 | 0.87 | 1 | 0.4 | 0.75 | 1 | 0.00024 | 3.3e-05 | 0 | 4.8e-05 | 0 | 8.8e-05 | 0.00012 | 0 | 0.00031 | 1.3 | 5 / 36 (14%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.1.10.3 | The Major Facilitator Superfamily (MFS) | 0.059 | 0.34 | 1 | 0.06 | 0.33 | 1 | 0.22 | 0.5 | 1 | 0.085 | 0.33 | 1 | 0.082 | 0.62 | 1 | 0.35 | 0.74 | 1 | 2e-04 | 3.8e-05 | 0 | 0.00011 | 0 | 0.00018 | 7.7e-05 | 0 | 2e-04 | -0.51 | 7 / 36 (19%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 5.5e-06 | 0 | 1.2e-05 | 2.7e-06 | 0 | 8.9e-06 | -1 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 2.A.86.1.2 | The Autoinducer-2 Exporter (AI-2E) Family (Formerly the PerM Family. TC #9.B.22) | 0.28 | 0.38 | 1 | 0.83 | 0.92 | 1 | 0.33 | 0.59 | 1 | 0.99 | 0.99 | 1 | 0.27 | 0.78 | 1 | 0.051 | 0.58 | 1 | 0.00059 | 4.9e-05 | 0 | 0.00012 | 0 | 0.00031 | 0.00013 | 0 | 0.00033 | 0.12 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 5.1e-06 | 0 | 1.7e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 1.B.25.1.13 | The Outer Membrane Porin (Opr) Family | 0.16 | 0.34 | 1 | 0.41 | 0.69 | 1 | 0.11 | 0.42 | 1 | 0.48 | 0.73 | 1 | 0.74 | 0.94 | 1 | 0.36 | 0.74 | 1 | 0.00031 | 3.5e-05 | 0 | 1e-04 | 0 | 0.00027 | 5.8e-05 | 0 | 0.00015 | -0.79 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 1.3e-05 | 0 | 3.5e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 1.B.14.9.5 | The Outer Membrane Receptor (OMR) Family | 0.083 | 0.34 | 1 | 0.035 | 0.28 | 1 | 0.13 | 0.43 | 1 | 0.11 | 0.36 | 1 | 0.19 | 0.72 | 1 | 0.45 | 0.79 | 1 | 0.00032 | 5.4e-05 | 0 | 0.00017 | 0 | 4e-04 | 9.7e-05 | 0 | 0.00026 | -0.81 | 6 / 36 (17%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 7.3e-06 | 0 | 2.4e-05 | 2.7e-06 | 0 | 8.9e-06 | -1.4 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 9.B.223.1.4 | The 4 TMS DUF2979 or YoaS (YoaS) Family | 0.51 | 0.57 | 1 | 0.57 | 0.77 | 1 | 0.8 | 0.91 | 1 | 0.91 | 0.97 | 1 | 0.8 | 0.94 | 1 | 1 | 1 | 1 | 0.00025 | 8.4e-05 | 0 | 0.00012 | 0 | 0.00016 | 6.1e-05 | 0 | 0.00011 | -0.98 | 12 / 36 (33%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 9.1e-05 | 0 | 0.00022 | 6.9e-05 | 0 | 2e-04 | -0.4 | 3 / 11 (27%) | 3 / 11 (27%) | 0.273 | 0.273 |
| 2.A.1.4.1 | The Major Facilitator Superfamily (MFS) | 0.05 | 0.34 | 1 | 0.042 | 0.29 | 1 | 0.046 | 0.37 | 1 | 0.054 | 0.29 | 1 | 0.25 | 0.77 | 1 | 0.15 | 0.67 | 1 | 5e-04 | 5.6e-05 | 0 | 0.00021 | 0 | 0.00053 | 5.8e-05 | 0 | 0.00015 | -1.9 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 1e-05 | 0 | 3.4e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.1.3.37 | The Major Facilitator Superfamily (MFS) | 0.9 | 0.93 | 1 | 0.47 | 0.73 | 1 | 0.76 | 0.9 | 1 | 0.34 | 0.63 | 1 | 0.63 | 0.91 | 1 | 0.85 | 0.94 | 1 | 0.00022 | 1.8e-05 | 0 | 1.7e-05 | 0 | 4.5e-05 | 0 | 0 | 0 | -Inf | 3 / 36 (8.3%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 3.5e-05 | 0 | 0.00012 | 1.3e-05 | 0 | 4.3e-05 | -1.4 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.21.2.1 | The Solute:Sodium Symporter (SSS) Family | 0.057 | 0.34 | 1 | 0.079 | 0.38 | 1 | 0.25 | 0.52 | 1 | 0.094 | 0.34 | 1 | 0.33 | 0.83 | 1 | 0.65 | 0.86 | 1 | 0.00024 | 4.6e-05 | 0 | 0.00014 | 0 | 0.00031 | 7.2e-05 | 0 | 0.00018 | -0.96 | 7 / 36 (19%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 1.1e-05 | 0 | 3.6e-05 | 2.7e-06 | 0 | 8.9e-06 | -2 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.D.2.1.1 | The Proton-translocating Transhydrogenase (PTH) Family | 0.44 | 0.51 | 1 | 0.19 | 0.49 | 1 | 0.72 | 0.88 | 1 | 0.71 | 0.87 | 1 | 0.16 | 0.69 | 1 | 0.62 | 0.85 | 1 | 0.00033 | 7.3e-05 | 0 | 0.00012 | 0 | 0.00031 | 0.00019 | 0 | 0.00046 | 0.66 | 8 / 36 (22%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 8.5e-06 | 0 | 2.1e-05 | 3.3e-05 | 0 | 7e-05 | 2 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 2.A.1.26.1 | The Major Facilitator Superfamily (MFS) | 0.75 | 0.8 | 1 | 0.9 | 0.96 | 1 | 0.49 | 0.74 | 1 | 0.2 | 0.5 | 1 | 0.59 | 0.91 | 1 | 0.26 | 0.72 | 1 | 0.00022 | 3e-05 | 0 | 5e-05 | 0 | 0.00013 | 5.8e-05 | 0 | 0.00015 | 0.21 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.5e-05 | 0 | 7.2e-05 | 5.4e-06 | 0 | 1.8e-05 | -2.2 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 9.B.125.1.1 | The AmpE/CobD (AmpE/CobD) Family | 0.32 | 0.41 | 1 | 0.16 | 0.46 | 1 | 0.79 | 0.91 | 1 | 0.33 | 0.63 | 1 | 0.9 | 0.98 | 1 | 0.19 | 0.68 | 1 | 0.00045 | 6.3e-05 | 0 | 0.00024 | 0 | 0.00062 | 5.8e-05 | 0 | 0.00015 | -2 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 1.9e-05 | 0 | 3.9e-05 | 0 | 0 | 0 | -Inf | 3 / 11 (27%) | 0 / 11 (0%) | 0.273 | 0 |
| 9.B.126.1.2 | The Putative Lipid IV Exporter (YhjD) Family | 0.53 | 0.59 | 1 | 0.86 | 0.94 | 1 | 0.58 | 0.79 | 1 | 0.11 | 0.36 | 1 | 0.47 | 0.87 | 1 | 0.57 | 0.84 | 1 | 6e-04 | 0.00046 | 0.00034 | 4e-04 | 0.00056 | 0.00042 | 0.00032 | 0.00024 | 0.00034 | -0.32 | 28 / 36 (78%) | 4 / 7 (57%) | 5 / 7 (71%) | 0.571 | 0.714 | 0.00047 | 0.00026 | 0.00059 | 0.00059 | 0.00038 | 0.00057 | 0.33 | 10 / 11 (91%) | 9 / 11 (82%) | 0.909 | 0.818 |
| 1.B.44.2.9 | The Probable Protein Translocating Porphyromonas gingivalis Porin (PorT) Family | 0.24 | 0.36 | 1 | 0.57 | 0.77 | 1 | 0.74 | 0.89 | 1 | 0.97 | 0.99 | 1 | 0.97 | 0.99 | 1 | 0.44 | 0.79 | 1 | 9.8e-05 | 1.4e-05 | 0 | 3e-05 | 0 | 5.1e-05 | 2.1e-05 | 0 | 4.6e-05 | -0.51 | 5 / 36 (14%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 1.2e-05 | 0 | 3.9e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 3.A.1.12.15 | The ATP-binding Cassette (ABC) Superfamily | 0.12 | 0.34 | 1 | 0.11 | 0.4 | 1 | 0.052 | 0.37 | 1 | 0.34 | 0.63 | 1 | 0.41 | 0.85 | 1 | 0.17 | 0.68 | 1 | 3e-04 | 6.7e-05 | 0 | 0.00019 | 0 | 0.00044 | 0.00011 | 0 | 0.00028 | -0.79 | 8 / 36 (22%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 5.5e-06 | 0 | 1.2e-05 | 2.5e-05 | 0 | 4.4e-05 | 2.2 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 2.A.3.2.1 | The Amino Acid-Polyamine-Organocation (APC) Family | 0.45 | 0.52 | 1 | 0.63 | 0.8 | 1 | 0.14 | 0.43 | 1 | 0.41 | 0.68 | 1 | 0.38 | 0.84 | 1 | 0.044 | 0.58 | 1 | 0.00036 | 4e-05 | 0 | 8.4e-05 | 0 | 0.00022 | 0.00011 | 0 | 0.00028 | 0.39 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 9.9e-06 | 0 | 2.5e-05 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 3.A.3.7.1 | The P-type ATPase (P-ATPase) Superfamily | 0.23 | 0.35 | 1 | 0.46 | 0.73 | 1 | 0.26 | 0.52 | 1 | 0.75 | 0.89 | 1 | 0.92 | 0.98 | 1 | 0.59 | 0.85 | 1 | 0.00036 | 6.9e-05 | 0 | 0.00017 | 0 | 0.00044 | 0.00014 | 0 | 0.00036 | -0.28 | 7 / 36 (19%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 5.5e-06 | 0 | 1.2e-05 | 2.6e-05 | 0 | 6.9e-05 | 2.2 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 2.A.1.36.3 | The Major Facilitator Superfamily (MFS) | 0.11 | 0.34 | 1 | 0.064 | 0.35 | 1 | 0.048 | 0.37 | 1 | 0.092 | 0.34 | 1 | 0.71 | 0.93 | 1 | 0.37 | 0.74 | 1 | 0.00046 | 5.1e-05 | 0 | 0.00015 | 0 | 4e-04 | 0.00011 | 0 | 0.00028 | -0.45 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 9.B.29.2.7 | The small 4-5 TMS Putative Permease (4-5PP) Family | 0.17 | 0.34 | 1 | 0.44 | 0.71 | 1 | 0.18 | 0.48 | 1 | 0.33 | 0.63 | 1 | 0.58 | 0.91 | 1 | 0.68 | 0.88 | 1 | 0.001 | 8.3e-05 | 0 | 0.00024 | 0 | 0.00062 | 0.00017 | 0 | 0.00046 | -0.5 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 1.1e-05 | 0 | 3.6e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 3.A.1.5.10 | The ATP-binding Cassette (ABC) Superfamily | 0.75 | 0.8 | 1 | 0.91 | 0.96 | 1 | 0.9 | 0.96 | 1 | 0.38 | 0.65 | 1 | 0.83 | 0.95 | 1 | 0.91 | 0.97 | 1 | 3e-04 | 1e-04 | 0 | 9.1e-05 | 0 | 0.00018 | 0.00013 | 0 | 0.00036 | 0.51 | 12 / 36 (33%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0.00013 | 0 | 0.00024 | 6e-05 | 0 | 8.2e-05 | -1.1 | 4 / 11 (36%) | 5 / 11 (45%) | 0.364 | 0.455 |
| 2.A.1.42.1 | The Major Facilitator Superfamily (MFS) | 0.18 | 0.34 | 1 | 0.48 | 0.73 | 1 | 0.24 | 0.52 | 1 | 0.48 | 0.73 | 1 | 0.93 | 0.98 | 1 | 0.49 | 0.82 | 1 | 0.00025 | 4.2e-05 | 0 | 1e-04 | 0 | 0.00027 | 8.7e-05 | 0 | 0.00023 | -0.2 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1.3e-05 | 0 | 3.5e-05 | 2.3 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 3.A.1.4.10 | The ATP-binding Cassette (ABC) Superfamily | 0.41 | 0.49 | 1 | 0.93 | 0.97 | 1 | 0.33 | 0.59 | 1 | 0.93 | 0.97 | 1 | 0.93 | 0.98 | 1 | 0.48 | 0.81 | 1 | 0.00012 | 2.4e-05 | 0 | 2.2e-05 | 0 | 5.2e-05 | 2.9e-05 | 0 | 7.7e-05 | 0.4 | 7 / 36 (19%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.8e-06 | 0 | 9.4e-06 | 4.3e-05 | 0 | 8.9e-05 | 3.9 | 1 / 11 (9.1%) | 3 / 11 (27%) | 0.0909 | 0.273 |
| 1.B.14.1.2 | The Outer Membrane Receptor (OMR) Family | 0.12 | 0.34 | 1 | 0.053 | 0.31 | 1 | 0.2 | 0.48 | 1 | 0.32 | 0.63 | 1 | 0.3 | 0.8 | 1 | 0.68 | 0.88 | 1 | 0.00068 | 0.00011 | 0 | 0.00034 | 0 | 0.00079 | 2e-04 | 0 | 0.00054 | -0.77 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 1.4e-05 | 0 | 3.7e-05 | 8.1e-06 | 0 | 2.7e-05 | -0.79 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 9.B.114.1.1 | The Vancomycin-sensitivity protein (SanA) Family | 0.18 | 0.34 | 1 | 0.54 | 0.76 | 1 | 0.12 | 0.43 | 1 | 0.61 | 0.82 | 1 | 0.81 | 0.95 | 1 | 0.33 | 0.73 | 1 | 0.00035 | 3.9e-05 | 0 | 1e-04 | 0 | 0.00027 | 5.8e-05 | 0 | 0.00015 | -0.79 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 2.7e-05 | 0 | 7e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 2.A.47.3.5 | The Divalent Anion:Na+ Symporter (DASS) Family | 0.28 | 0.38 | 1 | 0.77 | 0.88 | 1 | 0.94 | 0.98 | 1 | 0.67 | 0.86 | 1 | 0.52 | 0.87 | 1 | 0.79 | 0.91 | 1 | 8.2e-05 | 6.8e-06 | 0 | 1.2e-05 | 0 | 3.1e-05 | 1.9e-05 | 0 | 5.1e-05 | 0.66 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.6.2.2 | The Resistance-Nodulation-Cell Division (RND) Superfamily | 0.11 | 0.34 | 1 | 0.3 | 0.59 | 1 | 0.47 | 0.72 | 1 | 0.09 | 0.34 | 1 | 0.44 | 0.86 | 1 | 0.71 | 0.88 | 1 | 5e-04 | 0.00011 | 0 | 0.00027 | 0 | 0.00066 | 0.00028 | 0 | 0.00072 | 0.052 | 8 / 36 (22%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 5.5e-06 | 0 | 1.2e-05 | 8.1e-06 | 0 | 2.7e-05 | 0.56 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 2.A.6.2.12 | The Resistance-Nodulation-Cell Division (RND) Superfamily | 0.49 | 0.56 | 1 | 0.75 | 0.88 | 1 | 0.61 | 0.82 | 1 | 0.78 | 0.9 | 1 | 0.52 | 0.87 | 1 | 0.19 | 0.68 | 1 | 0.00071 | 0.00018 | 0 | 0.00038 | 0 | 0.00098 | 0.00032 | 0 | 0.00082 | -0.25 | 9 / 36 (25%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0.00011 | 0 | 0.00033 | 3.4e-05 | 0 | 7.8e-05 | -1.7 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 8.A.21.2.2 | The Stomatin/Podocin/Band 7/Nephrosis.2/SPFH (Stomatin) Family | 0.11 | 0.34 | 1 | 0.017 | 0.23 | 1 | 0.055 | 0.37 | 1 | 0.1 | 0.35 | 1 | 0.77 | 0.94 | 1 | 1 | 1 | 1 | 0.00036 | 3e-05 | 0 | 8.6e-05 | 0 | 0.00022 | 6.8e-05 | 0 | 0.00018 | -0.34 | 3 / 36 (8.3%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 2.A.3.1.6 | The Amino Acid-Polyamine-Organocation (APC) Family | 0.13 | 0.34 | 1 | 0.14 | 0.44 | 1 | 0.1 | 0.42 | 1 | 0.33 | 0.63 | 1 | 0.7 | 0.93 | 1 | 0.37 | 0.74 | 1 | 0.00038 | 6.3e-05 | 0 | 2e-04 | 0 | 0.00053 | 9.7e-05 | 0 | 0.00026 | -1 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1.3e-05 | 0 | 3.5e-05 | 2.3 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 2.A.9.3.1 | The Membrane Protein Insertase (YidC/Alb3/Oxa1) Family | 0.21 | 0.35 | 1 | 0.24 | 0.53 | 1 | 0.18 | 0.48 | 1 | 0.94 | 0.98 | 1 | 0.93 | 0.98 | 1 | 0.7 | 0.88 | 1 | 0.00053 | 8.8e-05 | 0 | 0.00029 | 0 | 0.00076 | 0.00012 | 0 | 0.00031 | -1.3 | 6 / 36 (17%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 5.5e-06 | 0 | 1.2e-05 | 2.6e-05 | 0 | 6.9e-05 | 2.2 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 1.C.39.13.1 | The Membrane Attack Complex/Perforin (MACPF) Family | 0.19 | 0.34 | 1 | 1 | 1 | 1 | 0.67 | 0.85 | 1 | 0.045 | 0.27 | 1 | 0.77 | 0.94 | 1 | 0.52 | 0.82 | 1 | 0.00018 | 1.5e-05 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 3 / 36 (8.3%) | 0 / 7 (0%) | 0 / 7 (0%) | 0 | 0 | 3.1e-05 | 0 | 7.6e-05 | 1.8e-05 | 0 | 6e-05 | -0.78 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 3.A.1.1.48 | The ATP-binding Cassette (ABC) Superfamily | 0.75 | 0.81 | 1 | 0.64 | 0.8 | 1 | 0.81 | 0.91 | 1 | 0.3 | 0.62 | 1 | 0.61 | 0.91 | 1 | 0.34 | 0.74 | 1 | 0.0021 | 0.0012 | 9.7e-05 | 0.00086 | 0 | 0.0012 | 0.0012 | 0 | 0.0017 | 0.48 | 21 / 36 (58%) | 3 / 7 (43%) | 3 / 7 (43%) | 0.429 | 0.429 | 0.0014 | 0.00011 | 0.0031 | 0.0014 | 1e-04 | 0.0021 | 0 | 9 / 11 (82%) | 6 / 11 (55%) | 0.818 | 0.545 |
| 2.A.3.2.2 | The Amino Acid-Polyamine-Organocation (APC) Family | 0.32 | 0.41 | 1 | 0.34 | 0.63 | 1 | 0.086 | 0.4 | 1 | 0.91 | 0.97 | 1 | 0.33 | 0.83 | 1 | 0.051 | 0.58 | 1 | 0.00076 | 8.5e-05 | 0 | 0.00019 | 0 | 0.00049 | 0.00024 | 0 | 0.00064 | 0.34 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 5.5e-06 | 0 | 1.2e-05 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 2.A.6.1.4 | The Resistance-Nodulation-Cell Division (RND) Superfamily | 0.18 | 0.34 | 1 | 0.93 | 0.97 | 1 | 0.12 | 0.43 | 1 | 0.31 | 0.63 | 1 | 0.93 | 0.98 | 1 | 0.12 | 0.65 | 1 | 0.00032 | 5.3e-05 | 0 | 1e-04 | 0 | 0.00027 | 0.00013 | 0 | 0.00033 | 0.38 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.8e-05 | 0 | 4.9e-05 | Inf | 0 / 11 (0%) | 3 / 11 (27%) | 0 | 0.273 |
| 3.A.11.1.2 | The Bacterial Competence-related DNA Transformation Transporter (DNA-T) Family | 0.91 | 0.93 | 1 | 0.46 | 0.72 | 1 | 0.78 | 0.91 | 1 | 0.38 | 0.65 | 1 | 0.66 | 0.92 | 1 | 0.93 | 0.98 | 1 | 0.00014 | 1.2e-05 | 0 | 1.2e-05 | 0 | 3.1e-05 | 0 | 0 | 0 | -Inf | 3 / 36 (8.3%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 2.3e-05 | 0 | 7.8e-05 | 6.8e-06 | 0 | 2.3e-05 | -1.8 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.15.2.1 | The Betaine/Carnitine/Choline Transporter (BCCT) Family | 0.16 | 0.34 | 1 | 0.27 | 0.56 | 1 | 0.12 | 0.43 | 1 | 0.28 | 0.6 | 1 | 0.72 | 0.94 | 1 | 0.38 | 0.75 | 1 | 0.00052 | 8.6e-05 | 0 | 0.00022 | 0 | 0.00058 | 0.00019 | 0 | 0.00051 | -0.21 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 3e-06 | 0 | 9.9e-06 | 1.3e-05 | 0 | 3.5e-05 | 2.1 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 2.A.66.1.4 | The Multidrug/Oligosaccharidyl-lipid/Polysaccharide (MOP) Flippase Superfamily | 0.36 | 0.45 | 1 | 0.16 | 0.46 | 1 | 0.52 | 0.75 | 1 | 0.76 | 0.89 | 1 | 0.12 | 0.66 | 1 | 0.63 | 0.85 | 1 | 0.00051 | 4.2e-05 | 0 | 5e-05 | 0 | 0.00013 | 0.00015 | 0 | 0.00041 | 1.6 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 8.1e-06 | 0 | 2.7e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 9.B.22.2.1 | The Leukotoxin Secretion Morphogenesis Protein C (MorC) Family | 0.15 | 0.34 | 1 | 0.49 | 0.75 | 1 | 0.24 | 0.52 | 1 | 0.12 | 0.38 | 1 | 0.95 | 0.98 | 1 | 0.57 | 0.83 | 1 | 5e-04 | 5.6e-05 | 0 | 0.00014 | 0 | 0.00036 | 0.00015 | 0 | 0.00038 | 0.1 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.1.2.20 | The Major Facilitator Superfamily (MFS) | 0.17 | 0.34 | 1 | 0.079 | 0.38 | 1 | 0.56 | 0.78 | 1 | 0.64 | 0.84 | 1 | 0.38 | 0.84 | 1 | 0.75 | 0.89 | 1 | 0.00029 | 4.1e-05 | 0 | 0.00013 | 0 | 0.00031 | 5.8e-05 | 0 | 0.00015 | -1.2 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 1.4e-05 | 0 | 3.7e-05 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 2.A.39.3.8 | The Nucleobase:Cation Symporter-1 (NCS1) Family | 0.25 | 0.36 | 1 | 0.58 | 0.77 | 1 | 0.65 | 0.84 | 1 | 0.7 | 0.87 | 1 | 0.65 | 0.92 | 1 | 0.51 | 0.82 | 1 | 0.00037 | 5.2e-05 | 0 | 0.00011 | 0 | 0.00026 | 0.00014 | 0 | 0.00036 | 0.35 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 1.1e-05 | 0 | 2.5e-05 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 1.B.18.3.1 | The Outer Membrane Auxiliary (OMA) Protein Family | 0.65 | 0.7 | 1 | 0.81 | 0.91 | 1 | 0.13 | 0.43 | 1 | 0.26 | 0.59 | 1 | 0.85 | 0.96 | 1 | 0.1 | 0.65 | 1 | 0.00031 | 5.1e-05 | 0 | 8.6e-05 | 0 | 0.00022 | 0.00014 | 0 | 0.00036 | 0.7 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.7e-05 | 0 | 5.3e-05 | 0 | 0 | 0 | -Inf | 3 / 11 (27%) | 0 / 11 (0%) | 0.273 | 0 |
| 1.A.8.3.1 | The Major Intrinsic Protein (MIP) Family | 0.37 | 0.45 | 1 | 0.9 | 0.96 | 1 | 0.38 | 0.63 | 1 | 0.61 | 0.82 | 1 | 0.62 | 0.91 | 1 | 0.65 | 0.86 | 1 | 0.00013 | 1.8e-05 | 0 | 3.4e-05 | 0 | 8.9e-05 | 2.9e-05 | 0 | 7.7e-05 | -0.23 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1.8e-05 | 0 | 4e-05 | 2.8 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 2.A.1.15.12 | The Major Facilitator Superfamily (MFS) | 0.07 | 0.34 | 1 | 0.02 | 0.23 | 1 | 0.062 | 0.37 | 1 | 0.036 | 0.27 | 1 | 0.26 | 0.78 | 1 | 0.45 | 0.79 | 1 | 0.00057 | 4.7e-05 | 0 | 0.00015 | 0 | 0.00035 | 9.7e-05 | 0 | 0.00026 | -0.63 | 3 / 36 (8.3%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 2.A.81.1.5 | The Aspartate:Alanine Exchanger (AAEx) Family | 0.099 | 0.34 | 1 | 0.48 | 0.73 | 1 | 0.18 | 0.48 | 1 | 0.065 | 0.3 | 1 | 0.45 | 0.86 | 1 | 0.13 | 0.65 | 1 | 0.00071 | 9.8e-05 | 0 | 0.00022 | 0 | 0.00048 | 0.00025 | 0 | 0.00067 | 0.18 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.1e-05 | 0 | 4.7e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 2.A.66.2.7 | The Multidrug/Oligosaccharidyl-lipid/Polysaccharide (MOP) Flippase Superfamily | 0.11 | 0.34 | 1 | 0.099 | 0.39 | 1 | 0.23 | 0.51 | 1 | 0.36 | 0.64 | 1 | 0.62 | 0.91 | 1 | 0.93 | 0.99 | 1 | 0.00035 | 6.9e-05 | 0 | 0.00022 | 0 | 0.00053 | 1e-04 | 0 | 0.00025 | -1.1 | 7 / 36 (19%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 8.5e-06 | 0 | 2.1e-05 | 1.1e-05 | 0 | 3.6e-05 | 0.37 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 4.A.6.1.1 | The PTS Mannose-Fructose-Sorbose (Man) Family | 0.016 | 0.34 | 1 | 0.018 | 0.23 | 1 | 0.019 | 0.37 | 1 | 0.0044 | 0.16 | 1 | 0.079 | 0.62 | 1 | 0.06 | 0.58 | 1 | 0.00028 | 7e-05 | 0 | 0.00022 | 3.2e-05 | 0.00043 | 0.00011 | 0 | 0.00028 | -1 | 9 / 36 (25%) | 4 / 7 (57%) | 2 / 7 (29%) | 0.571 | 0.286 | 3e-06 | 0 | 9.9e-06 | 1.6e-05 | 0 | 3.7e-05 | 2.4 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 1.B.54.1.9 | The Intimin/Invasin (Int/Inv) or Autotransporter-3 (AT-3) Family | 0.11 | 0.34 | 1 | 0.55 | 0.76 | 1 | 0.47 | 0.72 | 1 | 0.35 | 0.64 | 1 | 0.2 | 0.74 | 1 | 0.51 | 0.82 | 1 | 0.00037 | 4.1e-05 | 0 | 0.00015 | 0 | 4e-04 | 5.7e-05 | 0 | 0.00013 | -1.4 | 4 / 36 (11%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.7.7.2 | The Drug/Metabolite Transporter (DMT) Superfamily | 0.078 | 0.34 | 1 | 0.13 | 0.43 | 1 | 0.14 | 0.43 | 1 | 0.097 | 0.34 | 1 | 0.27 | 0.78 | 1 | 0.3 | 0.73 | 1 | 0.00023 | 3.8e-05 | 0 | 9.8e-05 | 0 | 0.00022 | 7.7e-05 | 0 | 2e-04 | -0.35 | 6 / 36 (17%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1e-05 | 0 | 3.4e-05 | 1.9 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.A.1.5.21 | The ATP-binding Cassette (ABC) Superfamily | 0.055 | 0.34 | 1 | 0.019 | 0.23 | 1 | 0.017 | 0.37 | 1 | 0.0024 | 0.15 | 1 | 0.088 | 0.62 | 1 | 0.065 | 0.58 | 1 | 0.00076 | 0.00015 | 0 | 0.00041 | 2.8e-05 | 0.001 | 0.00034 | 0 | 0.00089 | -0.27 | 7 / 36 (19%) | 4 / 7 (57%) | 2 / 7 (29%) | 0.571 | 0.286 | 0 | 0 | 0 | 8.1e-06 | 0 | 2.7e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 4.A.2.1.9 | The PTS Fructose-Mannitol (Fru) Family | 0.064 | 0.34 | 1 | 0.0043 | 0.22 | 1 | 0.042 | 0.37 | 1 | 0.021 | 0.26 | 1 | 0.05 | 0.62 | 1 | 0.3 | 0.73 | 1 | 6e-04 | 8.3e-05 | 0 | 0.00027 | 0 | 0.00066 | 0.00015 | 0 | 0.00041 | -0.85 | 5 / 36 (14%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.A.40.4.2 | The Nucleobase/Ascorbate Transporter (NAT) or Nucleobase:Cation Symporter-2 (NCS2) Family | 0.27 | 0.38 | 1 | 0.57 | 0.77 | 1 | 0.73 | 0.89 | 1 | 0.94 | 0.98 | 1 | 0.0041 | 0.36 | 1 | 0.029 | 0.58 | 1 | 0.00039 | 4.3e-05 | 0 | 1e-04 | 0 | 0.00027 | 0.00011 | 0 | 0.00028 | 0.14 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3e-06 | 0 | 9.9e-06 | 5.4e-06 | 0 | 1.8e-05 | 0.85 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.A.1.8.1 | The ATP-binding Cassette (ABC) Superfamily | 0.15 | 0.34 | 1 | 0.36 | 0.64 | 1 | 0.077 | 0.38 | 1 | 0.17 | 0.45 | 1 | 0.95 | 0.98 | 1 | 0.24 | 0.7 | 1 | 0.00084 | 0.00012 | 0 | 0.00031 | 0 | 8e-04 | 0.00028 | 0 | 0.00074 | -0.15 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 9.2e-06 | 0 | 2.2e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 2.A.40.1.2 | The Nucleobase/Ascorbate Transporter (NAT) or Nucleobase:Cation Symporter-2 (NCS2) Family | 0.13 | 0.34 | 1 | 1 | 1 | 1 | 0.46 | 0.71 | 1 | 0.046 | 0.27 | 1 | 0.7 | 0.93 | 1 | 0.32 | 0.73 | 1 | 0.00013 | 1.1e-05 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 3 / 36 (8.3%) | 0 / 7 (0%) | 0 / 7 (0%) | 0 | 0 | 3e-05 | 0 | 7.9e-05 | 5.3e-06 | 0 | 1.8e-05 | -2.5 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 2.A.1.7.14 | The Major Facilitator Superfamily (MFS) | 0.024 | 0.34 | 1 | 0.13 | 0.42 | 1 | 0.16 | 0.46 | 1 | 0.0046 | 0.16 | 1 | 0.4 | 0.85 | 1 | 0.39 | 0.75 | 1 | 0.00046 | 7.7e-05 | 0 | 0.00024 | 0 | 0.00052 | 0.00015 | 0 | 0.00035 | -0.68 | 6 / 36 (17%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.B.3.1.1 | The Sugar Porin (SP) Family | 0.084 | 0.34 | 1 | 0.21 | 0.5 | 1 | 0.073 | 0.38 | 1 | 0.061 | 0.3 | 1 | 0.41 | 0.85 | 1 | 0.13 | 0.65 | 1 | 0.00042 | 5.8e-05 | 0 | 0.00015 | 0 | 0.00035 | 0.00013 | 0 | 0.00033 | -0.21 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 1.3e-05 | 0 | 3.5e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 3.A.1.14.3 | The ATP-binding Cassette (ABC) Superfamily | 0.067 | 0.34 | 1 | 0.08 | 0.38 | 1 | 0.037 | 0.37 | 1 | 0.067 | 0.3 | 1 | 0.21 | 0.74 | 1 | 0.077 | 0.6 | 1 | 0.00071 | 0.00014 | 0 | 0.00044 | 0 | 0.0011 | 0.00015 | 0 | 0.00041 | -1.6 | 7 / 36 (19%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 7e-05 | 0 | 0.00018 | 4.8 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 9.B.20.1.4 | The Putative Mg2+ Transporter-C (MgtC) Family | 0.12 | 0.34 | 1 | 0.11 | 0.4 | 1 | 0.49 | 0.74 | 1 | 0.36 | 0.64 | 1 | 0.38 | 0.84 | 1 | 0.99 | 1 | 1 | 0.00031 | 3.4e-05 | 0 | 1e-04 | 0 | 0.00022 | 5.8e-05 | 0 | 0.00015 | -0.79 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 1.1e-05 | 0 | 3.6e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.A.2.1.1 | The Glycoside-Pentoside-Hexuronide (GPH):Cation Symporter Family | 0.46 | 0.53 | 1 | 0.11 | 0.4 | 1 | 0.27 | 0.54 | 1 | 0.5 | 0.75 | 1 | 0.31 | 0.8 | 1 | 0.62 | 0.85 | 1 | 7.2e-05 | 6e-06 | 0 | 1.7e-05 | 0 | 4.5e-05 | 0 | 0 | 0 | -Inf | 3 / 36 (8.3%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 6e-06 | 0 | 2e-05 | 2.7e-06 | 0 | 8.9e-06 | -1.2 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.66.1.48 | The Multidrug/Oligosaccharidyl-lipid/Polysaccharide (MOP) Flippase Superfamily | 0.22 | 0.35 | 1 | 0.42 | 0.7 | 1 | 0.91 | 0.96 | 1 | 0.72 | 0.87 | 1 | 0.23 | 0.76 | 1 | 0.037 | 0.58 | 1 | 0.00066 | 5.5e-05 | 0 | 0.00015 | 0 | 4e-04 | 0.00013 | 0 | 0.00033 | -0.21 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 4.A.1.2.3 | The PTS Glucose-Glucoside (Glc) Family | 0.17 | 0.34 | 1 | 0.95 | 0.98 | 1 | 0.95 | 0.99 | 1 | 0.72 | 0.87 | 1 | 0.14 | 0.66 | 1 | 0.21 | 0.69 | 1 | 0.00038 | 5.2e-05 | 0 | 0.00017 | 0 | 0.00045 | 9.3e-05 | 0 | 2e-04 | -0.87 | 5 / 36 (14%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 2.6e-06 | 0 | 8.5e-06 | 2.7e-06 | 0 | 8.9e-06 | 0.054 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.3.6.4 | The Amino Acid-Polyamine-Organocation (APC) Family | 0.021 | 0.34 | 1 | 0.018 | 0.23 | 1 | 0.021 | 0.37 | 1 | 0.0073 | 0.17 | 1 | 0.069 | 0.62 | 1 | 0.058 | 0.58 | 1 | 0.00019 | 2.6e-05 | 0 | 8.7e-05 | 0 | 0.00018 | 3.9e-05 | 0 | 1e-04 | -1.2 | 5 / 36 (14%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.1.14.2 | The Major Facilitator Superfamily (MFS) | 0.05 | 0.34 | 1 | 0.0035 | 0.22 | 1 | 0.021 | 0.37 | 1 | 0.037 | 0.27 | 1 | 0.019 | 0.56 | 1 | 0.098 | 0.65 | 1 | 0.00038 | 7.3e-05 | 0 | 0.00025 | 0 | 0.00057 | 0.00011 | 0 | 0.00028 | -1.2 | 7 / 36 (19%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 5.5e-06 | 0 | 1.2e-05 | 8.1e-06 | 0 | 2.7e-05 | 0.56 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 9.B.126.2.7 | The Putative Lipid IV Exporter (YhjD) Family | 0.23 | 0.35 | 1 | 1 | 1 | 1 | 0.9 | 0.96 | 1 | 0.047 | 0.27 | 1 | 0.71 | 0.93 | 1 | 0.7 | 0.88 | 1 | 0.00011 | 1.5e-05 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 5 / 36 (14%) | 0 / 7 (0%) | 0 / 7 (0%) | 0 | 0 | 1.6e-05 | 0 | 3e-05 | 3.3e-05 | 0 | 8.7e-05 | 1 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 2.A.124.1.1 | The Lysine Exporter (LysO) Family | 0.24 | 0.36 | 1 | 0.95 | 0.98 | 1 | 0.44 | 0.69 | 1 | 0.82 | 0.93 | 1 | 0.15 | 0.67 | 1 | 0.023 | 0.53 | 1 | 0.00056 | 4.6e-05 | 0 | 0.00012 | 0 | 0.00031 | 0.00012 | 0 | 0.00031 | 0 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 8.A.50.1.1 | The AcrZ RND auxillary subunit (AcrZ) Family | 0.28 | 0.38 | 1 | 0.37 | 0.65 | 1 | 0.99 | 0.99 | 1 | 0.38 | 0.66 | 1 | 0.032 | 0.62 | 1 | 0.36 | 0.74 | 1 | 0.00012 | 9.7e-06 | 0 | 1.7e-05 | 0 | 4.5e-05 | 2.9e-05 | 0 | 7.7e-05 | 0.77 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 5.A.3.3.2 | The Prokaryotic Molybdopterin-containing Oxidoreductase (PMO) Family | 0.017 | 0.34 | 1 | 0.00014 | 0.081 | 0.081 | 0.0015 | 0.35 | 0.83 | 0.00014 | 0.081 | 0.081 | 0.0024 | 0.31 | 1 | 0.016 | 0.46 | 1 | 8e-04 | 0.00011 | 0 | 0.00037 | 5.7e-05 | 0.00083 | 2e-04 | 0 | 0.00054 | -0.89 | 5 / 36 (14%) | 4 / 7 (57%) | 1 / 7 (14%) | 0.571 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 2.A.103.1.2 | The Bacterial Murein Precursor Exporter (MPE) Family | 0.43 | 0.51 | 1 | 0.24 | 0.53 | 1 | 0.36 | 0.6 | 1 | 0.43 | 0.69 | 1 | 0.088 | 0.62 | 1 | 0.13 | 0.65 | 1 | 0.00021 | 2.3e-05 | 0 | 3.4e-05 | 0 | 8.9e-05 | 7.7e-05 | 0 | 2e-04 | 1.2 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 2.7e-06 | 0 | 8.9e-06 | 0.054 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.C.1.4.2 | The TonB-ExbB-ExbD/TolA-TolQ-TolR Outer Membrane Receptor Energizers and Stabilizers (TonB/TolA) Family | 0.33 | 0.42 | 1 | 0.12 | 0.41 | 1 | 0.32 | 0.59 | 1 | 0.32 | 0.63 | 1 | 0.21 | 0.74 | 1 | 0.42 | 0.77 | 1 | 0.00064 | 0.00016 | 0 | 8.3e-05 | 0 | 0.00013 | 0.00063 | 2.7e-05 | 0.0013 | 2.9 | 9 / 36 (25%) | 3 / 7 (43%) | 4 / 7 (57%) | 0.429 | 0.571 | 6e-06 | 0 | 2e-05 | 6.3e-05 | 0 | 0.00021 | 3.4 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.66.1.23 | The Multidrug/Oligosaccharidyl-lipid/Polysaccharide (MOP) Flippase Superfamily | 0.23 | 0.36 | 1 | 0.48 | 0.73 | 1 | 0.57 | 0.79 | 1 | 0.9 | 0.97 | 1 | 0.63 | 0.91 | 1 | 0.63 | 0.85 | 1 | 0.00048 | 5.3e-05 | 0 | 0.00015 | 0 | 4e-04 | 0.00011 | 0 | 0.00028 | -0.45 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 5.1e-06 | 0 | 1.7e-05 | 5.4e-06 | 0 | 1.8e-05 | 0.082 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 1.B.14.6.1 | The Outer Membrane Receptor (OMR) Family | 0.91 | 0.93 | 1 | 0.77 | 0.88 | 1 | 0.36 | 0.6 | 1 | 0.63 | 0.83 | 1 | 0.98 | 0.99 | 1 | 0.39 | 0.75 | 1 | 0.00011 | 2.4e-05 | 0 | 1.3e-05 | 0 | 2.3e-05 | 1.3e-05 | 0 | 3.5e-05 | 0 | 8 / 36 (22%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 3e-05 | 0 | 9.9e-05 | 3.1e-05 | 0 | 5.1e-05 | 0.047 | 1 / 11 (9.1%) | 4 / 11 (36%) | 0.0909 | 0.364 |
| 4.D.1.1.13 | The Putative Vectorial Glycosyl Polymerization (VGP) Family | 0.3 | 0.39 | 1 | 0.28 | 0.57 | 1 | 0.84 | 0.92 | 1 | 0.94 | 0.98 | 1 | 0.11 | 0.65 | 1 | 0.64 | 0.86 | 1 | 0.00026 | 2.1e-05 | 0 | 3.4e-05 | 0 | 8.9e-05 | 6.8e-05 | 0 | 0.00018 | 1 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 9.B.104.1.1 | The Rhomboid Protease (Rhomboid) Family | 0.11 | 0.34 | 1 | 0.14 | 0.44 | 1 | 0.12 | 0.43 | 1 | 0.41 | 0.68 | 1 | 0.91 | 0.98 | 1 | 0.5 | 0.82 | 1 | 0.00021 | 1.8e-05 | 0 | 6.7e-05 | 0 | 0.00018 | 1.9e-05 | 0 | 5.1e-05 | -1.8 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 3.A.1.3.2 | The ATP-binding Cassette (ABC) Superfamily | 0.067 | 0.34 | 1 | 0.091 | 0.39 | 1 | 0.29 | 0.56 | 1 | 0.063 | 0.3 | 1 | 0.083 | 0.62 | 1 | 0.34 | 0.74 | 1 | 0.00021 | 4.1e-05 | 0 | 9.9e-05 | 0 | 0.00017 | 9.7e-05 | 0 | 0.00026 | -0.029 | 7 / 36 (19%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 5.5e-06 | 0 | 1.2e-05 | 2.7e-06 | 0 | 8.9e-06 | -1 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 2.A.7.5.3 | The Drug/Metabolite Transporter (DMT) Superfamily | 0.19 | 0.34 | 1 | 0.26 | 0.55 | 1 | 0.075 | 0.38 | 1 | 0.058 | 0.29 | 1 | 0.13 | 0.66 | 1 | 0.032 | 0.58 | 1 | 0.00013 | 1.1e-05 | 0 | 0 | 0 | 0 | 2.3e-05 | 0 | 6.1e-05 | Inf | 3 / 36 (8.3%) | 0 / 7 (0%) | 1 / 7 (14%) | 0 | 0.143 | 2.1e-05 | 0 | 4.8e-05 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 1.A.23.1.3 | The Small Conductance Mechanosensitive Ion Channel (MscS) Family | 0.082 | 0.34 | 1 | 0.07 | 0.36 | 1 | 0.033 | 0.37 | 1 | 0.062 | 0.3 | 1 | 0.15 | 0.66 | 1 | 0.067 | 0.58 | 1 | 0.00051 | 0.00011 | 0 | 0.00028 | 0 | 0.00066 | 0.00026 | 0 | 0.00069 | -0.11 | 8 / 36 (22%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 6e-06 | 0 | 2e-05 | 1.9e-05 | 0 | 3.3e-05 | 1.7 | 1 / 11 (9.1%) | 3 / 11 (27%) | 0.0909 | 0.273 |
| 9.B.22.1.2 | The Leukotoxin Secretion Morphogenesis Protein C (MorC) Family | 0.16 | 0.34 | 1 | 0.17 | 0.46 | 1 | 0.51 | 0.75 | 1 | 0.59 | 0.81 | 1 | 0.32 | 0.82 | 1 | 0.83 | 0.93 | 1 | 0.00044 | 7.3e-05 | 0 | 2e-04 | 0 | 0.00035 | 9.7e-05 | 0 | 0.00026 | -1 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 3.9e-05 | 0 | 0.00011 | 8.1e-06 | 0 | 2.7e-05 | -2.3 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 2.A.3.2.5 | The Amino Acid-Polyamine-Organocation (APC) Family | 0.097 | 0.34 | 1 | 0.033 | 0.27 | 1 | 0.17 | 0.46 | 1 | 0.091 | 0.34 | 1 | 0.22 | 0.75 | 1 | 0.7 | 0.88 | 1 | 0.00075 | 8.4e-05 | 0 | 0.00025 | 0 | 0.00062 | 0.00017 | 0 | 0.00046 | -0.56 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 1.B.71.1.5 | The Proteobacterial/Verrucomicrobial Porin (PVP) Family | 0.76 | 0.81 | 1 | 0.79 | 0.9 | 1 | 0.52 | 0.75 | 1 | 0.41 | 0.68 | 1 | 0.86 | 0.96 | 1 | 0.79 | 0.91 | 1 | 0.00034 | 6.7e-05 | 0 | 1e-04 | 0 | 0.00027 | 6.8e-05 | 0 | 0.00018 | -0.56 | 7 / 36 (19%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 4.4e-05 | 0 | 0.00011 | 6.7e-05 | 0 | 0.00012 | 0.61 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 3.A.1.19.1 | The ATP-binding Cassette (ABC) Superfamily | 0.13 | 0.34 | 1 | 0.16 | 0.46 | 1 | 0.12 | 0.43 | 1 | 0.32 | 0.63 | 1 | 0.68 | 0.92 | 1 | 0.73 | 0.88 | 1 | 0.00036 | 3e-05 | 0 | 1e-04 | 0 | 0.00027 | 4.8e-05 | 0 | 0.00013 | -1.1 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.1.2.14 | The Major Facilitator Superfamily (MFS) | 0.092 | 0.34 | 1 | 0.048 | 0.29 | 1 | 0.051 | 0.37 | 1 | 0.075 | 0.31 | 1 | 0.5 | 0.87 | 1 | 0.27 | 0.73 | 1 | 0.00041 | 4.6e-05 | 0 | 0.00015 | 0 | 4e-04 | 7.7e-05 | 0 | 2e-04 | -0.96 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 9.B.42.1.3 | The ExeAB (ExeAB) Secretin Assembly/Export Complex | 0.04 | 0.34 | 1 | 0.029 | 0.25 | 1 | 0.026 | 0.37 | 1 | 0.013 | 0.23 | 1 | 0.13 | 0.66 | 1 | 0.086 | 0.61 | 1 | 4e-04 | 5.6e-05 | 0 | 0.00017 | 0 | 0.00039 | 9.7e-05 | 0 | 0.00026 | -0.81 | 5 / 36 (14%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 0 | 0 | 0 | 1.1e-05 | 0 | 3.6e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 3.A.3.5.5 | The P-type ATPase (P-ATPase) Superfamily | 0.082 | 0.34 | 1 | 0.048 | 0.29 | 1 | 0.06 | 0.37 | 1 | 0.066 | 0.3 | 1 | 0.39 | 0.84 | 1 | 0.23 | 0.7 | 1 | 0.00042 | 4.7e-05 | 0 | 0.00017 | 0 | 0.00044 | 6.8e-05 | 0 | 0.00018 | -1.3 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.37.1.2 | The Monovalent Cation:Proton Antiporter-2 (CPA2) Family | 0.018 | 0.34 | 1 | 0.0063 | 0.22 | 1 | 0.016 | 0.37 | 1 | 0.012 | 0.22 | 1 | 0.027 | 0.62 | 1 | 0.044 | 0.58 | 1 | 0.00032 | 7e-05 | 0 | 0.00026 | 1.6e-05 | 0.00057 | 6.8e-05 | 0 | 0.00018 | -1.9 | 8 / 36 (22%) | 4 / 7 (57%) | 1 / 7 (14%) | 0.571 | 0.143 | 7.7e-06 | 0 | 2.6e-05 | 1.2e-05 | 0 | 2.7e-05 | 0.64 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 3.A.1.152.9 | The ATP-binding Cassette (ABC) Superfamily | 0.42 | 0.5 | 1 | 0.19 | 0.49 | 1 | 0.71 | 0.87 | 1 | 0.59 | 0.81 | 1 | 0.1 | 0.64 | 1 | 0.4 | 0.75 | 1 | 0.00047 | 0.00012 | 0 | 0.00024 | 0 | 0.00062 | 0.00028 | 0 | 0.00069 | 0.22 | 9 / 36 (25%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 1.7e-05 | 0 | 3.7e-05 | 3.8e-05 | 0 | 7.4e-05 | 1.2 | 3 / 11 (27%) | 3 / 11 (27%) | 0.273 | 0.273 |
| 2.A.76.1.1 | The Resistance to Homoserine/Threonine (RhtB) Family | 0.11 | 0.34 | 1 | 0.51 | 0.75 | 1 | 0.082 | 0.39 | 1 | 0.21 | 0.51 | 1 | 0.6 | 0.91 | 1 | 0.081 | 0.61 | 1 | 0.00023 | 3.9e-05 | 0 | 7e-05 | 0 | 0.00014 | 6.8e-05 | 0 | 0.00018 | -0.042 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 3.9e-05 | 0 | 7.7e-05 | Inf | 0 / 11 (0%) | 3 / 11 (27%) | 0 | 0.273 |
| 2.A.1.2.7 | The Major Facilitator Superfamily (MFS) | 0.25 | 0.36 | 1 | 0.4 | 0.68 | 1 | 0.95 | 0.99 | 1 | 0.69 | 0.87 | 1 | 0.16 | 0.69 | 1 | 0.77 | 0.9 | 1 | 0.00032 | 2.7e-05 | 0 | 5e-05 | 0 | 0.00013 | 7.7e-05 | 0 | 2e-04 | 0.62 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.66.4.3 | The Multidrug/Oligosaccharidyl-lipid/Polysaccharide (MOP) Flippase Superfamily | 0.19 | 0.34 | 1 | 0.17 | 0.47 | 1 | 0.56 | 0.78 | 1 | 0.75 | 0.89 | 1 | 0.51 | 0.87 | 1 | 0.11 | 0.65 | 1 | 5e-04 | 4.2e-05 | 0 | 0.00013 | 0 | 0.00036 | 7.7e-05 | 0 | 2e-04 | -0.76 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.A.1.10.2 | The Major Facilitator Superfamily (MFS) | 0.09 | 0.34 | 1 | 0.098 | 0.39 | 1 | 0.07 | 0.38 | 1 | 0.15 | 0.42 | 1 | 0.61 | 0.91 | 1 | 0.29 | 0.73 | 1 | 0.00045 | 4.9e-05 | 0 | 0.00017 | 0 | 0.00044 | 6.8e-05 | 0 | 0.00018 | -1.3 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 1e-05 | 0 | 3.4e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.95.1.7 | The 6 TMS Neutral Amino Acid Transporter (NAAT) Family | 0.19 | 0.34 | 1 | 0.11 | 0.4 | 1 | 0.21 | 0.48 | 1 | 0.76 | 0.89 | 1 | 0.032 | 0.62 | 1 | 0.069 | 0.58 | 1 | 0.00027 | 2.2e-05 | 0 | 5e-05 | 0 | 0.00013 | 6.4e-05 | 0 | 0.00013 | 0.36 | 3 / 36 (8.3%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 4.A.1.1.1 | The PTS Glucose-Glucoside (Glc) Family | 0.085 | 0.34 | 1 | 0.11 | 0.4 | 1 | 0.042 | 0.37 | 1 | 0.08 | 0.32 | 1 | 0.44 | 0.86 | 1 | 0.13 | 0.65 | 1 | 0.00062 | 8.7e-05 | 0 | 0.00026 | 0 | 0.00066 | 0.00015 | 0 | 0.00038 | -0.79 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.3e-05 | 0 | 6.8e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 2.A.1.5.1 | The Major Facilitator Superfamily (MFS) | 0.19 | 0.34 | 1 | 0.98 | 1 | 1 | 0.42 | 0.67 | 1 | 0.49 | 0.73 | 1 | 0.14 | 0.66 | 1 | 0.71 | 0.88 | 1 | 0.00033 | 2.7e-05 | 0 | 6.7e-05 | 0 | 0.00018 | 6.8e-05 | 0 | 0.00018 | 0.021 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.1.21.6 | The Major Facilitator Superfamily (MFS) | 0.87 | 0.9 | 1 | 0.77 | 0.88 | 1 | 0.42 | 0.67 | 1 | 0.49 | 0.74 | 1 | 0.49 | 0.87 | 1 | 0.27 | 0.73 | 1 | 0.00013 | 1.4e-05 | 0 | 2e-05 | 0 | 5.2e-05 | 1.9e-05 | 0 | 4e-05 | -0.074 | 4 / 36 (11%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 2.2e-05 | 0 | 7.3e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 3.A.1.3.3 | The ATP-binding Cassette (ABC) Superfamily | 0.39 | 0.47 | 1 | 0.9 | 0.96 | 1 | 0.54 | 0.76 | 1 | 0.64 | 0.84 | 1 | 0.64 | 0.92 | 1 | 0.27 | 0.73 | 1 | 0.00071 | 9.8e-05 | 0 | 0.00024 | 0 | 0.00062 | 0.00022 | 0 | 0.00059 | -0.13 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.5e-05 | 0 | 7.2e-05 | 5.4e-06 | 0 | 1.8e-05 | -2.2 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 3.A.1.9.1 | The ATP-binding Cassette (ABC) Superfamily | 0.61 | 0.66 | 1 | 0.26 | 0.55 | 1 | 0.2 | 0.48 | 1 | 0.41 | 0.68 | 1 | 0.23 | 0.76 | 1 | 0.14 | 0.65 | 1 | 0.00046 | 7.7e-05 | 0 | 0.00012 | 0 | 0.00031 | 0.00021 | 0 | 0.00056 | 0.81 | 6 / 36 (17%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.7e-05 | 0 | 7.3e-05 | 1.3e-05 | 0 | 3.5e-05 | -1.1 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 9.B.146.1.6 | The Putative Undecaprenyl-phosphate N-Acetylglucosaminyl Transferase (MurG) Family | 0.094 | 0.34 | 1 | 0.098 | 0.39 | 1 | 0.086 | 0.4 | 1 | 0.042 | 0.27 | 1 | 0.37 | 0.84 | 1 | 0.29 | 0.73 | 1 | 0.00059 | 6.6e-05 | 0 | 0.00018 | 0 | 0.00044 | 0.00015 | 0 | 0.00041 | -0.26 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.25.1.10 | The Alanine or Glycine:Cation Symporter (AGCS) Family | 0.14 | 0.34 | 1 | 0.46 | 0.72 | 1 | 0.1 | 0.42 | 1 | 0.28 | 0.6 | 1 | 0.93 | 0.98 | 1 | 0.26 | 0.72 | 1 | 3e-04 | 4.2e-05 | 0 | 1e-04 | 0 | 0.00027 | 8.7e-05 | 0 | 0.00023 | -0.2 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 1.6e-05 | 0 | 4.3e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 1.B.17.1.1 | The Outer Membrane Factor (OMF) Family | 0.024 | 0.34 | 1 | 0.019 | 0.23 | 1 | 0.058 | 0.37 | 1 | 0.0023 | 0.15 | 1 | 0.035 | 0.62 | 1 | 0.11 | 0.65 | 1 | 0.00055 | 6.1e-05 | 0 | 0.00018 | 0 | 0.00035 | 0.00014 | 0 | 0.00036 | -0.36 | 4 / 36 (11%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 2.A.1.3.2 | The Major Facilitator Superfamily (MFS) | 0.3 | 0.4 | 1 | 0.4 | 0.68 | 1 | 0.93 | 0.98 | 1 | 0.71 | 0.87 | 1 | 0.6 | 0.91 | 1 | 0.19 | 0.68 | 1 | 0.00048 | 6.6e-05 | 0 | 0.00019 | 0 | 0.00049 | 0.00014 | 0 | 0.00036 | -0.44 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 1e-05 | 0 | 2.5e-05 | 2.7e-06 | 0 | 8.9e-06 | -1.9 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 8.A.5.1.6 | The Voltage-gated K+ Channel β-subunit (Kvβ) Family | 0.54 | 0.6 | 1 | 0.49 | 0.74 | 1 | 0.83 | 0.92 | 1 | 0.31 | 0.63 | 1 | 0.27 | 0.78 | 1 | 0.53 | 0.82 | 1 | 0.00013 | 1.5e-05 | 0 | 1.7e-05 | 0 | 4.5e-05 | 3.9e-05 | 0 | 1e-04 | 1.2 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1e-05 | 0 | 3.4e-05 | 1.9 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.A.1.105.16 | The ATP-binding Cassette (ABC) Superfamily | 0.12 | 0.34 | 1 | 0.16 | 0.45 | 1 | 0.079 | 0.39 | 1 | 0.13 | 0.39 | 1 | 0.78 | 0.94 | 1 | 0.34 | 0.73 | 1 | 0.00052 | 5.8e-05 | 0 | 0.00017 | 0 | 0.00044 | 0.00012 | 0 | 0.00031 | -0.5 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 8.1e-06 | 0 | 2.7e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 3.A.1.2.25 | The ATP-binding Cassette (ABC) Superfamily | 0.097 | 0.34 | 1 | 0.061 | 0.33 | 1 | 0.14 | 0.43 | 1 | 0.06 | 0.29 | 1 | 0.1 | 0.64 | 1 | 0.21 | 0.69 | 1 | 0.00068 | 0.00017 | 0 | 0.00044 | 3.2e-05 | 0.0011 | 0.00039 | 0 | 0.001 | -0.17 | 9 / 36 (25%) | 4 / 7 (57%) | 1 / 7 (14%) | 0.571 | 0.143 | 1.6e-05 | 0 | 3.9e-05 | 1.5e-05 | 0 | 3.3e-05 | -0.093 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 9.B.34.1.4 | The Kinase/Phosphatase/Cyclic-GMP Synthase/Cyclic di-GMP Hydrolase (KPSH) Family | 0.076 | 0.34 | 1 | 0.052 | 0.31 | 1 | 0.075 | 0.38 | 1 | 0.06 | 0.29 | 1 | 0.31 | 0.8 | 1 | 0.2 | 0.69 | 1 | 0.00051 | 5.7e-05 | 0 | 0.00022 | 0 | 0.00058 | 6.8e-05 | 0 | 0.00018 | -1.7 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 9.B.169.4.2 | The Integral Membrane Protein (8 -10 TMSs) YeiB or DUF418 (YeiB) Family | 0.19 | 0.34 | 1 | 0.02 | 0.23 | 1 | 0.062 | 0.37 | 1 | 0.95 | 0.98 | 1 | 0.0015 | 0.31 | 0.84 | 0.0056 | 0.46 | 1 | 1e-04 | 1.1e-05 | 0 | 1.4e-05 | 0 | 3.2e-05 | 4.3e-05 | 0 | 9.2e-05 | 1.6 | 4 / 36 (11%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 9.B.97.1.1 | The Acyltransferase-3/Putative Acetyl-CoA Transporter (ATAT) Family | 0.17 | 0.34 | 1 | 0.37 | 0.65 | 1 | 0.17 | 0.46 | 1 | 0.33 | 0.63 | 1 | 0.26 | 0.78 | 1 | 0.92 | 0.98 | 1 | 0.00057 | 4.8e-05 | 0 | 0.00013 | 0 | 0.00036 | 0.00011 | 0 | 0.00028 | -0.24 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.7.3.19 | The Drug/Metabolite Transporter (DMT) Superfamily | 0.11 | 0.34 | 1 | 0.15 | 0.45 | 1 | 0.12 | 0.43 | 1 | 0.28 | 0.6 | 1 | 0.92 | 0.98 | 1 | 0.4 | 0.75 | 1 | 0.0011 | 9.3e-05 | 0 | 0.00035 | 0 | 0.00094 | 9.7e-05 | 0 | 0.00026 | -1.9 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 1.9e-05 | 0 | 6.2e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 9.B.14.3.6 | The Putative Heme Handling Protein (HHP) Family | 0.55 | 0.61 | 1 | 0.42 | 0.69 | 1 | 1 | 1 | 1 | 0.22 | 0.52 | 1 | 0.86 | 0.96 | 1 | 0.29 | 0.73 | 1 | 0.00023 | 3.8e-05 | 0 | 1e-04 | 0 | 0.00027 | 4.8e-05 | 0 | 0.00013 | -1.1 | 6 / 36 (17%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2e-05 | 0 | 4.8e-05 | 1.1e-05 | 0 | 3.6e-05 | -0.86 | 3 / 11 (27%) | 1 / 11 (9.1%) | 0.273 | 0.0909 |
| 3.D.1.1.1 | The H+ or Na+-translocating NADH Dehydrogenase (NDH) Family | 0.049 | 0.34 | 1 | 0.011 | 0.23 | 1 | 0.012 | 0.37 | 1 | 0.026 | 0.27 | 1 | 0.084 | 0.62 | 1 | 0.064 | 0.58 | 1 | 0.0011 | 0.00034 | 0 | 0.00099 | 6.4e-05 | 0.0023 | 0.00062 | 0 | 0.0016 | -0.68 | 11 / 36 (31%) | 4 / 7 (57%) | 2 / 7 (29%) | 0.571 | 0.286 | 1.7e-05 | 0 | 3.7e-05 | 5.1e-05 | 0 | 0.00011 | 1.6 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 8.A.46.2.2 | The Glycan-binding Protein (SusD) Family | 0.84 | 0.87 | 1 | 0.74 | 0.88 | 1 | 0.87 | 0.94 | 1 | 0.37 | 0.65 | 1 | 0.51 | 0.87 | 1 | 0.7 | 0.88 | 1 | 0.00013 | 1.8e-05 | 0 | 8.1e-06 | 0 | 2.1e-05 | 1.8e-05 | 0 | 4.7e-05 | 1.2 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.4e-05 | 0 | 7.9e-05 | 1.8e-05 | 0 | 5e-05 | -0.42 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 1.C.113.1.10 | The Hly III (Hly III) Family | 0.22 | 0.35 | 1 | 0.99 | 1 | 1 | 0.63 | 0.83 | 1 | 0.84 | 0.94 | 1 | 0.19 | 0.72 | 1 | 0.1 | 0.65 | 1 | 2e-04 | 2.2e-05 | 0 | 6.7e-05 | 0 | 0.00018 | 4.3e-05 | 0 | 1e-04 | -0.64 | 4 / 36 (11%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 3e-06 | 0 | 9.9e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.A.1.2.19 | The Major Facilitator Superfamily (MFS) | 0.057 | 0.34 | 1 | 0.053 | 0.31 | 1 | 0.035 | 0.37 | 1 | 0.12 | 0.38 | 1 | 0.16 | 0.69 | 1 | 0.09 | 0.62 | 1 | 0.00017 | 3.3e-05 | 0 | 9.8e-05 | 0 | 0.00022 | 2.9e-05 | 0 | 7.7e-05 | -1.8 | 7 / 36 (19%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 3e-06 | 0 | 9.9e-06 | 2.3e-05 | 0 | 6.8e-05 | 2.9 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 1.B.48.1.1 | The Curli Fiber Subunit Porin. CgsA. CsgG (CsgG) Family | 0.34 | 0.42 | 1 | 0.57 | 0.77 | 1 | 0.98 | 0.99 | 1 | 0.99 | 0.99 | 1 | 0.42 | 0.85 | 1 | 0.77 | 0.9 | 1 | 0.00042 | 4.7e-05 | 0 | 8.4e-05 | 0 | 0.00022 | 0.00013 | 0 | 0.00033 | 0.63 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1.6e-05 | 0 | 5.4e-05 | 2.6 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.1.21.1 | The Major Facilitator Superfamily (MFS) | 0.83 | 0.87 | 1 | 0.43 | 0.7 | 1 | 0.14 | 0.43 | 1 | 0.48 | 0.73 | 1 | 0.25 | 0.77 | 1 | 0.19 | 0.68 | 1 | 8e-04 | 0.00024 | 0 | 0.00011 | 0 | 2e-04 | 7.1e-05 | 0 | 0.00019 | -0.63 | 11 / 36 (31%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0.00018 | 0 | 0.00042 | 5e-04 | 0 | 0.0014 | 1.5 | 4 / 11 (36%) | 4 / 11 (36%) | 0.364 | 0.364 |
| 3.A.1.106.13 | The ATP-binding Cassette (ABC) Superfamily | 0.22 | 0.35 | 1 | 0.26 | 0.55 | 1 | 0.6 | 0.82 | 1 | 0.77 | 0.89 | 1 | 0.87 | 0.97 | 1 | 0.69 | 0.88 | 1 | 0.00054 | 9e-05 | 0 | 0.00025 | 0 | 0.00067 | 0.00018 | 0 | 0.00049 | -0.47 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 9.9e-06 | 0 | 2.5e-05 | 5.4e-06 | 0 | 1.8e-05 | -0.87 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 2.A.86.2.1 | The Autoinducer-2 Exporter (AI-2E) Family (Formerly the PerM Family. TC #9.B.22) | 0.14 | 0.34 | 1 | 0.21 | 0.51 | 1 | 0.13 | 0.43 | 1 | 0.35 | 0.64 | 1 | 0.73 | 0.94 | 1 | 0.62 | 0.85 | 1 | 0.00043 | 3.6e-05 | 0 | 0.00012 | 0 | 0.00031 | 5.8e-05 | 0 | 0.00015 | -1 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 3.A.1.3.29 | The ATP-binding Cassette (ABC) Superfamily | 0.15 | 0.34 | 1 | 0.069 | 0.36 | 1 | 0.23 | 0.51 | 1 | 0.31 | 0.63 | 1 | 0.28 | 0.78 | 1 | 0.69 | 0.88 | 1 | 0.00072 | 0.00012 | 0 | 0.00032 | 0 | 0.00079 | 0.00027 | 0 | 0.00072 | -0.25 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 8.5e-06 | 0 | 2.1e-05 | 8.1e-06 | 0 | 2.7e-05 | -0.07 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 2.A.26.1.10 | The Branched Chain Amino Acid:Cation Symporter (LIVCS) Family | 0.057 | 0.34 | 1 | 0.0065 | 0.22 | 1 | 0.059 | 0.37 | 1 | 0.031 | 0.27 | 1 | 0.07 | 0.62 | 1 | 0.38 | 0.75 | 1 | 0.00036 | 5e-05 | 0 | 0.00017 | 0 | 4e-04 | 8.7e-05 | 0 | 0.00023 | -0.97 | 5 / 36 (14%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 4.A.1.2.2 | The PTS Glucose-Glucoside (Glc) Family | 0.15 | 0.34 | 1 | 0.22 | 0.51 | 1 | 0.2 | 0.48 | 1 | 0.35 | 0.64 | 1 | 0.84 | 0.96 | 1 | 0.7 | 0.88 | 1 | 0.00059 | 8.2e-05 | 0 | 0.00024 | 0 | 0.00062 | 0.00016 | 0 | 0.00044 | -0.58 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1e-05 | 0 | 3.4e-05 | 1.9 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.A.1.5.32 | The ATP-binding Cassette (ABC) Superfamily | 0.44 | 0.52 | 1 | 0.36 | 0.64 | 1 | 0.42 | 0.67 | 1 | 0.056 | 0.29 | 1 | 0.21 | 0.74 | 1 | 0.27 | 0.72 | 1 | 7.7e-05 | 6.4e-06 | 0 | 0 | 0 | 0 | 9.7e-06 | 0 | 2.6e-05 | Inf | 3 / 36 (8.3%) | 0 / 7 (0%) | 1 / 7 (14%) | 0 | 0.143 | 8.8e-06 | 0 | 2.9e-05 | 6e-06 | 0 | 2e-05 | -0.55 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.A.7.10.1 | The Type IV (Conjugal DNA-Protein Transfer or VirB) Secretory Pathway (IVSP) Family | 0.13 | 0.34 | 1 | 0.47 | 0.73 | 1 | 0.26 | 0.52 | 1 | 0.84 | 0.94 | 1 | 0.92 | 0.98 | 1 | 0.47 | 0.81 | 1 | 0.00019 | 2.1e-05 | 0 | 6.8e-05 | 0 | 0.00018 | 1.5e-05 | 0 | 4e-05 | -2.2 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 1.5e-05 | 0 | 3.6e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 3.A.1.5.25 | The ATP-binding Cassette (ABC) Superfamily | 0.024 | 0.34 | 1 | 0.22 | 0.51 | 1 | 0.019 | 0.37 | 1 | 0.0071 | 0.17 | 1 | 0.061 | 0.62 | 1 | 0.0049 | 0.46 | 1 | 0.00021 | 5.3e-05 | 0 | 0 | 0 | 0 | 2.9e-05 | 0 | 5.3e-05 | Inf | 9 / 36 (25%) | 0 / 7 (0%) | 2 / 7 (29%) | 0 | 0.286 | 0.00012 | 0 | 0.00019 | 3.3e-05 | 0 | 8.7e-05 | -1.9 | 5 / 11 (45%) | 2 / 11 (18%) | 0.455 | 0.182 |
| 2.A.1.3.51 | The Major Facilitator Superfamily (MFS) | 0.18 | 0.34 | 1 | 0.68 | 0.83 | 1 | 0.084 | 0.39 | 1 | 0.42 | 0.68 | 1 | 0.67 | 0.92 | 1 | 0.25 | 0.71 | 1 | 0.00079 | 0.00011 | 0 | 0.00029 | 0 | 0.00076 | 0.00024 | 0 | 0.00064 | -0.27 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 2.2e-05 | 0 | 4.6e-05 | Inf | 0 / 11 (0%) | 3 / 11 (27%) | 0 | 0.273 |
| 9.B.51.1.1 | The Uncharacterized DUF202/YidH (YidH) Family | 0.03 | 0.34 | 1 | 0.027 | 0.25 | 1 | 0.038 | 0.37 | 1 | 0.027 | 0.27 | 1 | 0.13 | 0.66 | 1 | 0.13 | 0.65 | 1 | 0.00021 | 2.3e-05 | 0 | 8.4e-05 | 0 | 0.00018 | 2.9e-05 | 0 | 7.7e-05 | -1.5 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.42.1.3 | The Hydroxy/Aromatic Amino Acid Permease (HAAAP) Family | 0.12 | 0.34 | 1 | 0.23 | 0.51 | 1 | 0.14 | 0.43 | 1 | 0.2 | 0.51 | 1 | 0.51 | 0.87 | 1 | 0.3 | 0.73 | 1 | 0.00029 | 4.9e-05 | 0 | 0.00013 | 0 | 0.00031 | 9.7e-05 | 0 | 0.00026 | -0.42 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1.3e-05 | 0 | 3.5e-05 | 2.3 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 2.A.85.6.1 | The Aromatic Acid Exporter (ArAE) Family | 0.012 | 0.34 | 1 | 0.00031 | 0.086 | 0.17 | 0.0024 | 0.35 | 1 | 0.0012 | 0.15 | 0.68 | 5e-04 | 0.28 | 0.28 | 0.0041 | 0.46 | 1 | 0.00044 | 8.6e-05 | 0 | 0.00033 | 8.1e-05 | 0.00074 | 8.7e-05 | 0 | 0.00023 | -1.9 | 7 / 36 (19%) | 4 / 7 (57%) | 1 / 7 (14%) | 0.571 | 0.143 | 6e-06 | 0 | 2e-05 | 8.1e-06 | 0 | 2.7e-05 | 0.43 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.A.3.32.7 | The P-type ATPase (P-ATPase) Superfamily | 0.96 | 0.96 | 1 | 0.81 | 0.91 | 1 | 0.62 | 0.83 | 1 | 0.51 | 0.75 | 1 | 1 | 1 | 1 | 0.53 | 0.82 | 1 | 7e-04 | 0.00021 | 0 | 0.00027 | 0 | 0.00046 | 0.00025 | 0 | 0.00058 | -0.11 | 11 / 36 (31%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 3e-04 | 0 | 8e-04 | 7.4e-05 | 0 | 0.00016 | -2 | 4 / 11 (36%) | 3 / 11 (27%) | 0.364 | 0.273 |
| 2.A.118.1.8 | The Basic Amino Acid Antiporter (ArcD) Family | 0.14 | 0.34 | 1 | 0.27 | 0.56 | 1 | 0.047 | 0.37 | 1 | 0.47 | 0.73 | 1 | 0.97 | 0.99 | 1 | 0.13 | 0.65 | 1 | 0.00031 | 4.2e-05 | 0 | 0.00013 | 0 | 0.00036 | 4.8e-05 | 0 | 0.00013 | -1.4 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 2.3e-05 | 0 | 4.1e-05 | Inf | 0 / 11 (0%) | 3 / 11 (27%) | 0 | 0.273 |
| 3.A.1.17.10 | The ATP-binding Cassette (ABC) Superfamily | 0.17 | 0.34 | 1 | 0.52 | 0.75 | 1 | 0.12 | 0.43 | 1 | 0.42 | 0.68 | 1 | 0.59 | 0.91 | 1 | 0.47 | 0.81 | 1 | 0.00049 | 5.5e-05 | 0 | 0.00015 | 0 | 4e-04 | 0.00012 | 0 | 0.00031 | -0.32 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 9.5e-06 | 0 | 2.3e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 1.A.14.2.1 | The Calcium Transporter A (CaTA) (formerly the Testis-Enhanced Gene Transfer (TEGT) Family | 0.15 | 0.34 | 1 | 0.67 | 0.83 | 1 | 0.3 | 0.57 | 1 | 0.16 | 0.43 | 1 | 0.92 | 0.98 | 1 | 0.52 | 0.82 | 1 | 0.00034 | 3.7e-05 | 0 | 8.6e-05 | 0 | 0.00022 | 9.7e-05 | 0 | 0.00026 | 0.17 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.C.1.2.1 | The TonB-ExbB-ExbD/TolA-TolQ-TolR Outer Membrane Receptor Energizers and Stabilizers (TonB/TolA) Family | 0.15 | 0.34 | 1 | 0.11 | 0.4 | 1 | 0.2 | 0.48 | 1 | 0.26 | 0.59 | 1 | 0.79 | 0.94 | 1 | 0.95 | 1 | 1 | 0.00071 | 9.9e-05 | 0 | 0.00029 | 0 | 0.00076 | 0.00021 | 0 | 0.00056 | -0.47 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 2.7e-06 | 0 | 8.9e-06 | 0.054 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.A.1.5.24 | The ATP-binding Cassette (ABC) Superfamily | 0.073 | 0.34 | 1 | 0.094 | 0.39 | 1 | 0.18 | 0.48 | 1 | 0.015 | 0.24 | 1 | 0.5 | 0.87 | 1 | 0.68 | 0.88 | 1 | 0.00062 | 8.6e-05 | 0 | 0.00023 | 0 | 0.00057 | 0.00021 | 0 | 0.00054 | -0.13 | 5 / 36 (14%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 9.B.27.1.2 | The DedA or YdjX-Z (DedA) Family | 0.12 | 0.34 | 1 | 0.19 | 0.48 | 1 | 0.13 | 0.43 | 1 | 0.41 | 0.68 | 1 | 0.97 | 0.99 | 1 | 0.5 | 0.82 | 1 | 0.00028 | 2.4e-05 | 0 | 8.4e-05 | 0 | 0.00022 | 2.9e-05 | 0 | 7.7e-05 | -1.5 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 4.A.2.1.25 | The PTS Fructose-Mannitol (Fru) Family | 0.58 | 0.64 | 1 | 0.53 | 0.76 | 1 | 0.96 | 0.99 | 1 | 0.49 | 0.73 | 1 | 0.94 | 0.98 | 1 | 0.62 | 0.85 | 1 | 8.2e-05 | 6.8e-06 | 0 | 1.7e-05 | 0 | 4.5e-05 | 3.9e-06 | 0 | 1e-05 | -2.1 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 8.9e-06 | 0 | 3e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.A.1.6.2 | The Major Facilitator Superfamily (MFS) | 0.028 | 0.34 | 1 | 0.017 | 0.23 | 1 | 0.056 | 0.37 | 1 | 0.014 | 0.23 | 1 | 0.059 | 0.62 | 1 | 0.16 | 0.67 | 1 | 0.00098 | 8.2e-05 | 0 | 0.00035 | 0 | 0.00084 | 6.8e-05 | 0 | 0.00018 | -2.4 | 3 / 36 (8.3%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 1.B.14.3.1 | The Outer Membrane Receptor (OMR) Family | 0.13 | 0.34 | 1 | 0.26 | 0.55 | 1 | 0.17 | 0.46 | 1 | 0.22 | 0.52 | 1 | 0.5 | 0.87 | 1 | 0.33 | 0.73 | 1 | 0.00054 | 9e-05 | 0 | 0.00024 | 0 | 0.00057 | 0.00018 | 0 | 0.00049 | -0.42 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 5.1e-06 | 0 | 1.7e-05 | 2.3e-05 | 0 | 6.8e-05 | 2.2 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 2.A.46.1.9 | The Benzoate:H+ Symporter (BenE) Family | 0.25 | 0.36 | 1 | 0.62 | 0.79 | 1 | 0.31 | 0.58 | 1 | 0.99 | 0.99 | 1 | 0.69 | 0.93 | 1 | 0.73 | 0.88 | 1 | 0.00023 | 3.2e-05 | 0 | 8.4e-05 | 0 | 0.00022 | 5.8e-05 | 0 | 0.00015 | -0.53 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1.3e-05 | 0 | 3.5e-05 | 2.3 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 5.A.3.3.1 | The Prokaryotic Molybdopterin-containing Oxidoreductase (PMO) Family | 0.21 | 0.35 | 1 | 0.78 | 0.89 | 1 | 0.63 | 0.83 | 1 | 0.57 | 0.79 | 1 | 0.83 | 0.95 | 1 | 0.36 | 0.74 | 1 | 0.00076 | 0.00013 | 0 | 0.00032 | 0 | 0.00075 | 0.00026 | 0 | 0.00058 | -0.3 | 6 / 36 (17%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 4.6e-05 | 0 | 0.00014 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 2.A.3.1.15 | The Amino Acid-Polyamine-Organocation (APC) Family | 0.21 | 0.35 | 1 | 0.31 | 0.6 | 1 | 0.89 | 0.96 | 1 | 0.71 | 0.87 | 1 | 0.28 | 0.78 | 1 | 0.047 | 0.58 | 1 | 0.00063 | 5.3e-05 | 0 | 0.00015 | 0 | 4e-04 | 0.00012 | 0 | 0.00031 | -0.32 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 9.B.127.1.1 | The DUF2919 (PF11143) Family | 0.16 | 0.34 | 1 | 0.24 | 0.53 | 1 | 0.25 | 0.52 | 1 | 0.72 | 0.87 | 1 | 0.76 | 0.94 | 1 | 0.71 | 0.88 | 1 | 0.00014 | 1.9e-05 | 0 | 5.3e-05 | 0 | 0.00013 | 2.9e-05 | 0 | 7.7e-05 | -0.87 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 6.8e-06 | 0 | 2.3e-05 | 1.4 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.A.1.113.3 | The ATP-binding Cassette (ABC) Superfamily | 0.62 | 0.68 | 1 | 0.9 | 0.96 | 1 | 0.19 | 0.48 | 1 | 0.2 | 0.5 | 1 | 0.6 | 0.91 | 1 | 0.092 | 0.62 | 1 | 0.00039 | 6.5e-05 | 0 | 0.00013 | 0 | 0.00036 | 0.00015 | 0 | 0.00038 | 0.21 | 6 / 36 (17%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3e-05 | 0 | 7.3e-05 | 2.7e-06 | 0 | 8.9e-06 | -3.5 | 3 / 11 (27%) | 1 / 11 (9.1%) | 0.273 | 0.0909 |
| 2.A.1.1.114 | The Major Facilitator Superfamily (MFS) | 0.098 | 0.34 | 1 | 0.044 | 0.29 | 1 | 0.26 | 0.52 | 1 | 0.3 | 0.62 | 1 | 0.31 | 0.8 | 1 | 0.9 | 0.97 | 1 | 0.00038 | 4.2e-05 | 0 | 0.00015 | 0 | 0.00035 | 5.8e-05 | 0 | 0.00015 | -1.4 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 7.9e-06 | 0 | 2.6e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 5.B.7.1.1 | The YedZ (YedZ) Family | 0.23 | 0.36 | 1 | 0.38 | 0.65 | 1 | 0.99 | 0.99 | 1 | 0.86 | 0.95 | 1 | 0.32 | 0.83 | 1 | 0.057 | 0.58 | 1 | 0.00028 | 2.3e-05 | 0 | 6.7e-05 | 0 | 0.00018 | 4.8e-05 | 0 | 0.00013 | -0.48 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.A.2.2.1 | The Glycoside-Pentoside-Hexuronide (GPH):Cation Symporter Family | 0.11 | 0.34 | 1 | 0.21 | 0.51 | 1 | 0.13 | 0.43 | 1 | 0.041 | 0.27 | 1 | 0.024 | 0.61 | 1 | 0.055 | 0.58 | 1 | 0.00032 | 0.00021 | 9.8e-05 | 0.00012 | 0 | 0.00027 | 0.00019 | 0.00013 | 0.00018 | 0.66 | 24 / 36 (67%) | 2 / 7 (29%) | 6 / 7 (86%) | 0.286 | 0.857 | 0.00038 | 0.00013 | 0.00073 | 0.00012 | 1e-04 | 0.00013 | -1.7 | 7 / 11 (64%) | 9 / 11 (82%) | 0.636 | 0.818 |
| 1.B.22.1.3 | The Outer Bacterial Membrane Secretin (Secretin) Family | 0.16 | 0.34 | 1 | 0.029 | 0.25 | 1 | 0.15 | 0.43 | 1 | 0.4 | 0.68 | 1 | 0.16 | 0.69 | 1 | 0.49 | 0.82 | 1 | 0.00062 | 0.00015 | 0 | 0.00044 | 0 | 0.0011 | 3e-04 | 0 | 0.00079 | -0.55 | 9 / 36 (25%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 2.1e-05 | 0 | 4.1e-05 | 1.6e-05 | 0 | 3.7e-05 | -0.39 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 3.A.1.11.1 | The ATP-binding Cassette (ABC) Superfamily | 0.52 | 0.58 | 1 | 0.77 | 0.88 | 1 | 0.47 | 0.72 | 1 | 0.7 | 0.87 | 1 | 0.64 | 0.92 | 1 | 0.44 | 0.79 | 1 | 0.00074 | 8.2e-05 | 0 | 0.00015 | 0 | 4e-04 | 0.00019 | 0 | 0.00051 | 0.34 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 4.4e-05 | 0 | 0.00015 | 5.4e-06 | 0 | 1.8e-05 | -3 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.A.1.31.1 | The ATP-binding Cassette (ABC) Superfamily | 0.25 | 0.36 | 1 | 0.37 | 0.65 | 1 | 0.65 | 0.84 | 1 | 0.054 | 0.29 | 1 | 0.29 | 0.78 | 1 | 0.33 | 0.73 | 1 | 0.0016 | 0.0012 | 0.00087 | 0.00071 | 0.00068 | 0.00076 | 0.0012 | 0.00084 | 0.0013 | 0.76 | 28 / 36 (78%) | 4 / 7 (57%) | 4 / 7 (57%) | 0.571 | 0.571 | 0.0014 | 0.00077 | 0.002 | 0.0015 | 0.0012 | 0.0013 | 0.1 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 8.A.1.1.1 | The Membrane Fusion Protein (MFP) Family | 0.13 | 0.34 | 1 | 0.048 | 0.29 | 1 | 0.24 | 0.52 | 1 | 0.28 | 0.6 | 1 | 0.57 | 0.91 | 1 | 0.62 | 0.85 | 1 | 0.00047 | 5.3e-05 | 0 | 0.00017 | 0 | 0.00044 | 9.7e-05 | 0 | 0.00026 | -0.81 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.A.113.1.1 | The Nickel/cobalt Transporter (NicO) Family | 0.088 | 0.34 | 1 | 0.51 | 0.75 | 1 | 0.35 | 0.6 | 1 | 0.07 | 0.3 | 1 | 0.45 | 0.86 | 1 | 0.29 | 0.73 | 1 | 0.00055 | 6.1e-05 | 0 | 0.00014 | 0 | 0.00027 | 0.00016 | 0 | 0.00044 | 0.19 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 1e-05 | 0 | 3.4e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.40.5.1 | The Nucleobase/Ascorbate Transporter (NAT) or Nucleobase:Cation Symporter-2 (NCS2) Family | 0.28 | 0.38 | 1 | 0.099 | 0.39 | 1 | 0.16 | 0.45 | 1 | 0.94 | 0.98 | 1 | 0.02 | 0.56 | 1 | 0.025 | 0.55 | 1 | 4e-04 | 4.4e-05 | 0 | 1e-04 | 0 | 0.00027 | 0.00012 | 0 | 0.00031 | 0.26 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3e-06 | 0 | 9.9e-06 | 2.7e-06 | 0 | 8.9e-06 | -0.15 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.49.5.1 | The Chloride Carrier/Channel (ClC) Family | 0.052 | 0.34 | 1 | 0.075 | 0.38 | 1 | 0.069 | 0.37 | 1 | 0.034 | 0.27 | 1 | 0.24 | 0.77 | 1 | 0.17 | 0.67 | 1 | 0.00055 | 6.1e-05 | 0 | 0.00019 | 0 | 4e-04 | 0.00012 | 0 | 0.00031 | -0.66 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 8.1e-06 | 0 | 2.7e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.C.11.1.3 | The Pore-forming RTX Toxin (RTX-toxin) Family | 0.2 | 0.34 | 1 | 1 | 1 | 1 | 0.26 | 0.52 | 1 | 0.045 | 0.27 | 1 | 0.33 | 0.83 | 1 | 0.036 | 0.58 | 1 | 0.00021 | 1.7e-05 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 3 / 36 (8.3%) | 0 / 7 (0%) | 0 / 7 (0%) | 0 | 0 | 4.6e-05 | 0 | 0.00014 | 1e-05 | 0 | 3.4e-05 | -2.2 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 9.A.8.1.1 | The Ferrous Iron Uptake (FeoB) Family | 0.25 | 0.36 | 1 | 0.83 | 0.92 | 1 | 0.97 | 0.99 | 1 | 0.78 | 0.89 | 1 | 0.49 | 0.87 | 1 | 0.68 | 0.88 | 1 | 0.00046 | 1e-04 | 0 | 0.00024 | 0 | 0.00062 | 0.00023 | 0 | 0.00056 | -0.061 | 8 / 36 (22%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 1.3e-05 | 0 | 3.5e-05 | 2.1e-05 | 0 | 4.7e-05 | 0.69 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 3.A.1.105.17 | The ATP-binding Cassette (ABC) Superfamily | 0.38 | 0.45 | 1 | 0.26 | 0.55 | 1 | 0.32 | 0.59 | 1 | 0.71 | 0.87 | 1 | 0.19 | 0.72 | 1 | 0.24 | 0.7 | 1 | 0.00044 | 8.6e-05 | 0 | 0.00019 | 0 | 0.00049 | 0.00022 | 0 | 0.00053 | 0.21 | 7 / 36 (19%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 1.4e-05 | 0 | 3.7e-05 | 9.5e-06 | 0 | 2.3e-05 | -0.56 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 2.A.3.7.2 | The Amino Acid-Polyamine-Organocation (APC) Family | 0.17 | 0.34 | 1 | 0.1 | 0.4 | 1 | 0.14 | 0.43 | 1 | 0.94 | 0.98 | 1 | 0.19 | 0.72 | 1 | 0.23 | 0.7 | 1 | 0.00021 | 1.8e-05 | 0 | 6.8e-05 | 0 | 0.00018 | 0 | 0 | 0 | -Inf | 3 / 36 (8.3%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 3e-06 | 0 | 9.9e-06 | 1.1e-05 | 0 | 3.6e-05 | 1.9 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.1.2.21 | The Major Facilitator Superfamily (MFS) | 0.4 | 0.48 | 1 | 0.2 | 0.49 | 1 | 0.13 | 0.43 | 1 | 0.42 | 0.68 | 1 | 0.034 | 0.62 | 1 | 0.0077 | 0.46 | 1 | 0.00025 | 2.1e-05 | 0 | 3.4e-05 | 0 | 8.9e-05 | 6.8e-05 | 0 | 0.00018 | 1 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 4.A.2.1.11 | The PTS Fructose-Mannitol (Fru) Family | 0.035 | 0.34 | 1 | 0.0074 | 0.22 | 1 | 0.0044 | 0.35 | 1 | 0.0034 | 0.16 | 1 | 0.039 | 0.62 | 1 | 0.014 | 0.46 | 1 | 0.00069 | 0.00013 | 0 | 0.00042 | 1.6e-05 | 0.001 | 0.00023 | 0 | 0.00061 | -0.87 | 7 / 36 (19%) | 4 / 7 (57%) | 1 / 7 (14%) | 0.571 | 0.143 | 0 | 0 | 0 | 2.3e-05 | 0 | 5.6e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 4.A.2.1.2 | The PTS Fructose-Mannitol (Fru) Family | 0.15 | 0.34 | 1 | 0.3 | 0.59 | 1 | 0.23 | 0.51 | 1 | 0.3 | 0.62 | 1 | 0.4 | 0.84 | 1 | 0.29 | 0.73 | 1 | 0.00037 | 6.2e-05 | 0 | 0.00014 | 0 | 0.00031 | 0.00015 | 0 | 0.00038 | 0.1 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 7.3e-06 | 0 | 2.4e-05 | 1.2e-05 | 0 | 2.7e-05 | 0.72 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 2.A.17.1.3 | The Proton-dependent Oligopeptide Transporter (POT/PTR) Family | 0.18 | 0.34 | 1 | 0.12 | 0.42 | 1 | 0.5 | 0.74 | 1 | 0.63 | 0.83 | 1 | 0.45 | 0.86 | 1 | 0.91 | 0.97 | 1 | 0.00033 | 5.5e-05 | 0 | 0.00016 | 0 | 4e-04 | 9.7e-05 | 0 | 0.00026 | -0.72 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 1.2e-05 | 0 | 2.8e-05 | 2.7e-06 | 0 | 8.9e-06 | -2.2 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 9.B.158.1.2 | The Cut Copper Homeostasis (Cut) Family | 0.12 | 0.34 | 1 | 0.2 | 0.49 | 1 | 0.13 | 0.43 | 1 | 0.54 | 0.77 | 1 | 0.85 | 0.96 | 1 | 0.41 | 0.76 | 1 | 0.00028 | 2.3e-05 | 0 | 8.4e-05 | 0 | 0.00022 | 1.9e-05 | 0 | 5.1e-05 | -2.1 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 1e-05 | 0 | 3.4e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.A.30.1.1 | The H+- or Na+-translocating Bacterial Flagellar Motor/ExbBD Outer Membrane Transport Energizer (Mot/Exb) Superfamily | 0.011 | 0.34 | 1 | 0.043 | 0.29 | 1 | 0.035 | 0.37 | 1 | 0.0043 | 0.16 | 1 | 0.041 | 0.62 | 1 | 0.032 | 0.58 | 1 | 0.00019 | 3.2e-05 | 0 | 9.1e-05 | 1.6e-05 | 0.00014 | 4.8e-05 | 0 | 0.00013 | -0.92 | 6 / 36 (17%) | 4 / 7 (57%) | 1 / 7 (14%) | 0.571 | 0.143 | 0 | 0 | 0 | 1.6e-05 | 0 | 5.4e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.1.36.1 | The Major Facilitator Superfamily (MFS) | 0.095 | 0.34 | 1 | 0.11 | 0.4 | 1 | 0.14 | 0.43 | 1 | 0.21 | 0.51 | 1 | 0.71 | 0.93 | 1 | 0.44 | 0.79 | 1 | 0.00053 | 4.4e-05 | 0 | 0.00019 | 0 | 0.00049 | 3.9e-05 | 0 | 1e-04 | -2.3 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.1.60.2 | The Major Facilitator Superfamily (MFS) | 0.048 | 0.34 | 1 | 0.021 | 0.23 | 1 | 0.038 | 0.37 | 1 | 0.02 | 0.26 | 1 | 0.11 | 0.65 | 1 | 0.11 | 0.65 | 1 | 6e-04 | 6.7e-05 | 0 | 0.00025 | 0 | 0.00062 | 8.7e-05 | 0 | 0.00023 | -1.5 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.A.106.2.3 | The Calcium Load-activated Calcium Channel (CLAC) Family | 0.59 | 0.65 | 1 | 0.77 | 0.88 | 1 | 0.65 | 0.84 | 1 | 0.22 | 0.52 | 1 | 0.76 | 0.94 | 1 | 0.32 | 0.73 | 1 | 9.1e-05 | 1.3e-05 | 0 | 8.1e-06 | 0 | 2.1e-05 | 4.4e-06 | 0 | 1.2e-05 | -0.88 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.1e-05 | 0 | 4.6e-05 | 1.3e-05 | 0 | 4.3e-05 | -0.69 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 2.A.115.2.12 | The Novobiocin Exporter (NbcE) Family | 0.95 | 0.96 | 1 | 0.35 | 0.63 | 1 | 0.047 | 0.37 | 1 | 0.7 | 0.87 | 1 | 0.24 | 0.77 | 1 | 0.065 | 0.58 | 1 | 0.00049 | 0.00045 | 0.00023 | 0.00034 | 0.00024 | 0.00025 | 0.00026 | 0.00013 | 0.00031 | -0.39 | 33 / 36 (92%) | 7 / 7 (100%) | 5 / 7 (71%) | 1 | 0.714 | 0.00038 | 0.00021 | 0.00036 | 0.00072 | 0.00097 | 5e-04 | 0.92 | 10 / 11 (91%) | 11 / 11 (100%) | 0.909 | 1 |
| 2.A.23.1.1 | The Dicarboxylate/Amino Acid:Cation (Na+ or H+) Symporter (DAACS) Family | 0.067 | 0.34 | 1 | 0.079 | 0.38 | 1 | 0.16 | 0.46 | 1 | 0.034 | 0.27 | 1 | 0.26 | 0.77 | 1 | 0.44 | 0.79 | 1 | 5e-04 | 4.2e-05 | 0 | 0.00012 | 0 | 0.00026 | 9.7e-05 | 0 | 0.00026 | -0.31 | 3 / 36 (8.3%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 3.A.1.5.38 | The ATP-binding Cassette (ABC) Superfamily | 0.36 | 0.45 | 1 | 0.11 | 0.4 | 1 | 0.14 | 0.43 | 1 | 0.69 | 0.87 | 1 | 0.21 | 0.74 | 1 | 0.28 | 0.73 | 1 | 0.00015 | 1.3e-05 | 0 | 3.4e-05 | 0 | 9.1e-05 | 0 | 0 | 0 | -Inf | 3 / 36 (8.3%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 6e-06 | 0 | 2e-05 | 1.3e-05 | 0 | 4.5e-05 | 1.1 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.7.3.12 | The Drug/Metabolite Transporter (DMT) Superfamily | 0.29 | 0.39 | 1 | 0.82 | 0.91 | 1 | 0.53 | 0.76 | 1 | 0.68 | 0.86 | 1 | 0.83 | 0.95 | 1 | 0.49 | 0.82 | 1 | 0.00027 | 4.6e-05 | 0 | 7e-05 | 0 | 0.00018 | 0.00012 | 0 | 0.00031 | 0.78 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 2.9e-05 | 0 | 6.8e-05 | 3.5 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 1.B.1.1.21 | The General Bacterial Porin (GBP) Family | 0.19 | 0.34 | 1 | 0.98 | 1 | 1 | 0.42 | 0.67 | 1 | 0.49 | 0.73 | 1 | 0.14 | 0.66 | 1 | 0.71 | 0.88 | 1 | 0.00033 | 2.7e-05 | 0 | 6.7e-05 | 0 | 0.00018 | 6.8e-05 | 0 | 0.00018 | 0.021 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 4.A.4.1.1 | The PTS Glucitol (Gut) Family | 0.098 | 0.34 | 1 | 0.038 | 0.28 | 1 | 0.18 | 0.48 | 1 | 0.21 | 0.51 | 1 | 0.2 | 0.74 | 1 | 0.58 | 0.84 | 1 | 0.00039 | 7.5e-05 | 0 | 0.00024 | 0 | 0.00057 | 0.00012 | 0 | 0.00031 | -1 | 7 / 36 (19%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 1.4e-05 | 0 | 3.7e-05 | 5.4e-06 | 0 | 1.8e-05 | -1.4 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 2.A.6.2.7 | The Resistance-Nodulation-Cell Division (RND) Superfamily | 0.13 | 0.34 | 1 | 0.044 | 0.29 | 1 | 0.35 | 0.6 | 1 | 0.36 | 0.64 | 1 | 0.37 | 0.84 | 1 | 0.86 | 0.94 | 1 | 0.00082 | 0.00011 | 0 | 0.00037 | 0 | 0.00093 | 0.00019 | 0 | 0.00051 | -0.96 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 1.4e-05 | 0 | 3.7e-05 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 1.B.39.1.6 | The Bacterial Porin. OmpW (OmpW) Family | 0.088 | 0.34 | 1 | 0.64 | 0.8 | 1 | 0.47 | 0.72 | 1 | 0.2 | 0.51 | 1 | 0.74 | 0.94 | 1 | 0.53 | 0.82 | 1 | 0.00012 | 1.4e-05 | 0 | 2.8e-05 | 0 | 5e-05 | 3.9e-05 | 0 | 1e-04 | 0.48 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 9.B.27.2.1 | The DedA or YdjX-Z (DedA) Family | 0.28 | 0.38 | 1 | 0.014 | 0.23 | 1 | 0.048 | 0.37 | 1 | 0.95 | 0.98 | 1 | 0.0069 | 0.39 | 1 | 0.021 | 0.52 | 1 | 0.00025 | 2.1e-05 | 0 | 3.4e-05 | 0 | 8.9e-05 | 7.5e-05 | 0 | 0.00018 | 1.1 | 3 / 36 (8.3%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 9.B.126.2.1 | The Putative Lipid IV Exporter (YhjD) Family | 0.079 | 0.34 | 1 | 0.15 | 0.44 | 1 | 0.048 | 0.37 | 1 | 0.099 | 0.35 | 1 | 0.43 | 0.86 | 1 | 0.14 | 0.65 | 1 | 0.00044 | 6.1e-05 | 0 | 0.00017 | 0 | 4e-04 | 0.00013 | 0 | 0.00033 | -0.39 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 1.3e-05 | 0 | 3e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 4.A.5.1.1 | The PTS Galactitol (Gat) Family | 0.23 | 0.35 | 1 | 0.098 | 0.39 | 1 | 0.081 | 0.39 | 1 | 0.59 | 0.81 | 1 | 0.095 | 0.64 | 1 | 0.072 | 0.59 | 1 | 5e-04 | 8.3e-05 | 0 | 0.00017 | 0 | 0.00044 | 0.00024 | 0 | 0.00053 | 0.5 | 6 / 36 (17%) | 2 / 7 (29%) | 3 / 7 (43%) | 0.286 | 0.429 | 1.1e-05 | 0 | 3.6e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.A.5.5.1 | The Zinc (Zn2+)-Iron (Fe2+) Permease (ZIP) Family | 0.27 | 0.37 | 1 | 0.52 | 0.75 | 1 | 0.79 | 0.91 | 1 | 0.77 | 0.89 | 1 | 0.35 | 0.84 | 1 | 1 | 1 | 1 | 0.00034 | 2.8e-05 | 0 | 5e-05 | 0 | 0.00013 | 7.7e-05 | 0 | 2e-04 | 0.62 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 1e-05 | 0 | 3.4e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.95.1.5 | The 6 TMS Neutral Amino Acid Transporter (NAAT) Family | 0.23 | 0.35 | 1 | 0.28 | 0.56 | 1 | 0.83 | 0.92 | 1 | 0.63 | 0.83 | 1 | 0.5 | 0.87 | 1 | 0.11 | 0.65 | 1 | 0.00019 | 1.6e-05 | 0 | 5e-05 | 0 | 0.00013 | 2.9e-05 | 0 | 7.7e-05 | -0.79 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 5.A.2.1.2 | The Disulfide Bond Oxidoreductase B (DsbB) Family | 0.091 | 0.34 | 1 | 0.048 | 0.29 | 1 | 0.14 | 0.43 | 1 | 0.32 | 0.63 | 1 | 0.28 | 0.78 | 1 | 0.56 | 0.83 | 1 | 0.00059 | 9.8e-05 | 0 | 0.00037 | 0 | 0.00093 | 9.7e-05 | 0 | 0.00026 | -1.9 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 9.9e-06 | 0 | 2.5e-05 | 1e-05 | 0 | 3.4e-05 | 0.014 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 2.A.1.2.22 | The Major Facilitator Superfamily (MFS) | 0.04 | 0.34 | 1 | 0.0089 | 0.22 | 1 | 0.019 | 0.37 | 1 | 0.0056 | 0.16 | 1 | 0.045 | 0.62 | 1 | 0.054 | 0.58 | 1 | 0.00051 | 7.1e-05 | 0 | 0.00027 | 0 | 0.00066 | 9.7e-05 | 0 | 0.00026 | -1.5 | 5 / 36 (14%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 3.A.1.2.23 | The ATP-binding Cassette (ABC) Superfamily | 0.44 | 0.52 | 1 | 0.51 | 0.75 | 1 | 0.84 | 0.92 | 1 | 0.13 | 0.39 | 1 | 0.97 | 0.99 | 1 | 0.69 | 0.88 | 1 | 0.0033 | 0.0027 | 0.0019 | 0.0017 | 0.0018 | 0.0017 | 0.0026 | 0.0022 | 0.0031 | 0.61 | 29 / 36 (81%) | 5 / 7 (71%) | 4 / 7 (57%) | 0.714 | 0.571 | 0.0028 | 0.0014 | 0.0047 | 0.0033 | 0.0031 | 0.0025 | 0.24 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 9.B.318.1.1 | The YjgJ or DUF898 (DUF898) Family | 0.8 | 0.84 | 1 | 0.51 | 0.75 | 1 | 0.87 | 0.94 | 1 | 0.32 | 0.63 | 1 | 0.67 | 0.92 | 1 | 0.95 | 1 | 1 | 0.00018 | 1.5e-05 | 0 | 1.2e-05 | 0 | 3.1e-05 | 0 | 0 | 0 | -Inf | 3 / 36 (8.3%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 3.3e-05 | 0 | 0.00011 | 8.1e-06 | 0 | 2.7e-05 | -2 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.1.64.1 | The Major Facilitator Superfamily (MFS) | 0.0071 | 0.34 | 1 | 0.036 | 0.28 | 1 | 0.0015 | 0.35 | 0.82 | 0.0024 | 0.15 | 1 | 0.0055 | 0.36 | 1 | 0.00017 | 0.093 | 0.093 | 0.00021 | 7.7e-05 | 0 | 0 | 0 | 0 | 0.00011 | 3.1e-05 | 0.00017 | Inf | 13 / 36 (36%) | 0 / 7 (0%) | 4 / 7 (57%) | 0 | 0.571 | 0.00017 | 8e-05 | 0.00034 | 9.5e-06 | 0 | 2.3e-05 | -4.2 | 7 / 11 (64%) | 2 / 11 (18%) | 0.636 | 0.182 |
| 2.A.1.14.7 | The Major Facilitator Superfamily (MFS) | 0.17 | 0.34 | 1 | 0.98 | 1 | 1 | 0.5 | 0.74 | 1 | 0.19 | 0.49 | 1 | 0.84 | 0.96 | 1 | 0.55 | 0.83 | 1 | 0.00044 | 4.9e-05 | 0 | 1e-04 | 0 | 0.00027 | 0.00014 | 0 | 0.00036 | 0.49 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 8.1e-06 | 0 | 2.7e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.1.7.5 | The Major Facilitator Superfamily (MFS) | 0.069 | 0.34 | 1 | 0.021 | 0.23 | 1 | 0.063 | 0.37 | 1 | 0.033 | 0.27 | 1 | 0.16 | 0.69 | 1 | 0.31 | 0.73 | 1 | 0.0013 | 0.00011 | 0 | 4e-04 | 0 | 0.001 | 0.00016 | 0 | 0.00044 | -1.3 | 3 / 36 (8.3%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 9.B.32.1.1 | The DUF3302 or Pfam11742 (YibI) Family | 0.043 | 0.34 | 1 | 0.084 | 0.39 | 1 | 0.17 | 0.46 | 1 | 0.051 | 0.28 | 1 | 0.28 | 0.78 | 1 | 0.46 | 0.8 | 1 | 0.00028 | 2.3e-05 | 0 | 7e-05 | 0 | 0.00014 | 4.8e-05 | 0 | 0.00013 | -0.54 | 3 / 36 (8.3%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 2.A.10.1.2 | The 2-Keto-3-Deoxygluconate Transporter (KdgT) Family | 0.14 | 0.34 | 1 | 0.76 | 0.88 | 1 | 0.2 | 0.48 | 1 | 0.27 | 0.6 | 1 | 0.94 | 0.98 | 1 | 0.27 | 0.73 | 1 | 2e-04 | 2.8e-05 | 0 | 5.5e-05 | 0 | 0.00013 | 6.8e-05 | 0 | 0.00018 | 0.31 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 1.2e-05 | 0 | 2.7e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 9.B.50.1.1 | The Outer Membrane Beta-barrel Endoprotease. Omptin (Omptin) Family | 0.068 | 0.34 | 1 | 0.11 | 0.4 | 1 | 0.076 | 0.38 | 1 | 0.066 | 0.3 | 1 | 0.37 | 0.84 | 1 | 0.23 | 0.7 | 1 | 0.00048 | 5.4e-05 | 0 | 0.00015 | 0 | 0.00035 | 0.00011 | 0 | 0.00028 | -0.45 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 1e-05 | 0 | 3.4e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 4.C.1.1.4 | The Fatty Acid Transporter (FAT) Family | 0.32 | 0.41 | 1 | 0.15 | 0.44 | 1 | 0.26 | 0.52 | 1 | 0.97 | 0.99 | 1 | 0.13 | 0.66 | 1 | 0.22 | 0.7 | 1 | 0.00043 | 5.9e-05 | 0 | 0.00012 | 0 | 0.00031 | 0.00017 | 0 | 0.00035 | 0.5 | 5 / 36 (14%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 6e-06 | 0 | 2e-05 | 6.8e-06 | 0 | 2.3e-05 | 0.18 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.5.5.5 | The Zinc (Zn2+)-Iron (Fe2+) Permease (ZIP) Family | 0.28 | 0.38 | 1 | 0.55 | 0.76 | 1 | 0.83 | 0.92 | 1 | 0.55 | 0.78 | 1 | 0.37 | 0.84 | 1 | 0.77 | 0.9 | 1 | 0.00023 | 8.4e-05 | 0 | 0.00011 | 0 | 0.00015 | 0.00015 | 8.1e-05 | 0.00021 | 0.45 | 13 / 36 (36%) | 3 / 7 (43%) | 4 / 7 (57%) | 0.429 | 0.571 | 5.9e-05 | 0 | 0.00016 | 5e-05 | 0 | 8.8e-05 | -0.24 | 2 / 11 (18%) | 4 / 11 (36%) | 0.182 | 0.364 |
| 3.A.1.1.1 | The ATP-binding Cassette (ABC) Superfamily | 0.15 | 0.34 | 1 | 0.32 | 0.61 | 1 | 0.35 | 0.6 | 1 | 0.35 | 0.64 | 1 | 0.79 | 0.94 | 1 | 0.79 | 0.91 | 1 | 0.00074 | 0.00019 | 0 | 0.00052 | 0 | 0.0013 | 0.00034 | 0 | 0.00084 | -0.61 | 9 / 36 (25%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 2.4e-05 | 0 | 5.6e-05 | 3.7e-05 | 0 | 8.3e-05 | 0.62 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 9.B.121.1.3 | The AsmA (AsmA) Family | 0.24 | 0.36 | 1 | 0.3 | 0.59 | 1 | 0.76 | 0.9 | 1 | 0.51 | 0.75 | 1 | 0.42 | 0.86 | 1 | 0.9 | 0.97 | 1 | 0.00043 | 8.4e-05 | 0 | 2e-04 | 0 | 0.00048 | 0.00016 | 0 | 0.00044 | -0.32 | 7 / 36 (19%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 3.5e-05 | 0 | 0.00011 | 8.1e-06 | 0 | 2.7e-05 | -2.1 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 1.A.23.3.2 | The Small Conductance Mechanosensitive Ion Channel (MscS) Family | 0.35 | 0.44 | 1 | 0.54 | 0.76 | 1 | 0.76 | 0.9 | 1 | 0.74 | 0.88 | 1 | 0.77 | 0.94 | 1 | 0.29 | 0.73 | 1 | 6e-04 | 5e-05 | 0 | 0.00013 | 0 | 0.00036 | 8.7e-05 | 0 | 0.00023 | -0.58 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.2e-05 | 0 | 7.3e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 1.B.57.4.2 | The Legionella Major-Outer Membrane Protein (LM-OMP) Family | 0.34 | 0.42 | 1 | 0.39 | 0.67 | 1 | 0.48 | 0.73 | 1 | 0.66 | 0.85 | 1 | 0.43 | 0.86 | 1 | 0.57 | 0.83 | 1 | 2e-04 | 5.6e-05 | 0 | 7.6e-05 | 3.2e-05 | 0.00012 | 3.7e-05 | 0 | 6.5e-05 | -1 | 10 / 36 (28%) | 4 / 7 (57%) | 2 / 7 (29%) | 0.571 | 0.286 | 5.2e-05 | 0 | 0.00012 | 5.9e-05 | 0 | 0.00015 | 0.18 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 2.A.76.1.4 | The Resistance to Homoserine/Threonine (RhtB) Family | 0.38 | 0.45 | 1 | 0.88 | 0.95 | 1 | 0.81 | 0.91 | 1 | 0.57 | 0.79 | 1 | 0.48 | 0.87 | 1 | 0.75 | 0.9 | 1 | 0.00016 | 1.8e-05 | 0 | 3.4e-05 | 0 | 8.9e-05 | 3.9e-05 | 0 | 1e-04 | 0.2 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1e-05 | 0 | 3.4e-05 | 1.9 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.75.1.2 | The L-Lysine Exporter (LysE) Family | 0.1 | 0.34 | 1 | 0.85 | 0.93 | 1 | 0.97 | 0.99 | 1 | 0.13 | 0.39 | 1 | 0.79 | 0.94 | 1 | 0.88 | 0.96 | 1 | 0.00024 | 4.7e-05 | 0 | 9.8e-05 | 0 | 0.00022 | 0.00013 | 0 | 3e-04 | 0.41 | 7 / 36 (19%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 2.6e-06 | 0 | 8.5e-06 | 5.4e-06 | 0 | 1.8e-05 | 1.1 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 1.A.13.2.3 | The Epithelial Chloride Channel (E-ClC) Family | 0.22 | 0.35 | 1 | 0.28 | 0.57 | 1 | 0.84 | 0.92 | 1 | 0.82 | 0.93 | 1 | 0.51 | 0.87 | 1 | 0.11 | 0.65 | 1 | 0.00074 | 6.2e-05 | 0 | 0.00019 | 0 | 0.00049 | 0.00013 | 0 | 0.00033 | -0.55 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 5.1e-06 | 0 | 1.7e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 3.A.1.14.18 | The ATP-binding Cassette (ABC) Superfamily | 0.071 | 0.34 | 1 | 0.22 | 0.51 | 1 | 0.12 | 0.43 | 1 | 0.074 | 0.31 | 1 | 0.76 | 0.94 | 1 | 0.56 | 0.83 | 1 | 0.00072 | 1e-04 | 0 | 0.00031 | 0 | 0.00075 | 0.00019 | 0 | 0.00046 | -0.71 | 5 / 36 (14%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0 | 0 | 0 | 1.1e-05 | 0 | 3.6e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 3.A.1.15.3 | The ATP-binding Cassette (ABC) Superfamily | 0.27 | 0.37 | 1 | 0.45 | 0.72 | 1 | 0.53 | 0.76 | 1 | 0.12 | 0.38 | 1 | 0.2 | 0.74 | 1 | 0.45 | 0.79 | 1 | 0.00014 | 3.8e-05 | 0 | 2.3e-06 | 0 | 6.1e-06 | 2.1e-05 | 0 | 4.4e-05 | 3.2 | 10 / 36 (28%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 7.6e-05 | 0 | 0.00023 | 3.4e-05 | 0 | 5.1e-05 | -1.2 | 3 / 11 (27%) | 4 / 11 (36%) | 0.273 | 0.364 |
| 2.A.3.1.23 | The Amino Acid-Polyamine-Organocation (APC) Family | 0.13 | 0.34 | 1 | 0.14 | 0.44 | 1 | 0.1 | 0.42 | 1 | 0.33 | 0.63 | 1 | 0.7 | 0.93 | 1 | 0.37 | 0.74 | 1 | 0.00038 | 6.3e-05 | 0 | 2e-04 | 0 | 0.00053 | 9.7e-05 | 0 | 0.00026 | -1 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1.3e-05 | 0 | 3.5e-05 | 2.3 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 1.A.112.2.5 | The Cyclin M Mg2+ Exporter (CNNM) Family | 0.013 | 0.34 | 1 | 0.0052 | 0.22 | 1 | 0.019 | 0.37 | 1 | 0.0046 | 0.16 | 1 | 0.016 | 0.5 | 1 | 0.054 | 0.58 | 1 | 0.00025 | 4.9e-05 | 0 | 0.00016 | 1.6e-05 | 3e-04 | 6.8e-05 | 0 | 0.00018 | -1.2 | 7 / 36 (19%) | 4 / 7 (57%) | 1 / 7 (14%) | 0.571 | 0.143 | 5.1e-06 | 0 | 1.7e-05 | 8.1e-06 | 0 | 2.7e-05 | 0.67 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 9.B.97.7.1 | The Acyltransferase-3/Putative Acetyl-CoA Transporter (ATAT) Family | 0.23 | 0.35 | 1 | 0.52 | 0.75 | 1 | 0.78 | 0.91 | 1 | 0.76 | 0.89 | 1 | 0.21 | 0.74 | 1 | 0.033 | 0.58 | 1 | 0.00059 | 5e-05 | 0 | 0.00013 | 0 | 0.00036 | 0.00012 | 0 | 0.00031 | -0.12 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.A.114.1.1 | The Putative Peptide Transporter Carbon Starvation CstA (CstA) Family | 0.21 | 0.35 | 1 | 0.99 | 1 | 1 | 0.78 | 0.91 | 1 | 0.74 | 0.89 | 1 | 0.37 | 0.84 | 1 | 0.63 | 0.85 | 1 | 0.00049 | 0.00011 | 0 | 0.00029 | 0 | 0.00076 | 0.00023 | 0 | 0.00056 | -0.33 | 8 / 36 (22%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 8.1e-06 | 0 | 1.9e-05 | 1.8e-05 | 0 | 4.1e-05 | 1.2 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 2.A.1.1.115 | The Major Facilitator Superfamily (MFS) | 0.25 | 0.36 | 1 | 0.58 | 0.77 | 1 | 0.91 | 0.96 | 1 | 0.98 | 0.99 | 1 | 0.075 | 0.62 | 1 | 0.23 | 0.7 | 1 | 0.00024 | 3.3e-05 | 0 | 8.4e-05 | 0 | 0.00022 | 6.4e-05 | 0 | 0.00013 | -0.39 | 5 / 36 (14%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 3e-06 | 0 | 9.9e-06 | 1.1e-05 | 0 | 3.6e-05 | 1.9 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.1.10.1 | The Major Facilitator Superfamily (MFS) | 0.36 | 0.44 | 1 | 0.48 | 0.74 | 1 | 0.57 | 0.79 | 1 | 0.51 | 0.75 | 1 | 0.71 | 0.93 | 1 | 0.12 | 0.65 | 1 | 0.00054 | 6e-05 | 0 | 0.00017 | 0 | 0.00045 | 0.00012 | 0 | 0.00031 | -0.5 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 1.6e-05 | 0 | 3.9e-05 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 2.A.41.1.1 | The Concentrative Nucleoside Transporter (CNT) Family | 0.52 | 0.58 | 1 | 0.015 | 0.23 | 1 | 0.049 | 0.37 | 1 | 0.59 | 0.82 | 1 | 0.0058 | 0.36 | 1 | 0.019 | 0.52 | 1 | 0.00026 | 3.6e-05 | 0 | 3.4e-05 | 0 | 8.9e-05 | 0.00014 | 0 | 0.00033 | 2 | 5 / 36 (14%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 3e-06 | 0 | 9.9e-06 | 2.7e-06 | 0 | 8.9e-06 | -0.15 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.A.1.14.2 | The ATP-binding Cassette (ABC) Superfamily | 0.12 | 0.34 | 1 | 0.4 | 0.68 | 1 | 0.25 | 0.52 | 1 | 0.059 | 0.29 | 1 | 0.5 | 0.87 | 1 | 0.31 | 0.73 | 1 | 0.00086 | 9.5e-05 | 0 | 0.00023 | 0 | 0.00057 | 0.00025 | 0 | 0.00067 | 0.12 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.A.1.13.1 | The Voltage-gated Ion Channel (VIC) Superfamily | 0.32 | 0.41 | 1 | 0.65 | 0.81 | 1 | 0.67 | 0.85 | 1 | 0.97 | 0.99 | 1 | 0.41 | 0.85 | 1 | 0.95 | 1 | 1 | 3e-04 | 4.2e-05 | 0 | 8.4e-05 | 0 | 0.00022 | 0.00011 | 0 | 0.00028 | 0.39 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3e-06 | 0 | 9.9e-06 | 1.3e-05 | 0 | 3.5e-05 | 2.1 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 2.A.33.1.1 | The NhaA Na+:H+ Antiporter (NhaA) Family | 0.11 | 0.34 | 1 | 0.19 | 0.49 | 1 | 0.09 | 0.41 | 1 | 0.12 | 0.38 | 1 | 0.67 | 0.92 | 1 | 0.34 | 0.73 | 1 | 0.00061 | 6.8e-05 | 0 | 0.00019 | 0 | 0.00049 | 0.00015 | 0 | 0.00038 | -0.34 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 1e-05 | 0 | 3.4e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 3.A.1.5.41 | The ATP-binding Cassette (ABC) Superfamily | 0.12 | 0.34 | 1 | 0.015 | 0.23 | 1 | 0.022 | 0.37 | 1 | 0.28 | 0.6 | 1 | 0.098 | 0.64 | 1 | 0.092 | 0.62 | 1 | 0.00088 | 0.00027 | 0 | 0.00076 | 0 | 0.0018 | 5e-04 | 0 | 0.0013 | -0.6 | 11 / 36 (31%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 3.3e-05 | 0 | 7.3e-05 | 4.5e-05 | 0 | 7.4e-05 | 0.45 | 3 / 11 (27%) | 4 / 11 (36%) | 0.273 | 0.364 |
| 9.A.41.1.2 | The Capsular Polysaccharide Exporter (CPS-E) Family | 0.16 | 0.34 | 1 | 0.59 | 0.77 | 1 | 0.79 | 0.91 | 1 | 0.31 | 0.62 | 1 | 0.11 | 0.65 | 1 | 0.021 | 0.52 | 1 | 0.00058 | 8e-05 | 0 | 0.00022 | 0 | 0.00058 | 0.00019 | 0 | 0.00048 | -0.21 | 5 / 36 (14%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 2.6e-06 | 0 | 8.5e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.A.56.1.2 | The Tripartite ATP-independent Periplasmic Transporter (TRAP-T) Family | 0.038 | 0.34 | 1 | 0.13 | 0.42 | 1 | 0.12 | 0.43 | 1 | 0.011 | 0.22 | 1 | 0.14 | 0.66 | 1 | 0.12 | 0.65 | 1 | 0.00033 | 4.6e-05 | 0 | 0.00012 | 0 | 0.00022 | 0.00011 | 0 | 0.00028 | -0.13 | 5 / 36 (14%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 3.A.6.2.1 | The Type III (Virulence-related) Secretory Pathway (IIISP) Family | 0.79 | 0.84 | 1 | 0.17 | 0.47 | 1 | 0.51 | 0.75 | 1 | 0.33 | 0.63 | 1 | 0.08 | 0.62 | 1 | 0.48 | 0.81 | 1 | 0.00022 | 2.4e-05 | 0 | 2.3e-06 | 0 | 6.1e-06 | 0.00011 | 0 | 0.00028 | 5.6 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 9.5e-06 | 0 | 2.3e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 2.A.19.5.1 | The Ca2+:Cation Antiporter (CaCA) Family | 0.14 | 0.34 | 1 | 0.47 | 0.73 | 1 | 0.88 | 0.95 | 1 | 0.2 | 0.51 | 1 | 0.45 | 0.86 | 1 | 0.98 | 1 | 1 | 0.00054 | 6e-05 | 0 | 0.00014 | 0 | 0.00031 | 0.00016 | 0 | 0.00044 | 0.19 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 5.1e-06 | 0 | 1.7e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 4.A.7.1.1 | The PTS L-Ascorbate (L-Asc) Family | 0.63 | 0.68 | 1 | 0.28 | 0.57 | 1 | 0.78 | 0.91 | 1 | 0.28 | 0.6 | 1 | 0.7 | 0.93 | 1 | 0.67 | 0.88 | 1 | 0.00022 | 3.1e-05 | 0 | 8.4e-05 | 0 | 0.00022 | 9.7e-06 | 0 | 2.6e-05 | -3.1 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3.6e-05 | 0 | 0.00011 | 5.4e-06 | 0 | 1.8e-05 | -2.7 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 9.A.69.1.1 | The Intermembrane Phospholipid Translocase (IMPL-T) Family | 0.14 | 0.34 | 1 | 0.32 | 0.61 | 1 | 0.066 | 0.37 | 1 | 0.13 | 0.4 | 1 | 0.79 | 0.94 | 1 | 0.18 | 0.68 | 1 | 0.0011 | 0.00015 | 0 | 0.00039 | 0 | 0.001 | 0.00034 | 0 | 9e-04 | -0.2 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 1.8e-05 | 0 | 4.1e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 2.A.21.7.2 | The Solute:Sodium Symporter (SSS) Family | 0.096 | 0.34 | 1 | 0.09 | 0.39 | 1 | 0.11 | 0.42 | 1 | 0.23 | 0.55 | 1 | 0.38 | 0.84 | 1 | 0.35 | 0.74 | 1 | 0.00039 | 6.5e-05 | 0 | 2e-04 | 0 | 0.00048 | 9.7e-05 | 0 | 0.00026 | -1 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 7.3e-06 | 0 | 2.4e-05 | 1.3e-05 | 0 | 3.5e-05 | 0.83 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 2.A.85.1.1 | The Aromatic Acid Exporter (ArAE) Family | 0.18 | 0.34 | 1 | 0.71 | 0.86 | 1 | 0.84 | 0.92 | 1 | 0.14 | 0.4 | 1 | 0.74 | 0.94 | 1 | 0.73 | 0.88 | 1 | 0.00065 | 7.2e-05 | 0 | 0.00015 | 0 | 4e-04 | 0.00021 | 0 | 0.00056 | 0.49 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.36.6.11 | The Monovalent Cation:Proton Antiporter-1 (CPA1) Family | 0.21 | 0.35 | 1 | 0.73 | 0.87 | 1 | 0.9 | 0.96 | 1 | 0.15 | 0.41 | 1 | 0.63 | 0.91 | 1 | 0.61 | 0.85 | 1 | 0.00032 | 5.3e-05 | 0 | 6.4e-05 | 0 | 0.00013 | 0.00019 | 0 | 0.00051 | 1.6 | 6 / 36 (17%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 8.1e-06 | 0 | 2.7e-05 | 1.6 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 8.A.1.6.3 | The Membrane Fusion Protein (MFP) Family | 0.5 | 0.57 | 1 | 0.79 | 0.9 | 1 | 0.64 | 0.83 | 1 | 0.6 | 0.82 | 1 | 0.6 | 0.91 | 1 | 0.51 | 0.82 | 1 | 0.00042 | 4.6e-05 | 0 | 8.4e-05 | 0 | 0.00022 | 0.00011 | 0 | 0.00028 | 0.39 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.2e-05 | 0 | 7.3e-05 | 8.1e-06 | 0 | 2.7e-05 | -1.4 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.A.1.2.22 | The ATP-binding Cassette (ABC) Superfamily | 0.37 | 0.45 | 1 | 0.64 | 0.8 | 1 | 0.81 | 0.91 | 1 | 0.11 | 0.37 | 1 | 0.54 | 0.89 | 1 | 0.61 | 0.85 | 1 | 0.0032 | 0.0026 | 0.0021 | 0.0018 | 0.0012 | 0.002 | 0.0022 | 0.002 | 0.0022 | 0.29 | 30 / 36 (83%) | 5 / 7 (71%) | 5 / 7 (71%) | 0.714 | 0.714 | 0.0028 | 0.0021 | 0.0037 | 0.0033 | 0.0026 | 0.0026 | 0.24 | 10 / 11 (91%) | 10 / 11 (91%) | 0.909 | 0.909 |
| 1.C.126.1.2 | The HlyC HlyC) Family of Haemolysins | 0.091 | 0.34 | 1 | 0.018 | 0.23 | 1 | 0.057 | 0.37 | 1 | 0.12 | 0.38 | 1 | 0.72 | 0.94 | 1 | 0.97 | 1 | 1 | 0.00021 | 1.8e-05 | 0 | 5.3e-05 | 0 | 0.00013 | 3.9e-05 | 0 | 1e-04 | -0.44 | 3 / 36 (8.3%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 2.A.15.1.4 | The Betaine/Carnitine/Choline Transporter (BCCT) Family | 0.17 | 0.34 | 1 | 0.087 | 0.39 | 1 | 0.22 | 0.51 | 1 | 0.65 | 0.85 | 1 | 0.46 | 0.87 | 1 | 0.78 | 0.91 | 1 | 0.00049 | 0.00011 | 0 | 0.00035 | 0 | 0.00089 | 0.00017 | 0 | 0.00046 | -1 | 8 / 36 (22%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 1.3e-05 | 0 | 2.6e-05 | 1.3e-05 | 0 | 3.5e-05 | 0 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 2.A.7.3.2 | The Drug/Metabolite Transporter (DMT) Superfamily | 0.31 | 0.4 | 1 | 0.74 | 0.88 | 1 | 0.29 | 0.56 | 1 | 0.66 | 0.85 | 1 | 0.009 | 0.46 | 1 | 0.051 | 0.58 | 1 | 2e-04 | 2.2e-05 | 0 | 5e-05 | 0 | 0.00013 | 4.8e-05 | 0 | 0.00013 | -0.059 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3e-06 | 0 | 9.9e-06 | 5.4e-06 | 0 | 1.8e-05 | 0.85 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 8.A.4.1.1 | The Cytoplasmic Membrane-Periplasmic Auxiliary-2 (MPA2) Family | 0.23 | 0.35 | 1 | 0.62 | 0.8 | 1 | 0.31 | 0.58 | 1 | 0.76 | 0.89 | 1 | 0.65 | 0.92 | 1 | 0.76 | 0.9 | 1 | 0.00041 | 5.7e-05 | 0 | 0.00015 | 0 | 4e-04 | 0.00012 | 0 | 0.00031 | -0.32 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1.3e-05 | 0 | 3.5e-05 | 2.3 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 2.A.1.2.55 | The Major Facilitator Superfamily (MFS) | 0.087 | 0.34 | 1 | 0.034 | 0.27 | 1 | 0.35 | 0.6 | 1 | 0.2 | 0.51 | 1 | 0.13 | 0.66 | 1 | 0.79 | 0.91 | 1 | 0.00025 | 4.2e-05 | 0 | 0.00013 | 0 | 0.00026 | 6.8e-05 | 0 | 0.00018 | -0.93 | 6 / 36 (17%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 1.4e-05 | 0 | 3.7e-05 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 3.A.1.5.11 | The ATP-binding Cassette (ABC) Superfamily | 0.039 | 0.34 | 1 | 0.016 | 0.23 | 1 | 0.021 | 0.37 | 1 | 0.013 | 0.23 | 1 | 0.1 | 0.64 | 1 | 0.082 | 0.61 | 1 | 0.00067 | 9.3e-05 | 0 | 0.00031 | 0 | 7e-04 | 0.00015 | 0 | 0.00041 | -1 | 5 / 36 (14%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 0 | 0 | 0 | 1e-05 | 0 | 3.4e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 4.D.1.1.3 | The Putative Vectorial Glycosyl Polymerization (VGP) Family | 0.24 | 0.36 | 1 | 0.17 | 0.47 | 1 | 0.69 | 0.86 | 1 | 0.71 | 0.87 | 1 | 0.95 | 0.98 | 1 | 0.18 | 0.68 | 1 | 0.00047 | 5.2e-05 | 0 | 0.00019 | 0 | 0.00049 | 6.8e-05 | 0 | 0.00018 | -1.5 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 9.9e-06 | 0 | 2.5e-05 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 9.B.169.1.2 | The Integral Membrane Protein (8 -10 TMSs) YeiB or DUF418 (YeiB) Family | 0.083 | 0.34 | 1 | 0.02 | 0.23 | 1 | 0.1 | 0.42 | 1 | 0.2 | 0.51 | 1 | 0.11 | 0.64 | 1 | 0.47 | 0.81 | 1 | 0.00035 | 5.8e-05 | 0 | 0.00022 | 0 | 0.00053 | 6.8e-05 | 0 | 0.00018 | -1.7 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 5.5e-06 | 0 | 1.2e-05 | 2.7e-06 | 0 | 8.9e-06 | -1 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 3.A.1.19.4 | The ATP-binding Cassette (ABC) Superfamily | 0.19 | 0.34 | 1 | 0.12 | 0.41 | 1 | 0.52 | 0.75 | 1 | 0.45 | 0.71 | 1 | 0.5 | 0.87 | 1 | 0.82 | 0.93 | 1 | 0.00058 | 9.7e-05 | 0 | 0.00026 | 0 | 0.00066 | 0.00022 | 0 | 0.00059 | -0.24 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 8.1e-06 | 0 | 1.9e-05 | 2.7e-06 | 0 | 8.9e-06 | -1.6 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 9.B.223.1.9 | The 4 TMS DUF2979 or YoaS (YoaS) Family | 0.32 | 0.41 | 1 | 0.53 | 0.76 | 1 | 0.21 | 0.5 | 1 | 0.98 | 0.99 | 1 | 0.82 | 0.95 | 1 | 0.38 | 0.75 | 1 | 0.00023 | 1.9e-05 | 0 | 2.8e-05 | 0 | 7.5e-05 | 4.4e-06 | 0 | 1.2e-05 | -2.7 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 4.1e-05 | 0 | 0.00014 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.B.11.3.9 | The Outer Membrane Fimbrial Usher Porin (FUP) Family | 0.15 | 0.34 | 1 | 0.15 | 0.44 | 1 | 0.099 | 0.42 | 1 | 0.55 | 0.78 | 1 | 0.94 | 0.98 | 1 | 0.4 | 0.75 | 1 | 0.00097 | 0.00013 | 0 | 0.00047 | 0 | 0.0012 | 0.00018 | 0 | 0.00049 | -1.4 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3e-06 | 0 | 9.9e-06 | 2.2e-05 | 0 | 5e-05 | 2.9 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 9.A.69.1.2 | The Intermembrane Phospholipid Translocase (IMPL-T) Family | 0.017 | 0.34 | 1 | 0.0011 | 0.16 | 0.62 | 0.0049 | 0.35 | 1 | 0.0036 | 0.16 | 1 | 0.0028 | 0.31 | 1 | 0.01 | 0.46 | 1 | 0.00054 | 0.00012 | 0 | 0.00043 | 3.2e-05 | 0.00096 | 0.00017 | 0 | 4e-04 | -1.3 | 8 / 36 (22%) | 4 / 7 (57%) | 2 / 7 (29%) | 0.571 | 0.286 | 3e-06 | 0 | 9.9e-06 | 8.1e-06 | 0 | 2.7e-05 | 1.4 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.108.2.3 | The Iron/Lead Transporter (ILT) Family | 0.022 | 0.34 | 1 | 0.0043 | 0.22 | 1 | 0.02 | 0.37 | 1 | 0.0065 | 0.17 | 1 | 0.026 | 0.62 | 1 | 0.087 | 0.61 | 1 | 0.00032 | 5.4e-05 | 0 | 2e-04 | 0 | 0.00043 | 6.8e-05 | 0 | 0.00018 | -1.6 | 6 / 36 (17%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 2.7e-06 | 0 | 8.9e-06 | 0.054 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.7.3.61 | The Drug/Metabolite Transporter (DMT) Superfamily | 0.037 | 0.34 | 1 | 0.029 | 0.25 | 1 | 0.013 | 0.37 | 1 | 0.018 | 0.26 | 1 | 0.15 | 0.66 | 1 | 0.042 | 0.58 | 1 | 0.00038 | 6.3e-05 | 0 | 0.00021 | 0 | 0.00048 | 8.7e-05 | 0 | 0.00023 | -1.3 | 6 / 36 (17%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 0 | 0 | 0 | 1.8e-05 | 0 | 4.1e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 3.A.1.12.1 | The ATP-binding Cassette (ABC) Superfamily | 0.22 | 0.35 | 1 | 0.32 | 0.61 | 1 | 0.26 | 0.52 | 1 | 0.75 | 0.89 | 1 | 0.91 | 0.98 | 1 | 0.66 | 0.87 | 1 | 0.00059 | 0.00013 | 0 | 0.00036 | 0 | 0.00093 | 0.00027 | 0 | 0.00072 | -0.42 | 8 / 36 (22%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 1.1e-05 | 0 | 2.6e-05 | 1.6e-05 | 0 | 3e-05 | 0.54 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 3.A.3.32.6 | The P-type ATPase (P-ATPase) Superfamily | 0.99 | 0.99 | 1 | 0.83 | 0.92 | 1 | 0.59 | 0.8 | 1 | 0.61 | 0.82 | 1 | 0.79 | 0.94 | 1 | 0.73 | 0.89 | 1 | 0.00039 | 5.4e-05 | 0 | 4.1e-05 | 0 | 0.00011 | 2.6e-05 | 0 | 7e-05 | -0.66 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 6.7e-05 | 0 | 0.00022 | 6.7e-05 | 0 | 0.00015 | 0 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 2.A.7.3.18 | The Drug/Metabolite Transporter (DMT) Superfamily | 0.33 | 0.42 | 1 | 0.24 | 0.53 | 1 | 0.14 | 0.43 | 1 | 0.84 | 0.94 | 1 | 0.25 | 0.77 | 1 | 0.11 | 0.65 | 1 | 0.00042 | 3.5e-05 | 0 | 6.7e-05 | 0 | 0.00018 | 0.00011 | 0 | 0.00028 | 0.72 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3e-06 | 0 | 9.9e-06 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.A.3.7.3 | The Amino Acid-Polyamine-Organocation (APC) Family | 0.27 | 0.38 | 1 | 0.56 | 0.76 | 1 | 0.87 | 0.94 | 1 | 0.84 | 0.94 | 1 | 0.78 | 0.94 | 1 | 0.73 | 0.88 | 1 | 0.00029 | 4.8e-05 | 0 | 0.00011 | 0 | 0.00027 | 0.00012 | 0 | 0.00031 | 0.13 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 8.1e-06 | 0 | 1.9e-05 | 8.1e-06 | 0 | 2.7e-05 | 0 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 2.A.127.1.2 | The Enterobacterial Cardiolipin Transporter (CLT) Family | 0.11 | 0.34 | 1 | 0.18 | 0.48 | 1 | 0.045 | 0.37 | 1 | 0.11 | 0.37 | 1 | 0.66 | 0.92 | 1 | 0.15 | 0.65 | 1 | 0.00056 | 7.8e-05 | 0 | 0.00022 | 0 | 0.00058 | 0.00015 | 0 | 0.00041 | -0.55 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 1.6e-05 | 0 | 3.7e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 8.A.5.1.7 | The Voltage-gated K+ Channel β-subunit (Kvβ) Family | 0.28 | 0.38 | 1 | 0.95 | 0.99 | 1 | 0.75 | 0.9 | 1 | 0.99 | 0.99 | 1 | 0.53 | 0.88 | 1 | 0.8 | 0.91 | 1 | 0.00034 | 3.7e-05 | 0 | 8.4e-05 | 0 | 0.00022 | 8.7e-05 | 0 | 0.00023 | 0.051 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1e-05 | 0 | 3.4e-05 | 1.9 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 1.A.77.3.5 | The Mg2+/Ca2+ Uniporter (MCU) Family | 0.99 | 0.99 | 1 | 0.1 | 0.4 | 1 | 0.22 | 0.5 | 1 | 0.57 | 0.79 | 1 | 0.098 | 0.64 | 1 | 0.21 | 0.69 | 1 | 0.00011 | 3.4e-05 | 0 | 1.7e-05 | 0 | 3e-05 | 8.1e-05 | 3.1e-05 | 1e-04 | 2.3 | 11 / 36 (31%) | 2 / 7 (29%) | 4 / 7 (57%) | 0.286 | 0.571 | 2.7e-05 | 0 | 6.3e-05 | 2.2e-05 | 0 | 4.3e-05 | -0.3 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 1.B.1.1.3 | The General Bacterial Porin (GBP) Family | 0.2 | 0.34 | 1 | 0.89 | 0.96 | 1 | 0.21 | 0.48 | 1 | 0.6 | 0.82 | 1 | 0.6 | 0.91 | 1 | 0.53 | 0.82 | 1 | 0.00032 | 3.5e-05 | 0 | 8.4e-05 | 0 | 0.00022 | 7.7e-05 | 0 | 2e-04 | -0.13 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 1.3e-05 | 0 | 3.5e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 3.A.1.24.1 | The ATP-binding Cassette (ABC) Superfamily | 0.25 | 0.36 | 1 | 0.68 | 0.83 | 1 | 0.79 | 0.91 | 1 | 0.31 | 0.63 | 1 | 0.79 | 0.94 | 1 | 0.83 | 0.93 | 1 | 0.00049 | 6.9e-05 | 0 | 9.1e-05 | 0 | 0.00018 | 0.00024 | 0 | 0.00064 | 1.4 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 5.1e-06 | 0 | 1.7e-05 | 8.1e-06 | 0 | 2.7e-05 | 0.67 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.A.1.106.1 | The ATP-binding Cassette (ABC) Superfamily | 0.065 | 0.34 | 1 | 0.093 | 0.39 | 1 | 0.061 | 0.37 | 1 | 0.083 | 0.32 | 1 | 0.18 | 0.72 | 1 | 0.1 | 0.65 | 1 | 0.00026 | 5.1e-05 | 0 | 0.00014 | 0 | 0.00031 | 7.7e-05 | 0 | 2e-04 | -0.86 | 7 / 36 (19%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 5.1e-06 | 0 | 1.7e-05 | 2.4e-05 | 0 | 5.3e-05 | 2.2 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 1.E.19.1.15 | The Clostridium difficile TcdE Holin (TcdE Holin) Family | 0.21 | 0.35 | 1 | 0.74 | 0.88 | 1 | 0.38 | 0.63 | 1 | 0.063 | 0.3 | 1 | 1 | 1 | 1 | 0.27 | 0.72 | 1 | 0.00015 | 1.3e-05 | 0 | 2.3e-06 | 0 | 6.1e-06 | 0 | 0 | 0 | -Inf | 3 / 36 (8.3%) | 1 / 7 (14%) | 0 / 7 (0%) | 0.143 | 0 | 4.1e-05 | 0 | 0.00011 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 2.A.85.1.2 | The Aromatic Acid Exporter (ArAE) Family | 0.15 | 0.34 | 1 | 0.093 | 0.39 | 1 | 0.12 | 0.43 | 1 | 0.35 | 0.64 | 1 | 0.38 | 0.84 | 1 | 0.39 | 0.75 | 1 | 0.00052 | 1e-04 | 0 | 0.00029 | 0 | 0.00071 | 0.00019 | 0 | 0.00051 | -0.61 | 7 / 36 (19%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 8.5e-06 | 0 | 2.1e-05 | 1.6e-05 | 0 | 3.7e-05 | 0.91 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 3.A.1.140.1 | The ATP-binding Cassette (ABC) Superfamily | 0.082 | 0.34 | 1 | 0.48 | 0.73 | 1 | 0.38 | 0.63 | 1 | 0.073 | 0.31 | 1 | 0.71 | 0.93 | 1 | 0.96 | 1 | 1 | 0.00073 | 1e-04 | 0 | 0.00031 | 0 | 8e-04 | 2e-04 | 0 | 0.00048 | -0.63 | 5 / 36 (14%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 9.B.142.2.9 | The Integral membrane Glycosyltransferase family 39 (GT39) Family | 0.11 | 0.34 | 1 | 0.22 | 0.51 | 1 | 0.1 | 0.42 | 1 | 0.22 | 0.52 | 1 | 0.75 | 0.94 | 1 | 0.34 | 0.73 | 1 | 0.00046 | 5.1e-05 | 0 | 0.00015 | 0 | 4e-04 | 7.7e-05 | 0 | 2e-04 | -0.96 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.1e-05 | 0 | 6.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.C.105.2.8 | The Bacillus thuringiensis Vegetative Insecticidal Protein-3 (Vip3) Family | 0.34 | 0.43 | 1 | 0.7 | 0.85 | 1 | 0.58 | 0.79 | 1 | 0.14 | 0.4 | 1 | 0.93 | 0.98 | 1 | 0.45 | 0.79 | 1 | 0.00015 | 2.6e-05 | 0 | 2e-05 | 0 | 5.2e-05 | 3.9e-06 | 0 | 1e-05 | -2.4 | 6 / 36 (17%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 5.5e-05 | 0 | 0.00011 | 1.4e-05 | 0 | 4.5e-05 | -2 | 3 / 11 (27%) | 1 / 11 (9.1%) | 0.273 | 0.0909 |
| 3.A.1.106.20 | The ATP-binding Cassette (ABC) Superfamily | 0.065 | 0.34 | 1 | 0.093 | 0.39 | 1 | 0.061 | 0.37 | 1 | 0.083 | 0.32 | 1 | 0.18 | 0.72 | 1 | 0.1 | 0.65 | 1 | 0.00026 | 5.1e-05 | 0 | 0.00014 | 0 | 0.00031 | 7.7e-05 | 0 | 2e-04 | -0.86 | 7 / 36 (19%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 5.1e-06 | 0 | 1.7e-05 | 2.4e-05 | 0 | 5.3e-05 | 2.2 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 2.A.40.7.6 | The Nucleobase/Ascorbate Transporter (NAT) or Nucleobase:Cation Symporter-2 (NCS2) Family | 0.15 | 0.34 | 1 | 0.34 | 0.63 | 1 | 0.78 | 0.91 | 1 | 0.21 | 0.51 | 1 | 0.19 | 0.72 | 1 | 0.54 | 0.82 | 1 | 0.00053 | 7.4e-05 | 0 | 0.00017 | 0 | 0.00044 | 2e-04 | 0 | 0.00048 | 0.23 | 5 / 36 (14%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.B.38.4.2 | The Treponema Porin Major Surface Protein (TP-MSP) Family | 0.98 | 0.98 | 1 | 0.86 | 0.94 | 1 | 0.64 | 0.83 | 1 | 0.39 | 0.66 | 1 | 0.85 | 0.96 | 1 | 0.89 | 0.97 | 1 | 8e-05 | 1.1e-05 | 0 | 8.1e-06 | 0 | 2.1e-05 | 4.4e-06 | 0 | 1.2e-05 | -0.88 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 1.2e-05 | 0 | 3.9e-05 | 1.6e-05 | 0 | 3.8e-05 | 0.42 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 8.A.7.1.1 | The Phosphotransferase System Enzyme I (EI) Family | 0.14 | 0.34 | 1 | 0.46 | 0.72 | 1 | 0.22 | 0.5 | 1 | 0.12 | 0.38 | 1 | 0.96 | 0.98 | 1 | 0.56 | 0.83 | 1 | 0.00044 | 4.9e-05 | 0 | 0.00012 | 0 | 0.00031 | 0.00013 | 0 | 0.00033 | 0.12 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 9.B.156.1.1 | The Putative 4. 5. 8. 10 TMS Permease (4.5.8.10P) Family | 0.22 | 0.35 | 1 | 0.57 | 0.77 | 1 | 0.8 | 0.91 | 1 | 0.22 | 0.52 | 1 | 0.89 | 0.98 | 1 | 0.85 | 0.94 | 1 | 0.00029 | 3.2e-05 | 0 | 3.4e-05 | 0 | 5.8e-05 | 0.00013 | 0 | 0.00033 | 1.9 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.42.1.1 | The Hydroxy/Aromatic Amino Acid Permease (HAAAP) Family | 0.77 | 0.81 | 1 | 0.2 | 0.49 | 1 | 0.33 | 0.59 | 1 | 0.36 | 0.64 | 1 | 0.064 | 0.62 | 1 | 0.2 | 0.69 | 1 | 0.00025 | 3.4e-05 | 0 | 3.4e-05 | 0 | 8.9e-05 | 8.7e-05 | 0 | 0.00023 | 1.4 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.2e-05 | 0 | 7.3e-05 | 1.3e-05 | 0 | 3.5e-05 | -0.76 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 1.B.14.8.7 | The Outer Membrane Receptor (OMR) Family | 0.2 | 0.34 | 1 | 0.42 | 0.69 | 1 | 0.19 | 0.48 | 1 | 0.67 | 0.85 | 1 | 0.77 | 0.94 | 1 | 0.57 | 0.83 | 1 | 0.00054 | 7.5e-05 | 0 | 0.00022 | 0 | 0.00058 | 0.00014 | 0 | 0.00036 | -0.65 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1.8e-05 | 0 | 4.1e-05 | 2.8 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 4.A.3.2.4 | The PTS Lactose-N.N'-Diacetylchitobiose-β-glucoside (Lac) Family | 0.37 | 0.45 | 1 | 0.34 | 0.63 | 1 | 0.28 | 0.54 | 1 | 0.12 | 0.38 | 1 | 0.18 | 0.72 | 1 | 0.16 | 0.67 | 1 | 8.5e-05 | 7.1e-06 | 0 | 0 | 0 | 0 | 1.2e-05 | 0 | 3.1e-05 | Inf | 3 / 36 (8.3%) | 0 / 7 (0%) | 1 / 7 (14%) | 0 | 0.143 | 1.3e-05 | 0 | 4.3e-05 | 2.7e-06 | 0 | 8.8e-06 | -2.3 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.1.27.1 | The Major Facilitator Superfamily (MFS) | 0.11 | 0.34 | 1 | 0.067 | 0.36 | 1 | 0.2 | 0.48 | 1 | 0.31 | 0.63 | 1 | 0.34 | 0.83 | 1 | 0.68 | 0.88 | 1 | 0.00033 | 4.5e-05 | 0 | 0.00016 | 0 | 4e-04 | 5.8e-05 | 0 | 0.00015 | -1.5 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 7.7e-06 | 0 | 2.6e-05 | 2.7e-06 | 0 | 8.9e-06 | -1.5 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.D.1.9.2 | The H+ or Na+-translocating NADH Dehydrogenase (NDH) Family | 0.36 | 0.44 | 1 | 0.31 | 0.6 | 1 | 0.63 | 0.83 | 1 | 0.53 | 0.76 | 1 | 0.65 | 0.92 | 1 | 0.32 | 0.73 | 1 | 0.00088 | 0.00017 | 0 | 0.00047 | 0 | 0.0012 | 0.00032 | 0 | 0.00084 | -0.55 | 7 / 36 (19%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 3.3e-05 | 0 | 7.4e-05 | 2.6e-05 | 0 | 6.1e-05 | -0.34 | 3 / 11 (27%) | 2 / 11 (18%) | 0.273 | 0.182 |
| 1.A.29.1.4 | The Urea/Amide Channel (UAC) Family | 0.89 | 0.92 | 1 | 0.22 | 0.51 | 1 | 0.16 | 0.46 | 1 | 0.8 | 0.92 | 1 | 0.25 | 0.77 | 1 | 0.12 | 0.65 | 1 | 0.00017 | 7.7e-05 | 0 | 7.1e-05 | 1.6e-05 | 1e-04 | 1.9e-05 | 0 | 4e-05 | -1.9 | 16 / 36 (44%) | 4 / 7 (57%) | 2 / 7 (29%) | 0.571 | 0.286 | 9.8e-05 | 0 | 2e-04 | 9.6e-05 | 7.5e-05 | 0.00011 | -0.03 | 4 / 11 (36%) | 6 / 11 (55%) | 0.364 | 0.545 |
| 4.C.1.1.16 | The Fatty Acid Transporter (FAT) Family | 0.16 | 0.34 | 1 | 0.27 | 0.56 | 1 | 0.12 | 0.43 | 1 | 0.31 | 0.63 | 1 | 0.76 | 0.94 | 1 | 0.4 | 0.75 | 1 | 0.00047 | 7.9e-05 | 0 | 0.00021 | 0 | 0.00053 | 0.00017 | 0 | 0.00046 | -0.3 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 3e-06 | 0 | 9.9e-06 | 1.3e-05 | 0 | 3.5e-05 | 2.1 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 3.A.1.2.6 | The ATP-binding Cassette (ABC) Superfamily | 0.08 | 0.34 | 1 | 0.22 | 0.51 | 1 | 0.27 | 0.53 | 1 | 0.034 | 0.27 | 1 | 0.16 | 0.69 | 1 | 0.23 | 0.7 | 1 | 0.00052 | 8.6e-05 | 0 | 2e-04 | 0 | 0.00044 | 0.00023 | 0 | 0.00061 | 0.2 | 6 / 36 (17%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 6.8e-06 | 0 | 2.3e-05 | 1.4 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.1.6.10 | The Major Facilitator Superfamily (MFS) | 0.49 | 0.56 | 1 | 0.33 | 0.61 | 1 | 0.35 | 0.6 | 1 | 0.61 | 0.82 | 1 | 0.4 | 0.84 | 1 | 0.37 | 0.75 | 1 | 0.00042 | 4.6e-05 | 0 | 6.7e-05 | 0 | 0.00018 | 0.00015 | 0 | 0.00038 | 1.2 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 1.1e-05 | 0 | 3.6e-05 | 5.4e-06 | 0 | 1.8e-05 | -1 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 9.A.5.4.2 | The Putative Arginine Transporter (ArgW) Family | 0.32 | 0.41 | 1 | 0.23 | 0.52 | 1 | 0.71 | 0.88 | 1 | 0.94 | 0.98 | 1 | 0.098 | 0.64 | 1 | 0.58 | 0.84 | 1 | 0.00028 | 2.3e-05 | 0 | 3.4e-05 | 0 | 8.9e-05 | 7.7e-05 | 0 | 2e-04 | 1.2 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.7.6.1 | The Drug/Metabolite Transporter (DMT) Superfamily | 0.21 | 0.35 | 1 | 0.28 | 0.57 | 1 | 0.35 | 0.6 | 1 | 0.95 | 0.98 | 1 | 0.88 | 0.98 | 1 | 0.92 | 0.98 | 1 | 0.00037 | 4.2e-05 | 0 | 0.00013 | 0 | 0.00036 | 5.8e-05 | 0 | 0.00015 | -1.2 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 5.1e-06 | 0 | 1.7e-05 | 8.1e-06 | 0 | 2.7e-05 | 0.67 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.42.2.4 | The Hydroxy/Aromatic Amino Acid Permease (HAAAP) Family | 0.17 | 0.34 | 1 | 0.072 | 0.37 | 1 | 0.25 | 0.52 | 1 | 0.61 | 0.82 | 1 | 0.37 | 0.84 | 1 | 0.8 | 0.92 | 1 | 0.00026 | 4.4e-05 | 0 | 0.00013 | 0 | 0.00031 | 6.8e-05 | 0 | 0.00018 | -0.93 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 1e-05 | 0 | 2.5e-05 | 8.1e-06 | 0 | 2.7e-05 | -0.3 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 9.B.36.1.1 | The Acid Resistance Membrane Protein (HdeD) Family | 0.093 | 0.34 | 1 | 0.014 | 0.23 | 1 | 0.049 | 0.37 | 1 | 0.1 | 0.35 | 1 | 0.67 | 0.92 | 1 | 0.92 | 0.98 | 1 | 0.00028 | 2.3e-05 | 0 | 7e-05 | 0 | 0.00018 | 4.8e-05 | 0 | 0.00013 | -0.54 | 3 / 36 (8.3%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 2.A.20.1.2 | The Inorganic Phosphate Transporter (PiT) Family | 0.34 | 0.42 | 1 | 0.69 | 0.84 | 1 | 0.64 | 0.83 | 1 | 0.87 | 0.95 | 1 | 0.27 | 0.78 | 1 | 0.31 | 0.73 | 1 | 0.00048 | 6.7e-05 | 0 | 0.00017 | 0 | 0.00045 | 0.00013 | 0 | 0.00026 | -0.39 | 5 / 36 (14%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 2.2e-05 | 0 | 7.3e-05 | 1e-05 | 0 | 3.4e-05 | -1.1 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.3.7.5 | The Amino Acid-Polyamine-Organocation (APC) Family | 0.15 | 0.34 | 1 | 0.17 | 0.47 | 1 | 0.076 | 0.38 | 1 | 0.36 | 0.64 | 1 | 0.78 | 0.94 | 1 | 0.29 | 0.73 | 1 | 0.00067 | 0.00011 | 0 | 0.00034 | 0 | 0.00089 | 0.00019 | 0 | 0.00051 | -0.84 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 3e-06 | 0 | 9.9e-06 | 2.6e-05 | 0 | 6.9e-05 | 3.1 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 2.A.37.1.5 | The Monovalent Cation:Proton Antiporter-2 (CPA2) Family | 0.17 | 0.34 | 1 | 0.51 | 0.75 | 1 | 0.21 | 0.48 | 1 | 0.41 | 0.68 | 1 | 0.44 | 0.86 | 1 | 0.83 | 0.93 | 1 | 0.00044 | 3.6e-05 | 0 | 1e-04 | 0 | 0.00027 | 7.7e-05 | 0 | 2e-04 | -0.38 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 5.4e-06 | 0 | 1.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 9.B.143.2.2 | The 6 TMS DUF1275/Pf06912 (DUF1275) Family | 0.084 | 0.34 | 1 | 0.22 | 0.51 | 1 | 0.19 | 0.48 | 1 | 0.041 | 0.27 | 1 | 0.12 | 0.65 | 1 | 0.11 | 0.65 | 1 | 0.00014 | 3.5e-05 | 0 | 0 | 0 | 0 | 3.4e-05 | 0 | 5.9e-05 | Inf | 9 / 36 (25%) | 0 / 7 (0%) | 2 / 7 (29%) | 0 | 0.286 | 5.8e-05 | 0 | 0.00011 | 3.6e-05 | 0 | 6.8e-05 | -0.69 | 4 / 11 (36%) | 3 / 11 (27%) | 0.364 | 0.273 |
| 9.B.221.1.1 | The 5 TMS YgjV (YgjV) Family | 0.44 | 0.52 | 1 | 0.24 | 0.53 | 1 | 0.51 | 0.75 | 1 | 0.79 | 0.9 | 1 | 0.27 | 0.78 | 1 | 0.52 | 0.82 | 1 | 0.00036 | 4e-05 | 0 | 5e-05 | 0 | 0.00013 | 0.00014 | 0 | 0.00036 | 1.5 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1e-05 | 0 | 3.4e-05 | 1.9 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 2.A.1.35.1 | The Major Facilitator Superfamily (MFS) | 0.12 | 0.34 | 1 | 0.24 | 0.53 | 1 | 0.68 | 0.85 | 1 | 0.39 | 0.66 | 1 | 0.45 | 0.86 | 1 | 0.98 | 1 | 1 | 0.00024 | 2.7e-05 | 0 | 7e-05 | 0 | 0.00014 | 5.8e-05 | 0 | 0.00015 | -0.27 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 7.3e-06 | 0 | 2.4e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 3.A.1.101.1 | The ATP-binding Cassette (ABC) Superfamily | 0.043 | 0.34 | 1 | 0.018 | 0.23 | 1 | 0.057 | 0.37 | 1 | 0.028 | 0.27 | 1 | 0.17 | 0.72 | 1 | 0.33 | 0.73 | 1 | 0.00048 | 4e-05 | 0 | 0.00014 | 0 | 0.00031 | 6.8e-05 | 0 | 0.00018 | -1 | 3 / 36 (8.3%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 1.B.42.1.2 | The Outer Membrane Lipopolysaccharide Export Porin (LPS-EP) Family | 0.21 | 0.35 | 1 | 0.33 | 0.62 | 1 | 0.31 | 0.58 | 1 | 0.43 | 0.7 | 1 | 0.53 | 0.89 | 1 | 0.52 | 0.82 | 1 | 0.0013 | 0.00043 | 0 | 0.001 | 2.8e-05 | 0.0027 | 0.00095 | 0 | 0.0025 | -0.074 | 12 / 36 (33%) | 4 / 7 (57%) | 2 / 7 (29%) | 0.571 | 0.286 | 6e-05 | 0 | 0.00014 | 7.6e-05 | 0 | 0.00015 | 0.34 | 3 / 11 (27%) | 3 / 11 (27%) | 0.273 | 0.273 |
| 1.A.11.1.1 | The Ammonium Transporter Channel (Amt) Family | 0.18 | 0.34 | 1 | 0.58 | 0.77 | 1 | 0.23 | 0.51 | 1 | 0.37 | 0.65 | 1 | 0.21 | 0.74 | 1 | 0.84 | 0.93 | 1 | 0.00045 | 3.7e-05 | 0 | 1e-04 | 0 | 0.00027 | 8.7e-05 | 0 | 0.00023 | -0.2 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.E.52.1.1 | The Flp/Fap Pilin Putative Holin (FFPP-Hol) Family | 0.14 | 0.34 | 1 | 1 | 1 | 1 | 0.65 | 0.84 | 1 | 0.043 | 0.27 | 1 | 0.75 | 0.94 | 1 | 0.55 | 0.83 | 1 | 0.00011 | 1.2e-05 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 4 / 36 (11%) | 0 / 7 (0%) | 0 / 7 (0%) | 0 | 0 | 3e-05 | 0 | 7.5e-05 | 9.8e-06 | 0 | 2.4e-05 | -1.6 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 9.B.99.1.2 | The MipA-interacting Protein (MipA) Family | 0.07 | 0.34 | 1 | 0.099 | 0.39 | 1 | 0.06 | 0.37 | 1 | 0.092 | 0.34 | 1 | 0.39 | 0.84 | 1 | 0.2 | 0.69 | 1 | 0.00051 | 5.6e-05 | 0 | 0.00018 | 0 | 0.00044 | 7.7e-05 | 0 | 2e-04 | -1.2 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.1e-05 | 0 | 6.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.B.9.1.1 | The FadL Outer Membrane Protein (FadL) Family | 0.065 | 0.34 | 1 | 0.35 | 0.64 | 1 | 0.33 | 0.59 | 1 | 0.042 | 0.27 | 1 | 0.35 | 0.84 | 1 | 0.31 | 0.73 | 1 | 0.00037 | 4.1e-05 | 0 | 1e-04 | 0 | 0.00019 | 0.00011 | 0 | 0.00028 | 0.14 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 2.7e-06 | 0 | 8.9e-06 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.73.1.1 | The Short Chain Fatty Acid Uptake (AtoE) Family | 0.37 | 0.45 | 1 | 0.76 | 0.88 | 1 | 0.87 | 0.94 | 1 | 0.96 | 0.99 | 1 | 0.99 | 1 | 1 | 1 | 1 | 1 | 0.00026 | 4.3e-05 | 0 | 8.6e-05 | 0 | 0.00022 | 8.7e-05 | 0 | 0.00023 | 0.017 | 6 / 36 (17%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.2e-05 | 0 | 7.3e-05 | 9.2e-06 | 0 | 2.2e-05 | -1.3 | 1 / 11 (9.1%) | 2 / 11 (18%) | 0.0909 | 0.182 |
| 4.D.3.1.6 | The Glycan Glucosyl Transferase (OpgH) Family | 0.08 | 0.34 | 1 | 0.045 | 0.29 | 1 | 0.064 | 0.37 | 1 | 0.15 | 0.41 | 1 | 0.13 | 0.66 | 1 | 0.16 | 0.67 | 1 | 0.00059 | 0.00016 | 0 | 0.00044 | 0 | 0.00096 | 0.00032 | 0 | 0.00084 | -0.46 | 10 / 36 (28%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 1.6e-05 | 0 | 3e-05 | 3.1e-05 | 0 | 5.5e-05 | 0.95 | 3 / 11 (27%) | 3 / 11 (27%) | 0.273 | 0.273 |
| 2.A.96.1.1 | The Acetate Uptake Transporter (AceTr) Family | 0.056 | 0.34 | 1 | 0.26 | 0.55 | 1 | 0.37 | 0.63 | 1 | 0.071 | 0.3 | 1 | 0.39 | 0.84 | 1 | 0.58 | 0.84 | 1 | 0.00023 | 1.9e-05 | 0 | 5.1e-05 | 0 | 9.3e-05 | 4.8e-05 | 0 | 0.00013 | -0.087 | 3 / 36 (8.3%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 1.B.14.6.5 | The Outer Membrane Receptor (OMR) Family | 0.78 | 0.83 | 1 | 0.81 | 0.91 | 1 | 0.54 | 0.77 | 1 | 0.62 | 0.82 | 1 | 0.5 | 0.87 | 1 | 0.87 | 0.95 | 1 | 0.00042 | 3.5e-05 | 0 | 4.1e-06 | 0 | 1.1e-05 | 1.5e-05 | 0 | 4e-05 | 1.9 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 1e-04 | 0 | 0.00034 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.86.1.1 | The Autoinducer-2 Exporter (AI-2E) Family (Formerly the PerM Family. TC #9.B.22) | 0.2 | 0.34 | 1 | 0.95 | 0.98 | 1 | 0.73 | 0.89 | 1 | 0.5 | 0.75 | 1 | 0.89 | 0.98 | 1 | 0.58 | 0.84 | 1 | 0.00018 | 3.6e-05 | 0 | 7e-05 | 0 | 0.00014 | 1e-04 | 0 | 0.00025 | 0.51 | 7 / 36 (19%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 5.5e-06 | 0 | 1.2e-05 | 2.7e-06 | 0 | 8.9e-06 | -1 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 4.A.3.2.1 | The PTS Lactose-N.N'-Diacetylchitobiose-β-glucoside (Lac) Family | 0.24 | 0.36 | 1 | 0.14 | 0.44 | 1 | 0.44 | 0.7 | 1 | 0.69 | 0.86 | 1 | 0.98 | 0.99 | 1 | 0.31 | 0.73 | 1 | 0.00034 | 4.7e-05 | 0 | 0.00017 | 0 | 0.00045 | 5.8e-05 | 0 | 0.00015 | -1.6 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 8.1e-06 | 0 | 1.9e-05 | 2.7e-06 | 0 | 8.9e-06 | -1.6 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 3.A.1.122.1 | The ATP-binding Cassette (ABC) Superfamily | 0.14 | 0.34 | 1 | 0.048 | 0.29 | 1 | 0.33 | 0.59 | 1 | 0.36 | 0.64 | 1 | 0.11 | 0.65 | 1 | 0.62 | 0.85 | 1 | 0.00023 | 5.2e-05 | 0 | 0.00013 | 0 | 0.00026 | 0.00011 | 0 | 0.00028 | -0.24 | 8 / 36 (22%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 1.3e-05 | 0 | 2.6e-05 | 6.8e-06 | 0 | 2.3e-05 | -0.93 | 3 / 11 (27%) | 1 / 11 (9.1%) | 0.273 | 0.0909 |
| 3.A.1.3.4 | The ATP-binding Cassette (ABC) Superfamily | 0.029 | 0.34 | 1 | 0.069 | 0.36 | 1 | 0.024 | 0.37 | 1 | 0.0042 | 0.16 | 1 | 0.18 | 0.72 | 1 | 0.048 | 0.58 | 1 | 0.00064 | 0.00014 | 0 | 0.00039 | 1.6e-05 | 0.00087 | 3e-04 | 0 | 0.00074 | -0.38 | 8 / 36 (22%) | 4 / 7 (57%) | 2 / 7 (29%) | 0.571 | 0.286 | 0 | 0 | 0 | 2.6e-05 | 0 | 6.1e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 3.A.2.3.2 | The H+- or Na+-translocating F-type. V-type and A-type ATPase (F-ATPase) Superfamily | 0.92 | 0.93 | 1 | 0.53 | 0.76 | 1 | 0.81 | 0.91 | 1 | 0.44 | 0.7 | 1 | 0.91 | 0.98 | 1 | 0.53 | 0.82 | 1 | 0.00017 | 2.3e-05 | 0 | 3.2e-05 | 0 | 8.6e-05 | 1.3e-05 | 0 | 3.5e-05 | -1.3 | 5 / 36 (14%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 2.1e-05 | 0 | 4.6e-05 | 2.6e-05 | 0 | 8.6e-05 | 0.31 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 9.B.15.1.1 | The 4 TMS YbhQ (YbhQ) Family | 0.76 | 0.81 | 1 | 0.086 | 0.39 | 1 | 0.059 | 0.37 | 1 | 0.27 | 0.6 | 1 | 0.051 | 0.62 | 1 | 0.03 | 0.58 | 1 | 0.00016 | 1.8e-05 | 0 | 1.7e-05 | 0 | 4.5e-05 | 5.7e-05 | 0 | 0.00011 | 1.7 | 4 / 36 (11%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 1.1e-05 | 0 | 3.6e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 3.A.1.3.10 | The ATP-binding Cassette (ABC) Superfamily | 0.081 | 0.34 | 1 | 0.42 | 0.69 | 1 | 0.18 | 0.48 | 1 | 0.066 | 0.3 | 1 | 0.44 | 0.86 | 1 | 0.15 | 0.65 | 1 | 0.00043 | 5.9e-05 | 0 | 0.00014 | 0 | 0.00027 | 0.00015 | 0 | 0.00038 | 0.1 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 1.6e-05 | 0 | 3.7e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 2.A.38.1.1 | The K+ Transporter (Trk) Family | 0.036 | 0.34 | 1 | 0.0039 | 0.22 | 1 | 0.0039 | 0.35 | 1 | 0.0052 | 0.16 | 1 | 0.013 | 0.5 | 1 | 0.012 | 0.46 | 1 | 0.00046 | 0.00015 | 0 | 0.00041 | 8.1e-05 | 0.00091 | 0.00029 | 0 | 0.00077 | -0.5 | 12 / 36 (33%) | 5 / 7 (71%) | 2 / 7 (29%) | 0.714 | 0.286 | 1.4e-05 | 0 | 4e-05 | 3.6e-05 | 0 | 6.9e-05 | 1.4 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 5.A.3.1.2 | The Prokaryotic Molybdopterin-containing Oxidoreductase (PMO) Family | 0.19 | 0.34 | 1 | 0.66 | 0.82 | 1 | 0.35 | 0.6 | 1 | 0.4 | 0.68 | 1 | 0.81 | 0.95 | 1 | 0.47 | 0.81 | 1 | 0.00048 | 0.00013 | 0 | 3e-04 | 0 | 0.00075 | 3e-04 | 0 | 0.00079 | 0 | 10 / 36 (28%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 1.6e-05 | 0 | 4.3e-05 | 3.7e-05 | 0 | 6.3e-05 | 1.2 | 2 / 11 (18%) | 3 / 11 (27%) | 0.182 | 0.273 |
| 2.A.72.1.3 | The K+ Uptake Permease (KUP) Family | 0.088 | 0.34 | 1 | 0.54 | 0.76 | 1 | 0.18 | 0.48 | 1 | 0.034 | 0.27 | 1 | 0.34 | 0.83 | 1 | 0.12 | 0.65 | 1 | 9.4e-05 | 1.3e-05 | 0 | 0 | 0 | 0 | 7.7e-06 | 0 | 2e-05 | Inf | 5 / 36 (14%) | 0 / 7 (0%) | 1 / 7 (14%) | 0 | 0.143 | 3.1e-05 | 0 | 6.8e-05 | 6.8e-06 | 0 | 2.3e-05 | -2.2 | 3 / 11 (27%) | 1 / 11 (9.1%) | 0.273 | 0.0909 |
| 1.B.1.1.2 | The General Bacterial Porin (GBP) Family | 0.17 | 0.34 | 1 | 0.55 | 0.76 | 1 | 0.99 | 0.99 | 1 | 0.68 | 0.86 | 1 | 0.97 | 0.99 | 1 | 0.51 | 0.82 | 1 | 0.00026 | 5.7e-05 | 0 | 0.00015 | 0 | 0.00031 | 0.00011 | 0 | 0.00025 | -0.45 | 8 / 36 (22%) | 2 / 7 (29%) | 2 / 7 (29%) | 0.286 | 0.286 | 1.3e-05 | 0 | 2.7e-05 | 5.4e-06 | 0 | 1.8e-05 | -1.3 | 3 / 11 (27%) | 1 / 11 (9.1%) | 0.273 | 0.0909 |
| 2.A.34.1.1 | The NhaB Na+:H+ Antiporter (NhaB) Family | 0.31 | 0.4 | 1 | 0.96 | 0.99 | 1 | 0.32 | 0.59 | 1 | 0.93 | 0.97 | 1 | 0.82 | 0.95 | 1 | 0.19 | 0.68 | 1 | 0.00025 | 3.4e-05 | 0 | 7e-05 | 0 | 0.00018 | 9.7e-05 | 0 | 0.00026 | 0.47 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 5.5e-06 | 0 | 1.2e-05 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 2.A.28.2.6 | The Bile Acid:Na+ Symporter (BASS) Family | 0.17 | 0.34 | 1 | 0.083 | 0.39 | 1 | 0.3 | 0.57 | 1 | 0.057 | 0.29 | 1 | 0.011 | 0.46 | 1 | 0.043 | 0.58 | 1 | 0.00068 | 0.00047 | 0.00028 | 0.00022 | 0 | 4e-04 | 0.00037 | 0.00034 | 0.00039 | 0.75 | 25 / 36 (69%) | 3 / 7 (43%) | 5 / 7 (71%) | 0.429 | 0.714 | 0.00058 | 0.00039 | 0.0011 | 6e-04 | 0.00053 | 0.00054 | 0.049 | 9 / 11 (82%) | 8 / 11 (73%) | 0.818 | 0.727 |
| 3.A.1.11.9 | The ATP-binding Cassette (ABC) Superfamily | 0.14 | 0.34 | 1 | 0.91 | 0.96 | 1 | 0.56 | 0.78 | 1 | 0.08 | 0.32 | 1 | 0.59 | 0.91 | 1 | 0.33 | 0.73 | 1 | 0.00073 | 8.1e-05 | 0 | 0.00015 | 0 | 0.00035 | 0.00024 | 0 | 0.00064 | 0.68 | 4 / 36 (11%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 1.3e-05 | 0 | 4.5e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 2.A.72.1.1 | The K+ Uptake Permease (KUP) Family | 0.044 | 0.34 | 1 | 0.24 | 0.53 | 1 | 0.11 | 0.42 | 1 | 0.05 | 0.28 | 1 | 0.6 | 0.91 | 1 | 0.22 | 0.7 | 1 | 0.00034 | 8.6e-05 | 0 | 0.00025 | 0 | 0.00057 | 0.00016 | 0 | 0.00033 | -0.64 | 9 / 36 (25%) | 3 / 7 (43%) | 2 / 7 (29%) | 0.429 | 0.286 | 2.6e-06 | 0 | 8.5e-06 | 1.9e-05 | 0 | 3.8e-05 | 2.9 | 1 / 11 (9.1%) | 3 / 11 (27%) | 0.0909 | 0.273 |
| 4.A.7.1.2 | The PTS L-Ascorbate (L-Asc) Family | 0.15 | 0.34 | 1 | 0.28 | 0.56 | 1 | 0.14 | 0.43 | 1 | 0.35 | 0.64 | 1 | 0.81 | 0.95 | 1 | 0.58 | 0.84 | 1 | 0.0014 | 0.00011 | 0 | 0.00035 | 0 | 0.00094 | 0.00019 | 0 | 0.00051 | -0.88 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 2.1e-05 | 0 | 6.8e-05 | Inf | 0 / 11 (0%) | 1 / 11 (9.1%) | 0 | 0.0909 |
| 1.A.104.1.1 | The Proposed Flagellar Biosynthesis Na+ Channel. FlaH (FlaH) Family | 0.79 | 0.84 | 1 | 0.17 | 0.47 | 1 | 0.51 | 0.75 | 1 | 0.33 | 0.63 | 1 | 0.08 | 0.62 | 1 | 0.48 | 0.81 | 1 | 0.00022 | 2.4e-05 | 0 | 2.3e-06 | 0 | 6.1e-06 | 0.00011 | 0 | 0.00028 | 5.6 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 0 | 0 | 0 | 9.5e-06 | 0 | 2.3e-05 | Inf | 0 / 11 (0%) | 2 / 11 (18%) | 0 | 0.182 |
| 3.A.1.11.2 | The ATP-binding Cassette (ABC) Superfamily | 0.15 | 0.34 | 1 | 0.17 | 0.46 | 1 | 0.34 | 0.6 | 1 | 0.28 | 0.6 | 1 | 0.34 | 0.83 | 1 | 0.65 | 0.86 | 1 | 0.00079 | 0.00015 | 0 | 0.00041 | 0 | 0.001 | 0.00033 | 0 | 0.00087 | -0.31 | 7 / 36 (19%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 1.6e-05 | 0 | 3.9e-05 | 1.9e-05 | 0 | 6.2e-05 | 0.25 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 2.A.78.1.3 | The Branched Chain Amino Acid Exporter (LIV-E) Family | 0.32 | 0.41 | 1 | 0.77 | 0.88 | 1 | 0.55 | 0.78 | 1 | 0.75 | 0.89 | 1 | 0.5 | 0.87 | 1 | 0.12 | 0.65 | 1 | 0.00048 | 4e-05 | 0 | 1e-04 | 0 | 0.00027 | 8.7e-05 | 0 | 0.00023 | -0.2 | 3 / 36 (8.3%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 1e-05 | 0 | 3.4e-05 | 0 | 0 | 0 | -Inf | 1 / 11 (9.1%) | 0 / 11 (0%) | 0.0909 | 0 |
| 2.A.35.1.8 | The NhaC Na+:H+ Antiporter (NhaC) Family | 0.26 | 0.37 | 1 | 0.41 | 0.69 | 1 | 0.073 | 0.38 | 1 | 0.41 | 0.68 | 1 | 0.45 | 0.86 | 1 | 0.14 | 0.65 | 1 | 0.00027 | 9.6e-05 | 0 | 8.3e-05 | 8.5e-05 | 9.1e-05 | 4.9e-05 | 0 | 9.2e-05 | -0.76 | 13 / 36 (36%) | 4 / 7 (57%) | 3 / 7 (43%) | 0.571 | 0.429 | 3e-05 | 0 | 7.3e-05 | 2e-04 | 0 | 5e-04 | 2.7 | 2 / 11 (18%) | 4 / 11 (36%) | 0.182 | 0.364 |
| 3.A.1.5.3 | The ATP-binding Cassette (ABC) Superfamily | 0.14 | 0.34 | 1 | 0.23 | 0.53 | 1 | 0.13 | 0.43 | 1 | 0.16 | 0.43 | 1 | 0.2 | 0.73 | 1 | 0.14 | 0.65 | 1 | 0.00062 | 0.00014 | 0 | 0.00031 | 0 | 0.00075 | 0.00032 | 0 | 0.00084 | 0.046 | 8 / 36 (22%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 8.5e-06 | 0 | 2.1e-05 | 3.9e-05 | 0 | 8.8e-05 | 2.2 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 2.A.85.1.6 | The Aromatic Acid Exporter (ArAE) Family | 0.27 | 0.37 | 1 | 0.52 | 0.75 | 1 | 0.52 | 0.75 | 1 | 0.9 | 0.97 | 1 | 0.51 | 0.87 | 1 | 0.086 | 0.61 | 1 | 0.00061 | 6.8e-05 | 0 | 0.00019 | 0 | 0.00049 | 0.00015 | 0 | 0.00041 | -0.34 | 4 / 36 (11%) | 1 / 7 (14%) | 1 / 7 (14%) | 0.143 | 0.143 | 5.5e-06 | 0 | 1.2e-05 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
| 2.A.3.1.2 | The Amino Acid-Polyamine-Organocation (APC) Family | 0.063 | 0.34 | 1 | 0.015 | 0.23 | 1 | 0.051 | 0.37 | 1 | 0.027 | 0.27 | 1 | 0.16 | 0.69 | 1 | 0.32 | 0.73 | 1 | 8e-04 | 6.7e-05 | 0 | 0.00022 | 0 | 0.00053 | 0.00013 | 0 | 0.00033 | -0.76 | 3 / 36 (8.3%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 0 / 11 (0%) | 0 / 11 (0%) | 0 | 0 |
| 2.A.3.3.23 | The Amino Acid-Polyamine-Organocation (APC) Family | 0.12 | 0.34 | 1 | 1 | 1 | 1 | 0.33 | 0.59 | 1 | 0.046 | 0.27 | 1 | 0.64 | 0.92 | 1 | 0.16 | 0.67 | 1 | 2e-04 | 1.7e-05 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 3 / 36 (8.3%) | 0 / 7 (0%) | 0 / 7 (0%) | 0 | 0 | 4.8e-05 | 0 | 0.00012 | 6.8e-06 | 0 | 2.2e-05 | -2.8 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 1.B.55.1.1 | The Poly Acetyl Glucosamine Porin (PgaA) Family | 0.1 | 0.34 | 1 | 0.32 | 0.61 | 1 | 0.32 | 0.59 | 1 | 0.16 | 0.43 | 1 | 0.44 | 0.86 | 1 | 0.45 | 0.79 | 1 | 0.00049 | 6.8e-05 | 0 | 0.00017 | 0 | 0.00035 | 0.00016 | 0 | 0.00044 | -0.087 | 5 / 36 (14%) | 2 / 7 (29%) | 1 / 7 (14%) | 0.286 | 0.143 | 2.6e-06 | 0 | 8.5e-06 | 1e-05 | 0 | 3.4e-05 | 1.9 | 1 / 11 (9.1%) | 1 / 11 (9.1%) | 0.0909 | 0.0909 |
| 3.A.5.1.1 | The General Secretory Pathway (Sec) Family | 0.093 | 0.34 | 1 | 0.032 | 0.27 | 1 | 0.05 | 0.37 | 1 | 0.039 | 0.27 | 1 | 0.053 | 0.62 | 1 | 0.087 | 0.61 | 1 | 0.0012 | 0.00029 | 0 | 0.00078 | 4.8e-05 | 0.0019 | 0.00064 | 0 | 0.0017 | -0.29 | 9 / 36 (25%) | 4 / 7 (57%) | 1 / 7 (14%) | 0.571 | 0.143 | 1.8e-05 | 0 | 5.1e-05 | 3.4e-05 | 0 | 7.8e-05 | 0.92 | 2 / 11 (18%) | 2 / 11 (18%) | 0.182 | 0.182 |
| 3.D.6.1.4 | The Putative Ion (H+ or Na+)-translocating NADH:Ferredoxin Oxidoreductase (NFO or RNF) Family | 0.27 | 0.37 | 1 | 0.47 | 0.73 | 1 | 0.99 | 0.99 | 1 | 0.76 | 0.89 | 1 | 0.48 | 0.87 | 1 | 0.18 | 0.68 | 1 | 0.00086 | 0.00014 | 0 | 0.00045 | 0 | 0.0012 | 0.00024 | 0 | 0.00061 | -0.91 | 6 / 36 (17%) | 1 / 7 (14%) | 2 / 7 (29%) | 0.143 | 0.286 | 2.7e-05 | 0 | 7.3e-05 | 2.7e-06 | 0 | 8.9e-06 | -3.3 | 2 / 11 (18%) | 1 / 11 (9.1%) | 0.182 | 0.0909 |
| 4.A.1.2.4 | The PTS Glucose-Glucoside (Glc) Family | 0.069 | 0.34 | 1 | 0.048 | 0.29 | 1 | 0.36 | 0.6 | 1 | 0.082 | 0.32 | 1 | 0.086 | 0.62 | 1 | 0.6 | 0.85 | 1 | 0.00054 | 9e-05 | 0 | 0.00024 | 0 | 0.00047 | 0.00019 | 0 | 0.00051 | -0.34 | 6 / 36 (17%) | 3 / 7 (43%) | 1 / 7 (14%) | 0.429 | 0.143 | 1.6e-05 | 0 | 3.9e-05 | 0 | 0 | 0 | -Inf | 2 / 11 (18%) | 0 / 11 (0%) | 0.182 | 0 |
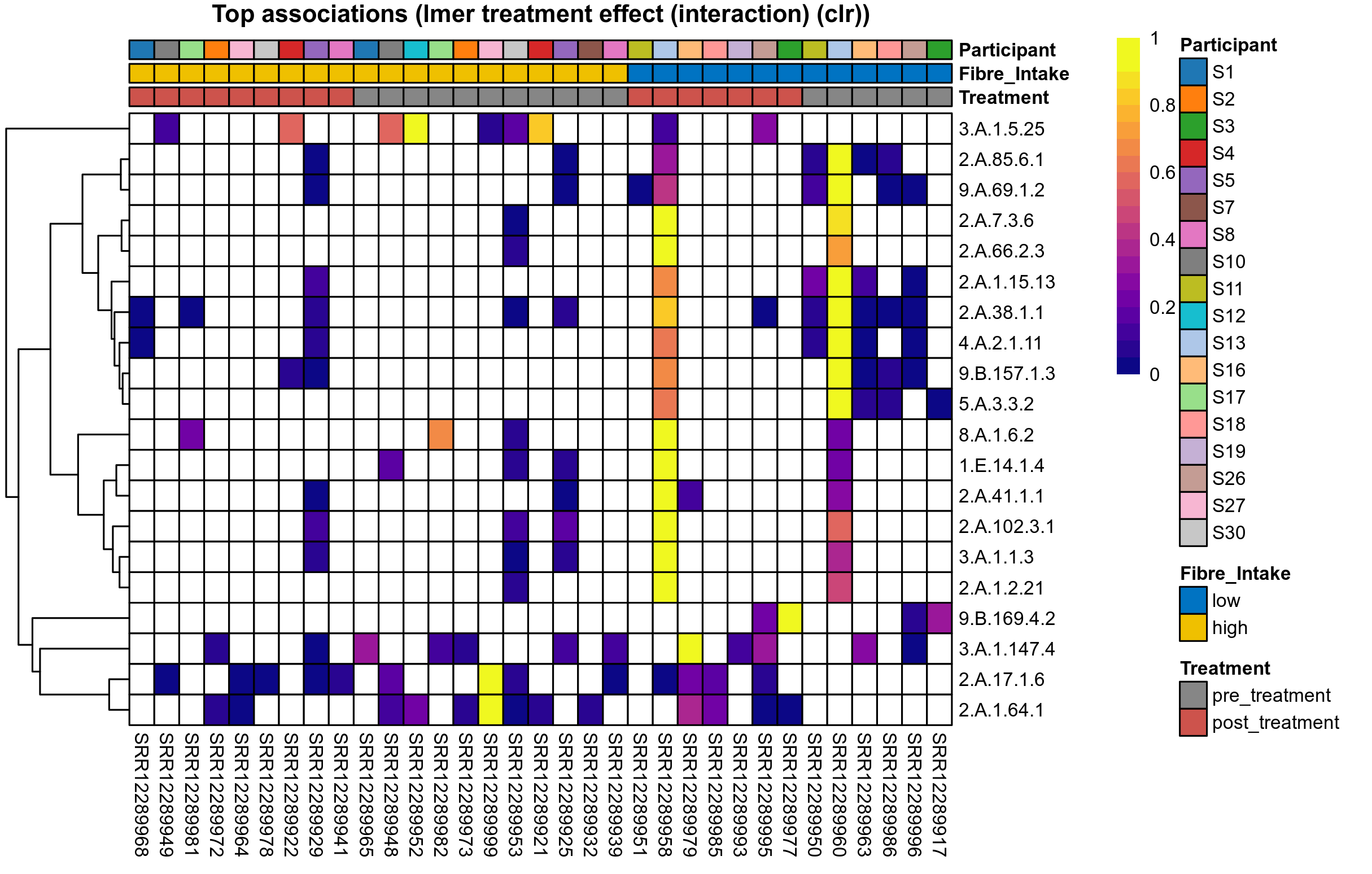
Click here to open full-sized image in new window.
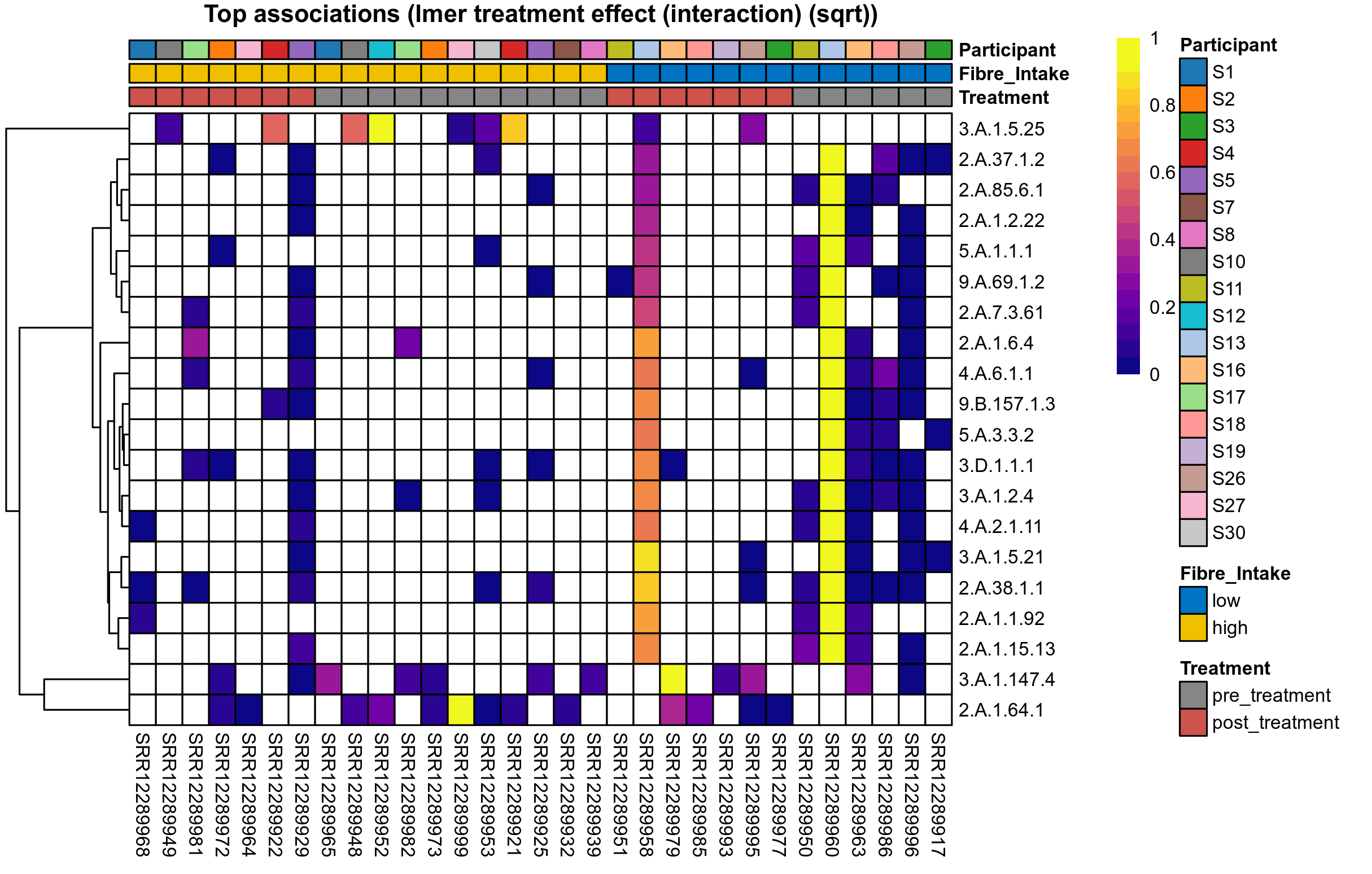
Click here to open full-sized image in new window.
P lmer study group effect (sqrt): Differentially abundant functions were identified by linear mixed effect regression without confounders. Random effects: Participant (intercept). Fixed effects: Fibre_Intake, Treatment, Fibre_Intake vs Treatment. Effect tested: Fibre_Intake.
P lmer study group effect (clr): Differentially abundant functions were identified by linear mixed effect regression without confounders. Random effects: Participant (intercept). Fixed effects: Fibre_Intake, Treatment, Fibre_Intake vs Treatment. Effect tested: Fibre_Intake.
P lmer time effect (sqrt): Differentially abundant functions were identified by linear mixed effect regression without confounders. Random effects: Participant (intercept). Fixed effects: Fibre_Intake, Treatment, Fibre_Intake vs Treatment. Effect tested: Treatment.
P lmer time effect (clr): Differentially abundant functions were identified by linear mixed effect regression without confounders. Random effects: Participant (intercept). Fixed effects: Fibre_Intake, Treatment, Fibre_Intake vs Treatment. Effect tested: Treatment.
P lmer treatment effect (interaction) (sqrt): Differentially abundant functions were identified by linear mixed effect regression without confounders. Random effects: Participant (intercept). Fixed effects: Fibre_Intake, Treatment, Fibre_Intake vs Treatment. Effect tested: Fibre_Intake vs Treatment.
P lmer treatment effect (interaction) (clr): Differentially abundant functions were identified by linear mixed effect regression without confounders. Random effects: Participant (intercept). Fixed effects: Fibre_Intake, Treatment, Fibre_Intake vs Treatment. Effect tested: Fibre_Intake vs Treatment.
The following plots present the distribution of the top most differentially abundant microbial functions across all applied statistical analysis. Plots are ordered alphabetically.